Abstract
Lateral root (LR) formation in vascular plants is regulated by auxin. The mechanisms of LR formation are not fully understood. Here, we have identified a novel recessive mutation in Arabidopsis thaliana, named fewer roots (fwr), that drastically reduces the number of LRs. Expression analyses of DR5::GUS, an auxin response reporter, and pLBD16::GUS, an LR initiation marker, suggested that FWR is necessary for the establishment of an auxin response maximum in LR initiation sites. We further identified that the fwr phenotypes are caused by a missense mutation in the GNOM gene, encoding an Arf-GEF (ADP ribosylation factor-GDP/GTP exchange factor), which regulates the recycling of PINs, the auxin efflux carriers. The fwr roots showed enhanced sensitivity to brefeldin A in a root growth inhibition assay, indicating that the fwr mutation reduces the Arf-GEF activity of GNOM. However, the other developmental processes except for LR formation appeared to be unaffected in the fwr mutant, indicating that fwr is a weaker allele of gnom compared with the other gnom alleles with pleiotropic phenotypes. The localization of PIN1–green fluorescent protein (GFP) appeared to be unaffected in the fwr roots but the levels of endogenous IAA were actually higher in the fwr roots than in the wild type. These results indicate that LR initiation is one of the most sensitive processes among GNOM-dependent developmental processes, strongly suggesting that GNOM is required for the establishment of the auxin response maximum for LR initiation, probably through the regulation of local and global auxin distribution in the root.
Keywords: Arabidopsis thaliana, Auxin, GNOM, Lateral root formation
Introduction
Lateral root (LR) formation is one of the post-embryonic developmental processes in vascular plants, which contributes to the establishment of the root system for efficient water and nutrient uptake from the soil, to support shoot development above the ground. Developmental events in LR formation include the priming and specification of LR founder cells, LR initiation (asymmetric cell divisions of LR founder cells), LR primordium development and LR emergence (Péret et al. 2009, De Rybel et al. 2010). In most dicot plants, LRs are initiated by the asymmetric cell divisions in the pericycle cells adjacent to the xylem pole. These newly divided cells develop as an LR primordium containing the LR meristem. The LR primordium emerges from the parent root tissues as a new LR. These developmental processes of LR formation are regulated by both endogenous and environmental signals (Malamy 2005, Péret et al. 2009).
Many physiological and genetic studies have shown that LR formation is regulated by several plant hormones, mainly by auxins (Fukaki et al. 2007, Fukaki and Tasaka 2009). Auxin biosynthesis, transport and signaling are important for many aspects in plant growth and developments, as well as for LR formation. Specifically, the mutations affecting endogenous auxin biosynthesis are known to affect LR formation. For example, the superroot1 (sur1)/rooty (rty)/aberrant lateral root formation1 (alf1) and sur2 mutants, which overproduce IAA, produce higher numbers of LRs (Boerjan et al. 1995, Celenza et al. 1995, King et al. 1995, Barlier et al. 2000), whereas the transport inhibitor response2 (tir2) and anthranilate synthase alpha subunit1 (asa1) mutants, which reduce IAA biosynthesis and contain less IAA, have decreased numbers of LRs (Sun et al. 2009, Yamada et al. 2009). In addition, mutants that are defective in auxin transport such as the auxin resistant1 (aux1), like-aux13 (lax3) and transport inhibitor response3 (tir3)/dark overexpression of CAB1 (doc1)/big/corymbosa (crm1) show the formation of fewer LRs (Ruegger et al. 1997, Casimiro et al. 2001, Gil et al. 2001, Swarup et al. 2001, Marchant et al. 2002, Paciorek et al. 2005, De Smet et al., 2007, Yamaguchi et al. 2007, Swarup et al. 2008). Furthermore, mutants that are defective in auxin signaling such as tir1 (transport inhibitor response1) in the auxin receptor F-box also have reduced LR formation (Ruegger et al. 1998).
In addition to these genes, several members of the AUXIN RESPONSE FACTOR (ARF) and Auxin/IAA (Aux/IAA) protein families, that regulate auxin-responsive transcription, also regulate LR formation. ARFs directly activate or repress the transcription of their target genes that contain the auxin response elements (AuxREs) in their promoter regions. In the absence of auxin, the Aux/IAA protein interacts with its partner ARF, thereby inactivating ARF activity. In the presence of auxin, the Aux/IAA protein is degraded through ubiquitination by the SCFTIR1/AFBs E3 ubiqutin ligase complex, thus permitting the activated ARF to regulate the target genes positively or negatively, resulting in the ARF-dependent auxin responses (reviewed in Hayashi 2012). Gain-of-function mutations in domain II of Aux/IAAs stabilize the protein in the presence of auxin, thereby constitutively inactivating ARF activity. Gain-of-function mutants in several Aux/IAA members were shown to inhibit LR formation (Fukaki et al. 2002, Uehara et al. 2008, reviewed in Overvoorde et al. 2010). Among them, solitary-root (slr), a gain-of-function mutant in IAA14, has no LR primordium initiation sites even in the presence of auxin, indicating that stabilized mutant IAA14 protein constitutively inactivates the ARFs responsible for LR initiation (Fukaki et al. 2002, Fukaki et al. 2005). On the other hand, the arf7 arf19 double mutant has almost no LRs, indicating that ARF7 and ARF19 positively regulate LR formation (Okushima et al. 2005, Wilmoth et al. 2005, Okushima et al. 2007). Recent molecular genetic analysis showed that the SLR/IAA14–ARF7–ARF19 auxin signaling module regulates LR initiation via the activation of several LATERAL ORGAN BOUNDARIES DOMAIN/ASYMMETRIC LEAVES2-LIKE (LBD/ASL) genes including LBD16/ASL18 (Okushima et al. 2007, Goh et al. 2012a). These LBD/ASL proteins regulate the establishment of asymmetry of LR founder cells prior to the asymmetric cell divisions for LR initiation (Goh et al. 2012a). In addition to the SLR/IAA14–ARF7–ARF19 module, other Aux/IAA–ARF signaling modules, including IAA28–ARFs, BODENLOS (BDL)/IAA12–MONOPTEROS (MP)/ARF5 and SUPPRESSOR OF HY2/SHORT HYPOCOTYL2 (SHY2)/IAA3–ARFs, take part in regulating the developmental steps during LR formation, from specification of LR founder cells (De Rybel et al. 2010), to establishment of asymmetry of LR founder cells (De Smet et al. 2010, Goh et al. 2012a), to LR primordium development and LR emergence (Swarup et al., 2008, Goh et al. 2012b). This suggests the existence of a rather complex feedback mechanism for LR formation through multiple Aux/IAA–ARF signaling modules.
The GNOM/EMBRYO DEFECTIVE30 (EMB30) encoding an ADP ribosylation factor (Arf)-GDP/GTP exchange factor (GEF), that regulates the Arf GTPase acting in vesicle trafficking, was originally reported to be required for embryogenesis (Mayer et al. 1991, Mayer et al. 1993, Shevell et al. 1994, Busch et al. 1996, Richter et al. 2010). Loss of GNOM led to improper cellular localization of PIN-FORMED1 (PIN1), an auxin efflux carrier, resulting in an embryo-lethal phenotype, indicating that GNOM plays an essential role for proper localization of PIN1 protein to the plasma membrane during embryogenesis (Steinmann et al. 1999). Partial loss-of-function gnom mutants, which can grow after germination, also have severe defects in auxin-regulated root and shoot development (Geldner et al. 2004). For example, gnomR5 has dwarf shoots, agravitropic roots, arrested root meristematic activity and defects in LR primordium initiation and development, suggesting that GNOM controls LR formation through the regulation of PIN1-dependent auxin transport as well as other auxin-regulated processes (Geldner et al. 2004). However, no gnom mutant allele has been reported to impair specifically LR initiation, and it remains unknown how GNOM regulates LR initiation, particularly in regards to the establishment of an auxin response maximum in LR initiation sites.
