Abstract
The mRNA-binding protein AUF1 regulates the expression of many key players in cancer including proto-oncogenes, regulators of apoptosis and the cell cycle, and pro-inflammatory cytokines, principally by directing the decay kinetics of their encoded mRNAs. Most studies support an mRNA-destabilizing role for AUF1, although other findings suggest additional functions for this factor. In this review, we explore how changes in AUF1 isoform distribution, subcellular localization, and post-translational protein modifications can influence the metabolism of targeted mRNAs. However, several lines of evidence also support a role for AUF1 in the initiation and/or development of cancer. Many AUF1-targeted transcripts encode products that control pro- and anti-oncogenic processes. Also, overexpression of AUF1 enhances tumorigenesis in murine models, and AUF1 levels are enhanced in some tumors. Finally, signaling cascades that modulate AUF1 function are deregulated in some cancerous tissues. Together, these features suggest that AUF1 may play a prominent role in regulating the expression of many genes that can contribute to tumorigenic phenotypes, and that this post-transcriptional regulatory control point may be subverted by diverse mechanisms in neoplasia.
Keywords: AUF1, alternative splicing, RNA turnover, RNA-binding protein, phosphorylation, cancer, review
2. INTRODUCTION
The need for cells to precisely control the extent and timing of protein production is met by the intricate array of mechanisms that regulate gene expression. This multi-faceted control circuitry can manipulate the utilization of genetic information at many levels, including transcription, RNA processing, RNA localization, translation, and protein degradation. One regulatory mechanism that facilitates very rapid cellular responses to a variety of stimuli is control of mRNA stability, which directly modulates the cytoplasmic concentrations of specific transcripts, and hence their potential to program protein synthesis, at minimal energetic cost (reviewed in Refs. 1, 2). In fact, microarray surveys of nascent and total RNA levels indicated that over half of all genes modulated by stress responses were principally regulated at the level of mRNA turnover (3).
Cells can degrade individual mRNA species at vastly different rates, which are largely controlled by the presence of specific cis-acting sequence and/or structural determinants within each transcript. Furthermore, the mRNA-stabilizing or -destabilizing activities of elements can be rapidly activated or inhibited in response to a plethora of intracellular and extrinsic signals. The best characterized cis-acting determinants of mRNA turnover are the AU-rich elements (AREs), a varied family of 3′-untranslated sequences that generally span 40–120 nucleotides and are dominated by uridylate residues, often containing repetitive AUUUA or similar motifs (reviewed in Ref. 4). Recent bioinformatics surveys have indicated that as many as 5–8 % of all genes may contain ARE-like sequences (5), but they are disproportionally frequent in transcripts that encode factors contributing to the development and progression of cancer including oncoproteins, cell cycle regulators, growth factors, and inflammatory cytokines. Among different mRNAs, AREs can vary widely in sequence context. For example, AREs from mRNAs encoding cytokines or lymphokines such as granulocyte macrophage-colony stimulating factor (GM-CSF) or interleukin-3 are normally fairly short (40–60 nt) and include several overlapping AUUUA motifs. By contrast, AREs from proto-oncogene mRNAs like c-fos or c-myc typically have a few dispersed pentamers (or none at all) within a larger U-rich background (reviewed in Ref. 4). However, for homologous mRNAs, the ARE sequence can be more highly conserved than the coding region, indicating a strong pressure to maintain distinct features of each ARE (6, 7). For example, the 34 nt core sequence of the ARE from tumor necrosis factor α (TNFα) mRNA is identical between humans and other mammals as diverse as sheep and whales (8).
In cells, AREs are recognized by a diverse population of trans-acting factors collectively termed ARE-binding proteins (ARE-BPs). Recruitment of ARE-BPs to ARE-containing mRNAs can positively or negatively regulate their decay kinetics or translational efficiency (reviewed in Refs. 9–11). For example, members of the Hu family of proteins including the ubiquitously expressed HuR or neuron-specific homologues HuC, HuD, and Hel-N1 stabilize ARE-containing mRNA substrates (12–17). Conversely, binding of tristetraprolin (TTP) (18, 19) and the KH-type splicing regulatory protein (KSRP) are closely associated with acceleration of mRNA decay (20, 21). A separate subpopulation of ARE-BPs is represented by T-cell intracellular antigen 1 (TIA-1) and TIA-1-related protein (TIAR), which are associated with translational repression of targeted mRNAs (reviewed in Refs. 9, 10).
AU-rich element RNA-binding factor 1 (AUF1) was the first ARE-BP to be purified and cloned (22, 23), and in the nearly two decades since its discovery has revealed a rich array of biochemical and functional detail. Normally associated with destabilization of mRNA substrates, other studies have indicated mRNA-stabilizing roles and even indirect control of translation. Recent work has also linked perturbation of AUF1 expression with cancer and inflammatory dysfunction, likely involving aberrant post-transcriptional control of selected ARE-containing mRNAs. In this review, we will first summarize our current knowledge of the mRNA targeting mechanisms of AUF1 and the functional consequences of these interactions. Subsequently, we will describe links between AUF1 and the development and progression of cancer, involving atypical regulation of (a) relative levels of AUF1 isoforms, (b) subcellular localization of AUF1, and (c) post-translational modifications that modify biochemical properties of this factor.