In this study, we have isolated a new LR mutant, fewer roots (fwr) in Arabidopsis thaliana, in which LR initiation is strongly inhibited. The fwr mutation dramatically decreases the number of LR founder cells with an auxin response maximum for LR initiation. In addition, we provide evidence that the fwr LR phenotype was caused by a missense mutation in the GNOM gene. Our results indicate that LR initiation is one of the most sensitive processes among the GNOM-dependent developmental processes, and strongly suggest that GNOM is required for the establishment of an auxin response maximum for LR initiation, probably through the regulation of local and global auxin distribution.
Results
The fwr mutation decreases the frequency of LR primordium initiation sites in Arabidopsis thaliana
fwr was isolated from ethyl methanesulfonate (EMS)-mutagenized A. thaliana M2 seedlings (Fig. 1A) as a single recessive mutant line that had fewer LRs. In 10-day-old seedlings, the primary root length of the fwr mutant was slightly shorter than that of the wild type, but the number of emerged LRs was dramatically reduced in fwr, compared with that of the wild type (Fig. 1A–C). Similarly, the total number of emerged LRs plus non-emerged LR primordia of the 10-day-old seedlings was also reduced in the fwr primary roots, compared with that in the wild-type primary roots (Fig. 1D). In 15-day-old seedlings, LRs were formed in the fwr mutant but the number of emerged LRs was still reduced in fwr, compared with that of the wild type [wild type, 63.4 ± 7.6 (mean ± SD); fwr, 20.1 ± 5.4 (mean ± SD), 13 < n < 17]. These results indicate that the fwr mutation decreases the number of LRs while allowing limited formation of LRs. When LR primordia were formed in the fwr, no obvious stage-specific developmental arrests were observed except that the transition from Stage I to Stage II was slightly affected (Fig. 1E). In addition, the organization of the fwr LR primordia appeared to be the same as that of the wild type (Supplementary Fig. S1). These observations indicate that the fwr mutation specifically reduces the frequency of LR initiation but does not drastically affect LR primordium development and LR emergence.
Fig. 1.
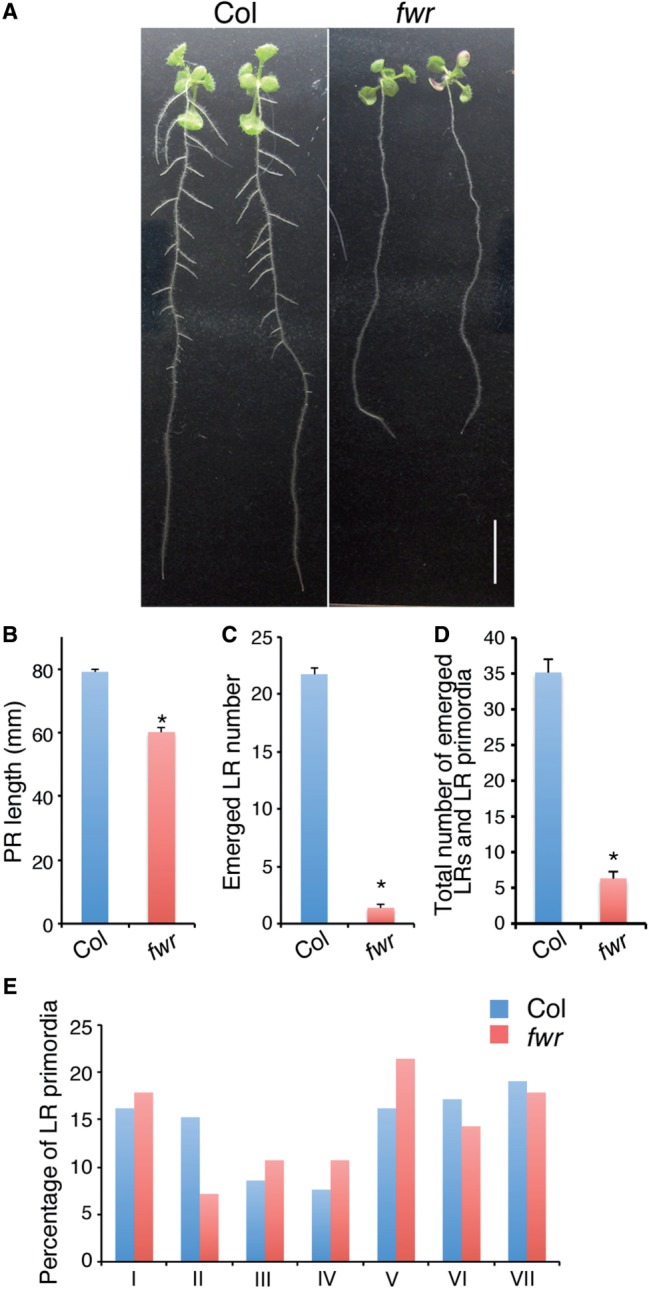
Phenotype of the fewer roots (fwr) mutant plants. (A) Ten-day-old wild-type (Col) and fwr mutant seedlings. Scale bar = 10 mm. (B–D) Primary root (PR) length (B), emerged lateral root (LR) number (C), total number of emerged LRs and LR primordia of 10-day-old wild-type (Col) and fwr mutant seedlings (D). The error bars represent the SEM (n = 25). Asterisks indicate a statistical difference (*P < 0.01, Student’s t-test). (E) Percentage of LR primordia at given developmental stages in 8-day-old wild-type (Col) and fwr mutant seedlings. Stage classification of LR primordia is based on Malamy and Benfey (1997). Total numbers of LR primordia are 105 in 14 Col seedlings and 28 in 31 fwr mutant seedlings, respectively.
To examine further whether the fwr mutation affects the identity of xylem pole pericycle from which LRs are initiated, we investigated the expression of the xylem pole pericycle-specific marker J0121 in the fwr mutant roots. J0121 was expressed in the xylem pole pericycle of the fwr mutant roots to the same levels as in the wild type (Fig. 2), suggesting that the identity of xylem pole pericycle is not affected in the fwr mutants.
Fig. 2.
Cell identity of the xylem pole pericycle in the fwr mutant seedling. (A–D) Expression of the J0121 enhancer trap line that expresses GFP at the xylem pole pericycle of Col (A, B) and fwr (C, D) seedlings. Scale bar = 50 µm.
In the fwr mutants, both root hair formation and root gravitropic response appeared to be unaffected, and leaf and flower development and embryogenesis also appeared not to be severely impaired (data not shown), although the rosette leaf shape was slightly narrow and curled downwards compared with that of the wild type (Supplementary Fig. S2). This indicates that the fwr mutation mainly affects post-embryonic root development, specifically LR formation.
The fwr mutation inhibits the establishment of an auxin response maximum in LR initiation sites
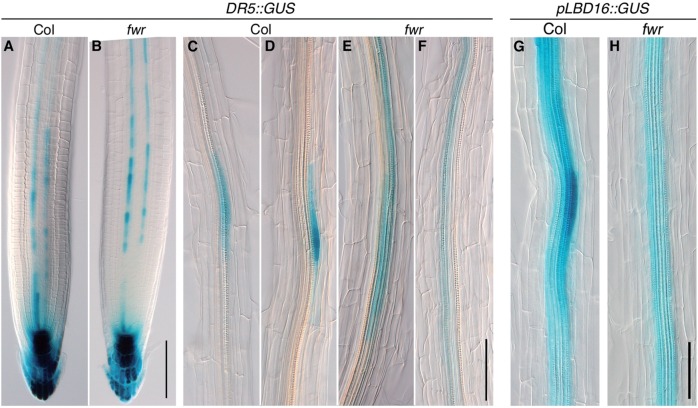
To investigate whether the fwr mutation affects the auxin response maximum in the xylem pole pericycle, which is the site of LR initiation, we examined the expression of a reporter GUS (β-glucuronidase) gene construct under the control of the DR5 promoter, DR5::GUS, to monitor the auxin response maximum in both fwr and the wild type (Ulmasov et al. 1997, Benková et al. 2003). The 5-day-old wild-type seedlings had several LR initiation sites and LR primordia marked by a strong DR5::GUS activity (Fig. 3C, D; Supplementary Fig. S3). In contrast, the 5-day-old fwr mutant seedlings had almost no LR initiation sites, and distinct DR5::GUS activity was not observed in the xylem pole pericycle (Fig. 3E, F; Supplementary Fig. S3). These observations suggest that the fwr mutation led to decreased frequency of LR initiation with the auxin response maximum at the xylem pole pericycle, thereby decreasing the number of LRs. In order to determine upon which developmental step, during the LR initiation, the fwr mutation exerts its inhibitory effect, we examined the expression of the LBD16/ASL18 gene, which is the direct target of ARF7 and ARF19 for LR initiation (Okushima et al. 2007). It is reported that LBD16/ASL18 expression occurs in the LR founder cells with an auxin response maximum (Goh et al. 2012a). In the 5-day-old seedlings, strong pLBD16::GUS activity was observed in the LR initiation sites at the xylem pole pericycle in the wild-type background (Fig. 3G; Supplementary Fig. S4), whereas almost no LR initiation sites with distinct pLBD16::GUS activity could be observed in the fwr seedlings, although weak GUS activity was observed along the root stele which was also shown in the wild type (Fig. 3H; Supplementary Fig. S4). This indicates that the LBD16/ASL18 gene was not adequately activated in the xylem pole pericycle of the 5-day-old fwr mutant seedlings. On the other hand, when LR primordia were formed in the 10-day-old fwr mutant seedlings, pLBD16::GUS was normally expressed in the LR initiation sites and the developing LR primordia as observed in the wild type (Supplementary Fig. S1), although the number of LR primordia was reduced in the fwr mutant (data not shown). Taken together, these observations indicate that the fwr mutation inhibits the establishment of an auxin response maximum in LR initiation sites.
Fig. 3.
The fwr mutation inhibits the establishment of an auxin response maximum for LR initiation. (A, B) Expression of DR5::GUS in the root tip of Col (A) and fwr (B) seedlings [5 days after germination (DAG)]. (C–F) Expression of DR5::GUS in the mature root region of Col (C, D) and fwr (E, F) seedlings (5 DAG). (G, H) Expression pattern of pLBD16::GUS in the Col (G) and fwr (H) roots. Scale bars = 100 µm.
In the root tip, similar DR5::GUS expression patterns were detected in both fwr and the wild type except that GUS activity in the LR cap region tended to be absent or lower in fwr (16 cases /20 samples) than in the wild type (3 cases /20 samples) (Fig. 3A, B). This suggests that the fwr mutation may reduce auxin transport or signaling in the root tip region. Taken together, the fwr mutation affects the auxin response pattern in the root, resulting in the reduced LR phenotype.
The fwr mutant LR phenotype can be restored by low concentrations of auxins
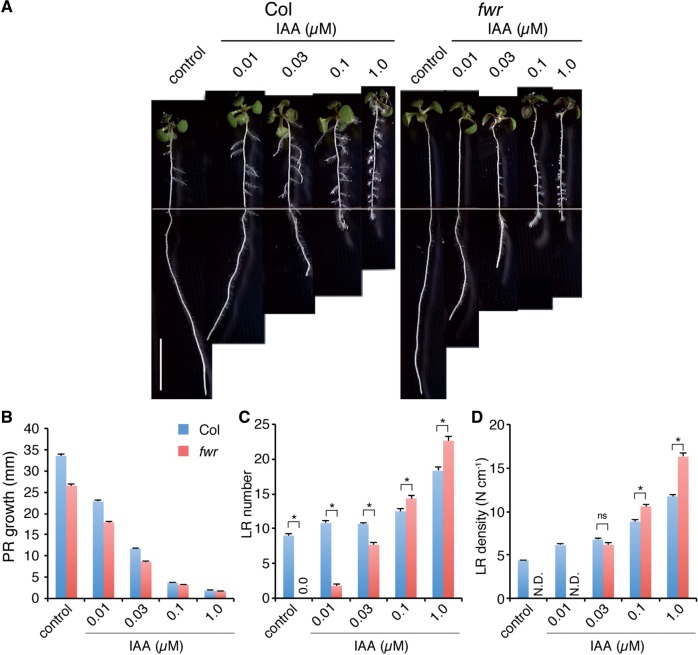
Based on the fwr LR phenotype, we hypothesized that the fwr mutation might affect auxin responses. In order to investigate whether the fwr mutation changes the auxin responsiveness in the roots, we examined the effect of exogenous auxin on primary root growth and LR formation. When 4-day-old seedlings grown on auxin-free media were transferred onto auxin [IAA or naphthylphthalamic acid (NAA)]-containing media and incubated for an additional 3 d, the effect of exogenously applied auxin on primary root growth was found to be similar in both the fwr mutant and the wild type (Fig. 4A, B; Supplementary Fig. S5). In the wild type, LR formation was induced depending on the concentration of exogenous auxin, and the LR density (the number of LRs per portion of the primary root where LRs are present) increased with auxin treatment (Fig. 4A, C, D; Supplementary Fig. S5). In the fwr mutant, the auxin-induced LR formation was observed with either 0.03 µM IAA or 0.03 µM NAA and the LR density was restored to wild-type levels (Fig. 4C, D; Supplementary Fig. S5). Interestingly, higher concentrations of auxin (either 0.1 µM IAA, 1.0 µM IAA or 1.0 µM NAA) increased the LR density in the fwr mutant significantly more than in the wild type (Fig. 4C, D; Supplementary Fig. S5). These results indicate that the fwr mutant remains responsive to low concentrations of exogenous auxin to form LRs but shows enhanced sensitivity to higher concentrations of exogenous auxin, strongly suggesting that the fwr mutation may attenuate auxin distribution for LR formation.
Fig. 4.
Auxin inhibition of primary root growth and auxin induction of LR formation in Col and fwr mutant seedlings. (A) Four-day-old Col and fwr mutant seedlings were transferred onto IAA-free (control) or IAA-containing media, and incubated for an additional 3 d. Scale bar = 10 mm. Representative seedlings are shown for each treatment. (B) Root elongation of seedlings on IAA-containing media for 72 h. The error bars represent the SEM (n = 25). (C) LR number of seedlings grown on IAA-containing media for 72 h. The values for fwr mutants on hormone-free media (control) are 0.0 ± 0.0 (mean ± SEM). The error bars represent the SEM. The asterisks indicate a statistical difference between Col and the fwr mutant (*P < 0.01 by Student’s t-test). ns, not significant. n = 25. (D) LR density of seedlings on IAA-containing media for 72 h. The values of fwr mutants on hormone-free (control) and 0.01 µM IAA media is not determined due to a limited number of LR primordia (N.D.). The error bars represent the SEM. The asterisks indicate a statistical difference between Col and the fwr mutant (*P < 0.01 by Student’s t-test). ns, not significant. n = 25. Experiments were repeated twice, and similar results were obtained in each experiment.
fwr is a new weak gnom allele that specifically inhibits LR initiation
To isolate the FWR gene, detailed mapping using the F2 generation of fwr (accession Columbia) and accession Landsberg erecta was performed. The FWR gene was identified within an approximately 120 kb genomic region of a bacterial artificial chromosome (BAC) clone (F7A19) of chromosome 1 (Supplementary Fig. S6). After sequencing the genes annotated in this region, we found that the fwr mutant genome has a single base pair substitution in the At1g13980 gene, which encodes GNOM/EMB30, an Arf-GEF that functions in vesicle trafficking and is required for embryogenesis (Mayer et al. 1991, Mayer et al. 1993, Shevell et al. 1994, Busch et al. 1996). This mutation caused a missense mutation (C to T) in the open reading frame that changes the 1,005th serine to phenyalanine in the HDS2 (Homology Downstream of SEC7 2) domain (Fig. 5A; Supplementary Fig. S6). Interestingly, this serine residue is conserved among several GNOM-related proteins, AtGNL1 and AtGNL2 in Arabidopsis, and CeGBF1 and HsGBF1 in animals (Mouratou et al. 2005) (Fig. 5B), suggesting the functional importance of this residue. Expression analysis by semi-quantitative reverse transcription–PCR (RT–PCR) showed that mutant GNOM mRNAs were expressed in the fwr mutant seedlings, suggesting that the mutant GNOM protein is expressed in the fwr mutant (Fig. 5D). To confirm genetically that the fwr is a mutant allele of the GNOM gene, the allelism test using a T-DNA insertion line of GNOM, SALK_103014, gnomSALK_103014 was performed. The gnomSALK_103014 line showed the aberrant seedling lethal phenotype in which shoot and root apical meristems were not produced, similar to that of gnom/emb30 (Fig. 5C; Mayer et al. 1991, Mayer et al. 1993). All heterozygous plants of the fwr allele and the gnomSALK_103014 gnomSALK_103014 allele had a shorter primary root without any LRs, which is a more severe phenotype than fwr but a milder phenotype than gnomSALK_103014, indicating that fwr is genetically allelic to gnomSALK_103014 (Fig. 5C). In addition, we also performed a molecular complementation test by introducing the genomicGNOM-GFP construct into the fwr mutant. The fwr mutant plants expressing the GNOM–green fluorescent protein (GFP) fusion protein under the control of the native GNOM promoter (genomicGNOM-GFP/fwr) produced wild-type levels of LRs (Supplementary Fig. S7). The primary root growth was also restored in the genomicGNOM-GFP/fwr seedlings (Supplementary Fig. S7). Furthermore, we confirmed that the subcellular localization of GNOM–GFP fluorescence was observed at the specific compartments closely associated and partially overlapping with FM4-64-labeled endosomes in these transgenic root cells, consistent with previous reports on the subcellular localization of GNOM (Supplementary Fig. S7; Geldner et al. 2003). These results indicate that GNOM–GFP functionally rescued the fwr phenotype for both LR formation and primary root growth. To examine whether the fwr mutation affects the subcellular localization of GNOM–GFP, we also produced transgenic plants expressing the gnomfwr–GFP fusion protein under the control of the native GNOM promoter. Interestingly, the subcellular localization of gnomfwr–GFP fluorescence was mainly observed at the plasma membrane, which is different from that of GNOM–GFP (Supplementary Fig. S7). This suggests that the fwr mutation might affect the subcellular localization of GNOM. However, the gnomfwr–GFP fusion protein was functional as well as the GNOM-GFP in the gnomSALK_103014 mutant background, suggesting that the addition of GFP to the C-terminus of the gnomfwr mutant protein might rescue the function (data not shown).
Fig. 5.
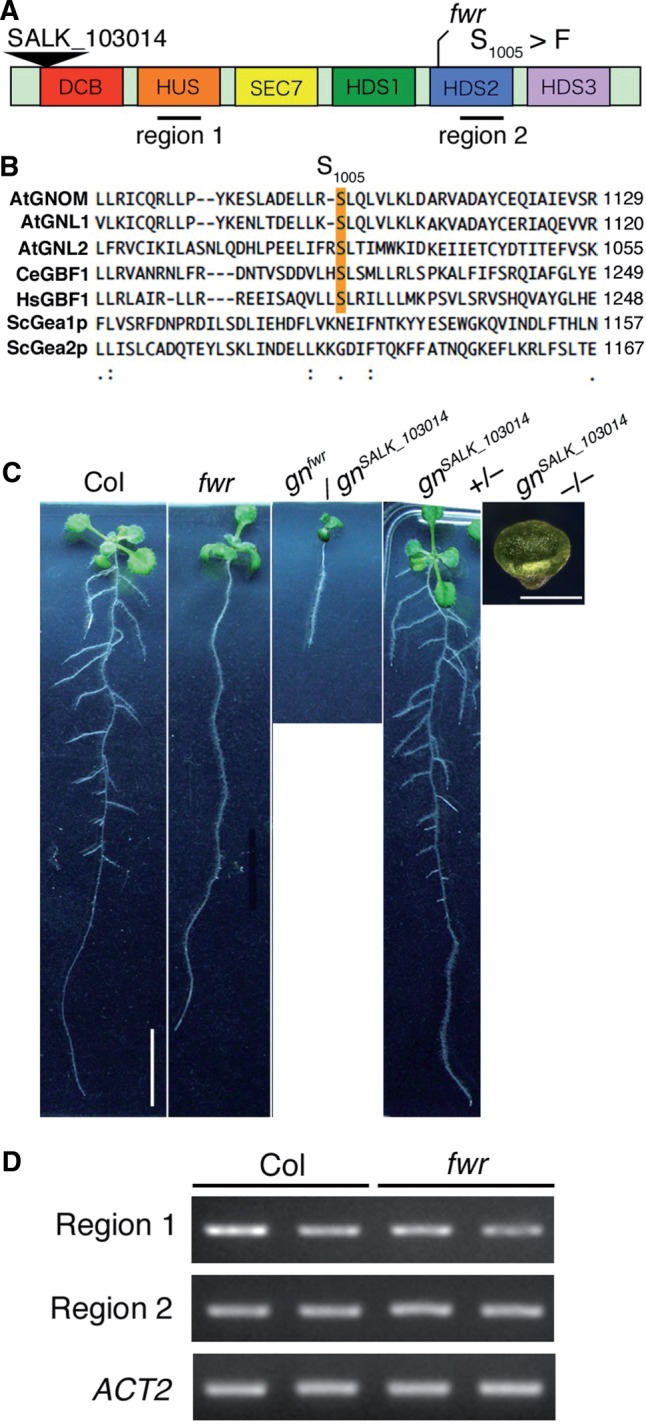
The FWR gene encodes GNOM protein. (A) Protein structure of GNOM (GN) and mutation point of fwr. GN has several characteristic domains: DCB, dimerization/cyclophilin-binding domain; HUS, homology upstream of Sec7 domain; Sec7, Sec7 domain; HDS, homology downstream of Sec7 domain. fwr has a single nucleotide mutation in HDS2 that caused Ser1,005Phe alteration. The black triangle represents the insertion site of T-DNA in the SALK_103014 line. (B) Amino acid sequence alignment of the flanking region of the fwr mutation of Gea/GNOM/GBF family members. Budding yeast Gea1p and Gea2p (ScGea1p and ScGea2p), Caenorhabditis elegans GBF1 (CeGBF1), human GBF1 (HsGBF1) and Arabidopsis GNL1 and GNL2. Sequences were aligned using ClutstalW. The mutated residue (Ser1,005Phe) in fwr is highlighted in orange. (C) Allelism test between the fwr mutant and the GNOM knockout T-DNA mutant (SALK_103014). Scale bars indicate 10 mm (Col) and 2 mm (gnomSALK_103014), respectively. (D) Accumulation of GNOM mRNA in Col and fwr mutant seedling roots. The black lines in (A) indicate the amplified regions by RT–PCR. The expression of the ACT2 gene was used as a control.
It is reported that several partial loss-of-function gnom mutant alleles affect most of the growth and developmental processes (Koizumi et al. 2000, Geldner at al. 2004, Richter et al. 2010). gnomR5, one of the weak gnom alleles, has a pleiotropic phenotype including dwarf shoots, agravitropic roots, arrested root meristematic activity and defects in LR primordium initiation and development (Geldner et al. 2004). However, no gnom mutant allele has been reported specifically to impair LR initiation. It is noted that the embryogenesis, shoot development and fertility were not drastically affected by the fwr mutation. These results indicate that fwr is a new partial loss-of-function mutant allele of the GNOM gene, which mainly inhibits GNOM’s function in LR initiation.
The fwr mutation confers hypersensitivity to BFA-mediated root growth inhibition
To examine whether the fwr mutation affects the Arf-GEF activity of GNOM, we examined the sensitivity of the fwr mutant to brefeldin A (BFA) that inhibits the Arf-GEF activity in vesicle transport. When 4-day-old seedlings grown on BFA-free media were transferred onto BFA-containing media and incubated for an additional 3 d, the primary root growth of the fwr mutant was more inhibited at either 1 or 5 µM BFA, compared with that of the wild type, indicating that the fwr mutation confers enhanced sensitivity to BFA-mediated root growth inhibition (Fig. 6). This result indicates that the fwr mutation reduces the Arf-GEF activity of GNOM, thereby confirming that fwr is a weak gnom allele.
Fig. 6.
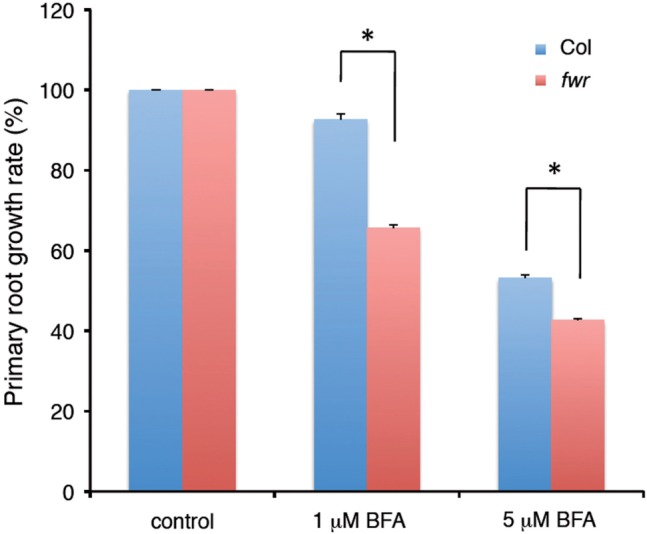
BFA inhibition of primary root growth in Col and fwr mutant seedlings. Four-day-old seedlings grown on MS medium were transferred to medium containing different concentrations of BFA. Root growth after 3 d with each concentration of BFA was measured. For each genotype, the inhibition of root growth relative to the growth on unsupplemented medium (mock) was shown. Root increment (elongation) of mock-treated seedlings during 3 d was 25.1 ± 0.5 (mean ± SEM) mm for wild-type Col, and 22.1 ± 0.9 mm for fwr. The error bars represent the SEM. The asterisks indicate a statistical difference between Col and the fwr mutant (*P < 0.0001 by Student’s t test). ns, not significant. n = 17. Experiments were repeated twice, and similar results were obtained in each experiment.
Localization of PIN1–GFP in the fwr mutant roots
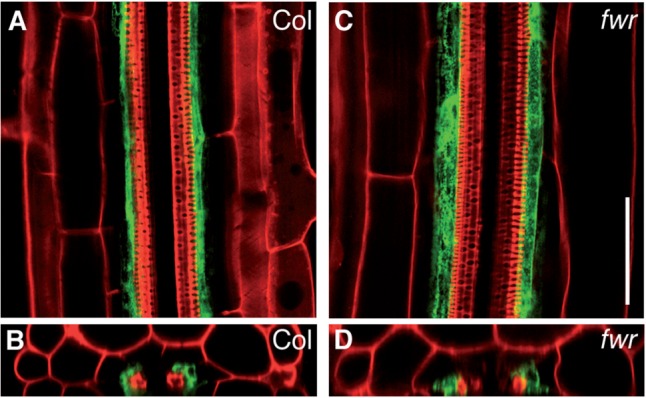
GNOM is known to regulate the recycling of PIN1, an auxin efflux carrier, thereby affecting the subcellular localization of PIN1 at the basal plasma membrane, which is important for several auxin-dependent growth and developmental processes (Steinmann et al. 1999, Geldner et al. 2001, Geldner et al. 2003). To determine whether the fwr mutation affects the localization of PIN1 in the root, we genetically crossed the PIN1–GFP line (Friml et al. 2003) into the fwr mutant and observed PIN1–GFP fluorescence in the fwr roots. As shown in Fig. 7, PIN1–GFP was localized at the basal membrane of the root stele cells in fwr as observed in the wild type (Fig. 7A). In the Stage I LR primordia and later stages, no distinct difference in PIN1–GFP localization was observed between the wild type and fwr (Fig. 7B, C), which suggest that at least PIN1-mediated auxin transport for LR formation is dramatically unaffected by the fwr mutation.
Fig. 7.
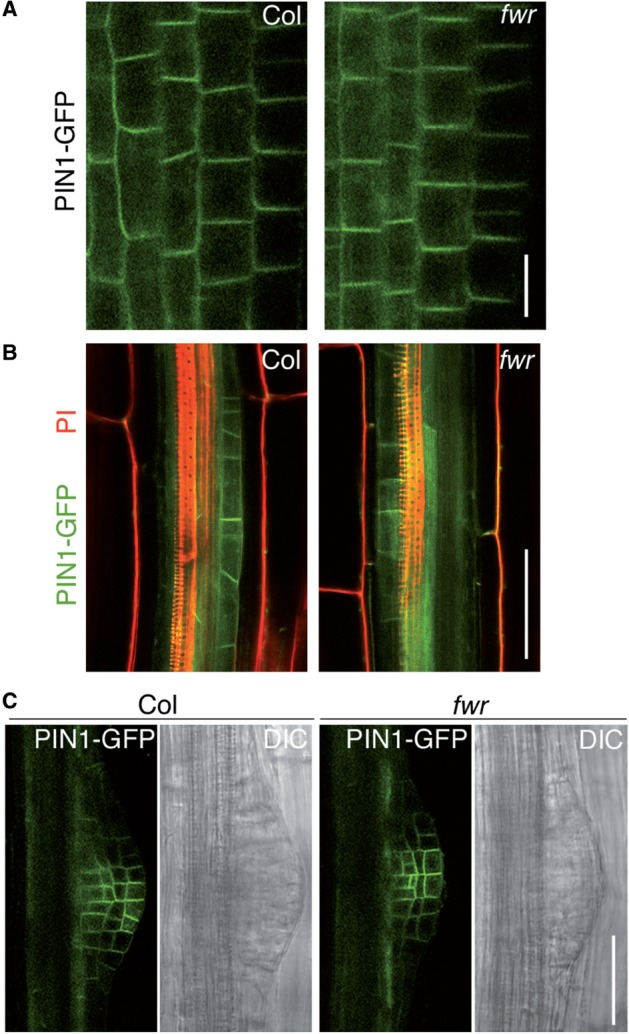
Subcellular localization of PIN1–GFP in Col and fwr mutant seedling. (A–C) Subcellular localization of PIN1–GFP in the stele cells of the root tip (A), Stage I LR primordium (B) and Stage IV LR primordium (C). Scale bar in (A) = 10 µm. Scale bar in (B, C) = 50 µm.
The fwr mutation increases the endogenous IAA level in roots
PIN1 is necessary for polar auxin transport from the shoots toward the roots (Okada et al. 1991). As this process is important for LR formation (Reed et al. 1998, Casimiro et al. 2001), it is possible that the fwr mutation impairs PIN1-dependent polar auxin transport from the shoots toward the roots, thereby inhibiting LR formation. To examine whether the fwr mutation in GNOM decrease the levels of endogenous IAA in the roots, we measured endogenous IAA levels in the roots of 5-day-old wild-type and fwr seedlings by LC-ESI-MS/MS (liquid chromatography-electrospray ionization-tandem mass spectroscopy) analysis (see the Materials and Methods). Interestingly, the level of endogenous IAA in the roots of fwr seedlings is much higher than that in wild-type seedlings (Fig. 8). These results indicate that that GNOM/FWR negatively control the endogenous IAA levels in roots and that the fwr mutation inhibits LR initiation without decreasing the total levels of endogenous IAA in the roots.
Fig. 8.
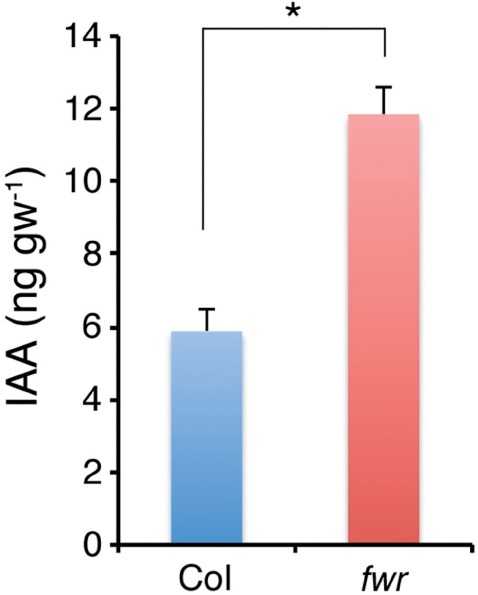
IAA levels in Col and fwr mutant seedlings. IAA levels (ng g−1 FW) in the roots of the 5-day-old Col and fwr mutant seedlings are shown (n = 3). Error bars indicate the SEM. The asterisk indicates a statistical difference (P < 0.001 by a two-sided t-test). Experiments were repeated twice, and similar results were obtained in each experiment.
Discussion
In this study, we have identified the fwr mutant in Arabidopsis that is specifically defective in LR initiation and we demonstrated that fwr is a weak mutant allele of gnom, which revealed a more critical role for GNOM in LR initiation. Our results indicated that LR initiation is one of the most sensitive developmental processes among those regulated by GNOM, and strongly suggest that GNOM is required for the establishment of an auxin response maximum for LR initiation, probably through the regulation of local and global auxin distribution.
GNOM is required for the establishment of an auxin response maximum for LR initiation
GNOM is known to be required for embryogenesis and post-embryogenic development (Mayer et al. 1991, Mayer et al. 1993, Geldner et al. 2004, Wolters et al. 2011). The use of gnomR5, a weak mutant allele, has shown that GNOM plays a role in the establishment of an auxin gradient in the developing LR primordia (Geldner et al. 2004). The multiple mutations among the PIN members including PIN1, PIN3, PIN4 and PIN7 also showed the seedling-lethal phenotype in which the auxin-induced LR primordium development was also altered; aberrant pericycle cell divisions occurred, indicating that PIN-dependent auxin transport is important for proper LR primordium development (Benková et al. 2003). Although the gnomR5 mutation blocks the expression of the ARABIDOPSIS CRINKY 4 (ACR4) gene in the xylem pole pericycle, that is known as a marker gene for the LR initiation site (De Smet et al. 2008), it is difficult to distinguish this from the possibility that the gnomR5 LR phenotypes might be due to the secondary effects of gnomR5 primary growth arrest because gnomR5 had pleiotropic phenotypes both in the shoots and in the roots, in which the primary root growth was inhibited to 20–30% of the wild-type level along with reduced root meristematic activity (Geldner et al. 2004). In contrast, the fwr mutant had a strong phenotype in LR initiation but not in LR primordium development, LR emergence or other growth and developmental processes. While the gnomR5 allele exhibited aberrant LR primordium development in response to exogenous NAA at 0.1 µM, that showed multilayered dividing pericycle cells with altered PIN1 localization (Geldner et al. 2004), the fwr mutant did not show similar aberrant LR primordium in response to exogenous auxin treatments at 0.1 µM NAA (data not shown). These observations indicate that LR initiation is more sensitive to the activity of GNOM than to other developmental processes. Measurements of endogenous auxin and physiological analysis with exogenous auxin suggested that the fwr mutation might have reduced the availability of endogenous auxin for LR initiation in spite of the increased levels of IAA in the roots. This strongly suggests that the fwr mutation affects local and global auxin distribution for the establishment of an auxin response maximum in the LR founder cells. It is possible that GNOM mediates auxin transport from the adjacent tissues towards the LR founder cells. It will be necessary to determine which tissues/cells and which components contribute to such local and global auxin transport for LR initiation.
Our results indicate that GNOM is necessary for LR initiation, most probably for the step for the establishment of an auxin response maximum in the LR founder cells. It was reported that GNOM regulates LR primordium development probably through the regulation of PIN1 relocalization from Stage I to Stage II (Geldner et al. 2004). As the gnomR5 seedlings had no LR initiation events in the absence of exogenous auxin and showed aberrant formation of LR primordia in the presence of exogenous auxin, it was postulated that GNOM regulates PIN1 localization for the initial anticlinal cell divisions of the pericycle toward Stage I, and also in periclinal cell divisions from Stage I to Stage II during LR formation (Geldner et al. 2004). Kleine-Vehn et al. (2008) demonstrated that PIN1 is localized at both the apical and basal sides of the divided pericycle cells at Stage I and also at the lateral side of the inner layer cells at Stage II. However, the location of PIN1 in the LR founder cells before the first anticlinal, asymmetric cell divisions of the xylem pole pericycle is unknown, because of the difficulty in locating the protein before Stage I. The role played by PIN1 before Stage I, for the specification of LR founder cells, or the establishment of an auxin response maximum in the LR founder cells, remains unknown. The pin1 loss-of-function mutation did not alter LR density (Laskowski et al. 2008), strongly suggesting the redundant functions of PIN members in LR primordium initiation and development. It is unknown whether GNOM regulates the localization of other PIN members, thereby mediating auxin transport from the adjacent tissues toward the LR founder cells. Previous results on the gnomR5 mutant (Geldner et al. 2004) and our results on the fwr mutant suggest that GNOM probably regulates PIN-mediated auxin transport to establish the auxin response maximum in the LR founder cells, thereby activating auxin signaling modules to induce LR initiation, although we do not exclude the possibility that the other proteins regulated by GNOM also regulate auxin distribution for LR initiation.
The gnomfwr protein acts in a dose-dependent manner and is sufficient for embryogenesis and post-embryonic shoot development
In our allelism test using a T-DNA insertion line of GNOM, SALK_103014, gnomSALK_103014, all F1 plants from the cross between fwr and gnomSALK_103014 showed a more severe phenotype than fwr whereas the F1 plants from the cross between the wild type and gnomSALK_103014 showed the wild-type phenotype (Fig. 5C). Although the accumulation of gnomfwr mutant protein in both the fwr/fwr homozygote and fwr/gnomSALK_103014 heterozygote was not determined in this study, the genetic analysis suggests that the fwr (gnomfwr) mutant gene acts in a dose-dependent manner in the absence of the wild-type GNOM gene. It is possible that the level of gnomfwr mutant protein in the fwr/fwr homozygote was sufficient to support shoot development and primary root growth but insufficient for normal levels of LR initiation. It is also possible that the fwr mutation in the HDS2 domain affects the activity or stability of the GNOM protein. Further analysis of the gnomfwr mutant protein will reveal the mechanism of how GNOM functions in regulating LR initiation.
The fwr mutant is not embryonic/seedling lethal and it produces leaves, flowers and seeds as well as the wild type, indicating that the gnomfwr mutation does not impair the molecular function of GNOM in embryogenesis and shoot development. This suggests that GNOM-dependent vesicle transport for root development, particularly LR initiation, is more sensitive to the alteration of GNOM activity. It is unknown how the HDS domains in the GNOM protein control the activity of the protein, but our results and previous results indicate that these domains may be involved in determining GNOM activity. The mizukussei2 (miz2) is another gnom mutant allele that specifically inhibits root hydrotropism (Miyazawa et al. 2009). The miz2 mutation caused an amino acid substitution located in the region between the HDS1 and HDS2 domains in the C-terminal half of GNOM. Interestingly, the miz2 mutation does not cause any other developmental defects, indicating that it specifically affects GNOM’s activity in the regulation of hydrotropism (Miyazawa et al. 2009). The miz2 mutation was also reported to have no effect on the cellular localization of PIN1 (Miyazawa et al. 2009), suggesting that GNOM regulates hydrotropism without involving PIN1. The mutant phenotypes are totally different between miz2 and fwr, suggesting that the HDS domains of the GNOM C-terminal region affect specifically its function in LR initiation and root hydrotropism.
In this study, we have provided evidence showing that GNOM functions in the specific step of LR initiation, which is controlling the establishment of an auxin response maximum to activate LR initiation. It is possible that GNOM regulates local and global auxin distribution, thereby affecting auxin availability for the establishment of an auxin response maximum in the LR founder cells. It remains unknown whether GNOM regulates this step through the control of cellular localization of several PIN members, or, as in the case of the miz2 mutant, that vesicle transport of unknown components other than PIN members may be involved in LR initiation. In A. thaliana, several genes that are unrelated to auxin physiology, such as ABERRANT LATERAL ROOT FORMATION4 (ALF4; Didonato et al. 2004) and REDUCED LATERAL ROOT FORMATION (RLF; Ikeyama et al. 2010), have been shown to be involved in LR initiation as well. It would be of interest to examine the relationship between GNOM and these regulators and to study how LR founder cells with an auxin response maximum are established in the xylem pole pericycle. Analysis with the use of the fwr mutant allele could prove to be helpful to unravel the genetic and molecular mechanisms of GNOM-mediated LR formation as well as other growth and developmental processes in higher plants.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana used in this study were Columbia (Col) and Landsberg erecta (Ler) accessions. The DR5::GUS seeds were kindly provided by Tom Guilfoyle (University of Missouri, USA). The PIN1-GFP seeds were kindly provided by Jiří Friml (Ghent University, Belgium). The pLBD16::GUS line was described by Okushima et al. (2007). The T-DNA insertion line (gnomSALK_103014) in the GNOM (SALK_103014) seeds was obtained from the Arabidopsis Biological Resource Center (ABRC). The J0121 line was obtained from the Nottingham Arabidopsis Stock Centre (NASC). The fwr mutant was isolated from the EMS-mutagenized M2 Col seeds obtained from LEHLE SEEDS (http://www.arabidopsis.com/). The fwr mutant was backcrossed more than three times before the analysis. Seeds were surface-sterilized and plated on Murashige and Skoog (MS) medium containing 1.0% sucrose solidified with 0.5% gellan gum as described previously (Fukaki et al. 2002). Plants were grown at 23°C under continuous light. The number of LRs and root lengths were determined using a dissecting microscope and ImageJ software (NIH).
Mapping of the FWR gene
DNA from individual wild-type F2 plants from a cross between fwr (Col) and the wild type (Ler) was isolated and analyzed for co-segregation with various cleaved amplified polymorphic sequence (CAPS) and simple sequence length polymorphism (SSLP) markers (Konieczny and Ausubel 1993, Bell and Ecker 1994). Primers used for mapping are listed in Supplementary Table S1. Polymorphisms between Col and Ler ecotypes were found with the use of a dCAPS (F7A19C) marker and an SSLP (F7A19) marker on chromosome 1, respectively (Supplementary Fig. S4). Of the 2,920 chromosomes examined, four recombination events were found between the F7A19C marker and the fwr mutation. Of the 3,042 chromosomes examined, two recombination events were found between F7A19 and the fwr mutation. These markers mapped the fwr mutation to within about a 120 kb genomic region in the BAC clone F7A19. The At1g13980 gene encoding GNOM in this region was amplified from the fwr mutant genomic DNA with several pairs of primers based on the genomic sequence containing the At1g13980 gene. PCR products were purified and directly sequenced by standard methods using these PCR primers and additional internal primers.
Vector construction and plant transformation
Primers used for plasmid construction of GNOM-GFP are listed in Supplementary Table S2. The genomic GNOM-GFP line was constructed by the technique of fluorescent tagging of full-length proteins (Tian et al. 2004). The cDNA encoding GFP was inserted in-frame immediately preceding the stop codon of the GNOM gene-coding sequence. The complete sequence includes 2.4 kb of the 5′- and 0.6 kb of the 3′-flanking sequences of GNOM. The amplified sequence was subcloned into pENTR D-TOPO (Invitrogen) and then transferred to pGWB501 (Nakagawa et al. 2007) by Gateway technology (Invitrogen). The genomic GNOMfwr-GFP was produced by PCR-based site-directed mutagenesis. The transformation of Arabidopsis plants was performed via floral dipping using Agrobacterium tumefaciens (strain C58MP90) (Clough and Bent 1998).
RNA extraction and RT–PCR analysis
Total RNA was isolated from 5-day-old root tissues using an RNeasy Plant mini kit (Qiagen). First-strand cDNA was synthesized from 1 µg of total RNA with a ReverTra Ace qPCR RT kit (TOYOBO). Transcripts were quantified by PCR using 1/30 (GNOM) or 1/100 (ACT2) of the resulting cDNA as template, respectively. Transcripts were amplified by 30 cycles of PCR with gene-specific primers. Primers used for RT–PCR are listed in Supplementary Table S3.
Microscopic analysis
GUS staining, fixation and whole-mount clearing preparation of roots were performed essentially as described previously (Malamy and Benfey 1997), and samples were observed with a Leica DM6000 microscope equipped with Nomarski optics (Leica Microsystems). For confocal microscopy, roots were counterstained with propidium iodide (10 µg ml–1) and analyzed with an Olympus FV1000 confocal microscope. For labeling with FM4-64, the roots were soaked in MS liquid medium plus 4 µM FM4-64 on ice for 10 min. Samples were washed twice, incubated for 15 min at 23°C and then observed under a confocal laser scanning microscope.
Measurement of endogenous IAA
Fresh plant tissues (30–40 mg) were homogenized in 80% acetonitrile/1% acetic acid/H2O (v/v, 0.5 ml) containing [13C6]IAA (Cambridge Isotope Laboratories) as an internal standard by using a Tissue Lyser (Qiagen. The extract was centrifuged at 14,000×g for 10 min and the supernatant was transferred to a glass tube (5 ml). The pellet was further washed with 80% acetonitrile containing 1% acetic acid (v/v) and centrifuged. The supernatants were combined and evaporated by a Speed Vac (Thermo Scientific). The extract was dissolved in 1% acetic acid and loaded in a Waters Oasis WAX extraction cartridge (1 ml). After washing with 1% acetic acid (1 ml) and 80% acetonitrile (2 ml), IAA was eluted with 80% acetonitrile/1% acetic acid/H2O (1 ml). After evaporation by a Speed Vac, the IAA fraction was re-dissolved in 1% acetic acid (20 µl) and injected into an LC-ESI-MS/MS apparatus 6410 (Agilent) equipped with a ZORBAX Eclipse XDB-C18 column (Agilent). Endogenous IAA levels were quantified as previously described (Yoshimoto et al. 2009).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan [Grants in Aid to H.F. for Scientific Research on Priority Areas (Plant Meristems, grant No. 19060006)].
Supplementary Material
Acknowledgments
We wish to thank Tom Guilfoyle (University of Missouri, Missouri, USA) and Jiří Friml (Ghent University, Belgium) for providing the materials. We thank Masao Tasaka (Nara Institute of Science and Technology) for supporting the mutant screening. We thank Yoshie Okamoto for technical assistance.
Glossary
Abbreviations
- ARF
AUXIN RESPONSE FACTOR
- Arf
ADP ribosylation factor
- Aux/IAA
Auxin/IAA
- BAC
bacterial artificial chromosome
- BFA
brefeldin A
- EMS
ethyl methanesulfonate
- fwr
fewer roots
- GEF
GDP/GTP exchange factor
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- LC-ESI-MS/MS
liquid chromatography-electrospray ionization-tandem mass spectroscopy
- LBD16/ASL18
LATERAL ORGAN BOUNDARIES DOMAIN16/ASYMMETRIC LEAVES2-LIKE18
- LR
lateral root
- MS
Murashige and Skoog
- NAA
naphthylphthalamic acid
- PIN
PIN-FORMED
- RT–PCR
reverse transcription–PCR
- SLR
SOLITARY-ROOT.
References
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, et al. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl Acad. Sci. USA. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Benková E, Micchniewicz M, Sauer M, Teichmann T, Seifertova D, Jürgens G, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera M-T, Delarue M, Beeckman T, Dewitte W, Bellini C, et al. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch M, Mayer U, Jürgens G. Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol. Gen. Genet. 1996;250:681–691. doi: 10.1007/BF02172979. [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, et al. Auxin transport promotes Arabidopsis lateral root inhibition. Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisa PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 2010;20:1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher EH, et al. Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl Acad. Sci. USA. 2010;107:2705–2710. doi: 10.1073/pnas.0915001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322:594–597. doi: 10.1126/science.1160158. [DOI] [PubMed] [Google Scholar]
- DiDonato RJ, Arbuckle E, Buker S, Sheets J, Tobar J, Totong R, et al. Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J. 2004;37:340–353. doi: 10.1046/j.1365-313x.2003.01964.x. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 2005;44:382–395. doi: 10.1111/j.1365-313X.2005.02537.x. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Okushima Y, Tasaka M. Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 2007;256:111–137. doi: 10.1016/S0074-7696(07)56004-3. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M. Hormone interactions during lateral root formation. Plant Mol. Biol. 2009;69:437–449. doi: 10.1007/s11103-008-9417-2. [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof Y-D, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Geldner N, Richter S, Vieten A, Marquardt S, Torres-Ruiz RA, Mayer U, et al. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development. 2004;131:389–400. doi: 10.1242/dev.00926. [DOI] [PubMed] [Google Scholar]
- Gil P, Dewey E, Friml J, Zhao Y, Snowden KC, Putterill J, et al. BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 2001;15:1985–1997. doi: 10.1101/gad.905201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development. 2012a;139:883–893. doi: 10.1242/dev.071928. [DOI] [PubMed] [Google Scholar]
- Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple Aux/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signaling. Philos. Trans. R. Soc. B: Biol. Sci. 2012b;367:1461–1468. doi: 10.1098/rstb.2011.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. The interaction and integration of auxin signaling components. Plant Cell Physiol. 2012;53:965–975. doi: 10.1093/pcp/pcs035. [DOI] [PubMed] [Google Scholar]
- Ikeyama Y, Tasaka M, Fukaki H. RLF, a cytochrome b5-like heme/steroid binding domain protein, controls lateral root formation independently of ARF7/19-mediated auxin signaling in Arabidopsis thaliana. Plant J. 2010;62:865–875. doi: 10.1111/j.1365-313X.2010.04199.x. [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell. 1995;7:2023–2037. doi: 10.1105/tpc.7.12.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe P, Sauer M, Brewer PB, Wisniewska J, Paciorek T, et al. ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 2008;18:526–531. doi: 10.1016/j.cub.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Koizumi K, Sugiyama M, Fukuda H. A series of novel mutants of Arabidopsis thaliana that are defective in the formation of continuous vascular network: calling the auxin signal flow canalization hypothesis into question. Development. 2000;127:3197–3204. doi: 10.1242/dev.127.15.3197. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, et al. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 2008;6:e307. doi: 10.1371/journal.pbio.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005;28:67–77. doi: 10.1111/j.1365-3040.2005.01306.x. [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, et al. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell. 2002;14:589–597. doi: 10.1105/tpc.010354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Misera S, Jürgens G. Mutations affecting body organization in the Arabidopsis embryo. Nature. 1991;353:402–407. [Google Scholar]
- Mayer U, Buttner G, Jürgens G. Apical–basal pattern formation in the Arabidopsis embryo: studies on the role of the gnom gene. Development. 1993;117:149–162. [Google Scholar]
- Miyazawa Y, Takahashi A, Kobayashi A, Kaneyasu T, Fujii N, Takahashi H. GNOM-mediated vesicular trafficking plays an essential role in hydrotropism of Arabidopsis roots. Plant Physiol. 2009;149:835–840. doi: 10.1104/pp.108.131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouratou B, Biou V, Joubert A, Cohen J, Shields DJ, Geldner N, et al. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics. 2005;6:20. doi: 10.1186/1471-2164-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010;2:a001537. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, Kleine-Vehn J, et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435:1251–1256. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- Péret B, Rybel BD, Casimiro I, Benková E, Swarup R, Laplaze L, et al. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 1998;118:1369–1378. doi: 10.1104/pp.118.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Anders N, Wolters H, Beckmann H, Thomann A, Heinrich R, et al. Role of the GNOM gene in Arabidopsis apical–basal patterning—from mutant phenotype to cellular mechanism of protein action. Eur. J. Cell Biol. 2010;89:138–144. doi: 10.1016/j.ejcb.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, et al. Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell. 1997;9:745–757. doi: 10.1105/tpc.9.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell DE, Leu WM, Gillmor CS, Xia G, Feldmann KA, Chua NH. EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell. 1994;77:1051–1062. doi: 10.1016/0092-8674(94)90444-8. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, et al. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell. 2009;21:1495–1511. doi: 10.1105/tpc.108.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, et al. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001;15:2648–2653. doi: 10.1101/gad.210501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Peret B, Yang Y, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, et al. High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 2004;135:25–38. doi: 10.1104/pp.104.040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1025–1038. doi: 10.1093/pcp/pcn079. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- Wolters H, Anders N, Geldner N, Gavidia R, Jürgens G. Coordination of apical and basal embryo development revealed by tissue-specific GNOM functions. Development. 2011;138:117–126. doi: 10.1242/dev.059147. [DOI] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009;151:168–179. doi: 10.1104/pp.109.138859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Suzuki M, Fukaki H, Morita-Terao M, Tasaka M, Komeda Y. CRM1/BIG-mediated auxin action regulates Arabidopsis inflorescence development. Plant Cell Physiol. 2007;48:1275–1290. doi: 10.1093/pcp/pcm094. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.