3. FUNCTIONS OF AUF1 IN POST-TRANSCRIPTIONAL CONTROL OF GENE EXPRESSION
AUF1 is encoded by a single copy gene comprised of 10 exons on chromosome 4 (4q21), and is expressed as a family of four protein isoforms generated by alternative pre-mRNA splicing of exons 2 and 7 (24, 25). The AUF1 proteins are named according to their apparent molecular masses as p37AUF1, p40AUF1, p42AUF1, and p45AUF1. The two largest isoforms contain sequences encoded by exon 7 while p40AUF1 and p45AUF1 both contain the exon 2-encoded domain (Fig. 1) (25). All isoforms include two centrally-positioned, tandemly arranged RNA recognition motifs (RRMs) which mediate RNA binding (26). The general organization of an RRM is a β-α-β-β-α-β RNA binding platform of anti-parallel β-sheets backed by the α-helices (27). Structures of individual AUF1 RRM domains resolved by NMR are largely consistent with this overall tertiary fold (28, 29). The protein domain encoded by exon 2 is immediately N-terminal to the RRMs; interactions or interference between this domain and RNA substrates may contribute to the decreased ARE-binding affinity reported for p40AUF1 and p45AUF1 relative to AUF1 isoforms lacking this domain (25). Downstream of the RRMs, all AUF1 isoforms include a glutamine-rich region that may also contribute to ARE binding affinity (26). The exon 7-encoded domain contained within p42AUF1 and p45AUF1 is located proximal to the C-terminus, and plays a role in the subcellular distribution of this protein (described in section 6).
Figure 1.
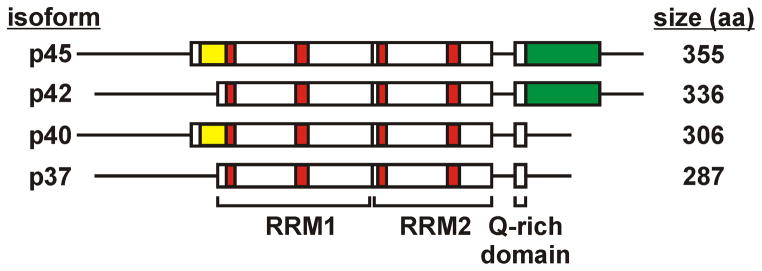
Schematic of AUF1 isoforms generated by alternative splicing. Both p42 AUF1 and p45AUF1 contain the 49 amino acid region encoded by exon 7 (green) while p40 AUF1 and p45 AUF1 contain the 19 amino acid region encoded by exon 2 (yellow). All isoforms contain two RRMs (RNA recognition motifs) each composed of characteristic RNP-2 and RNP-1 boxes respectively (red) followed by a Gln-rich domain. The isoform names are listed at left and number of amino acids at right.
3.1. Substrate selectivity of AUF1
Early biochemical analyses with recombinant p37AUF1 demonstrated high affinity binding (low nM Kd) to AREs from c-fos, c-myc, and TNFα mRNAs (30, 31), all prototypical ARE sequences containing multiple AUUUA motifs. More recently, comparative sequence analyses of AUF1-associated cellular mRNAs recovered by ribonucleoprotein immunopreipitation (RNP-IP) supported a preference for AU-rich sequences. Based on these studies, a loose consensus motif enriched in A and U residues (79% A+U) spanning 29–39 nucleotides was identified in 75% of all mRNA targets of AUF1 (32). However, since 25% of AUF1-associated mRNAs lacked this sequence, it is possible that AUF1 recruitment may also be mediated by distinct families of mRNA sequences, may involve higher-order RNA structures, and/or be facilitated by ancillary RNA-binding factors. Further analyses of AUF1-associated RNA populations supported roles for these proteins in both nuclear and cytoplasmic RNA metabolic processes. First, both nuclear and cytoplasmic AUF1 associated with target RNAs. Second, AUF1 bound to both mature mRNAs and pre-mRNAs containing the target motif (32), implying that AUF1 may be loaded onto some RNA substrates at co-transcriptional or early post-transcriptional pre-mRNA processing steps.
The possibilities that: (i) AUF1 loads early in nuclear RNA processing, and (ii) selected AUF1 isoforms may escort processed mRNAs during nucleocytoplasmic transport (described in section 6.1) suggest a stable RNA:protein interface. However, in vitro AUF1 binding to RNA substrates is highly dynamic in solution. For example, the dissociative half-time for a p37AUF1 complex with the TNFα ARE is approximately 10 seconds (31). Since the equilibrium dissociation constant for p37AUF1 binding to this RNA substrate is approximately 1 nM (33), the on-rate for this interaction must approach 108 M−1s−1, very rapid but well within the diffusion limit of 109–1010 M−1s−1 (34). The rapid dynamics of AUF1 binding to RNA substrates observed in vitro present some interesting functional possibilities for AUF1 in cellular environments. One possibility is that AUF1 may require ancillary binding factors or post-translational modifications to stabilize the AUF1:mRNA complex in cells. Consistent with this option, several AUF1 isoforms are phosphorylated in cells (23, 35), and independent labs have identified a variety of AUF1-interacting proteins (described below). Conversely, AUF1 function could be coupled to re-iterative cycles of RNA binding and release. For example, local RNA conformations resulting from transient AUF1 binding to AREs (discussed below) may obstruct or enhance access of other trans-acting factors, including RNA-binding proteins or miRNA-RISC complexes, to proximal sites. Furthermore, dynamic interactions between AUF1 and RNA substrates would permit rapid changes in RNA occupancy in response to alterations in the cellular environment.
In addition to binding RNA, ongoing research indicates that DNA-targeted events can be regulated by AUF1. All AUF1 isoforms have high affinity for the G-rich strand of the telomeric repeat sequence (36). AUF1 binding to these sequences destabilizes local guanosine quadruplex structures (37), which may enhance telomerase access and thus promote telomere maintenance. Other studies have provided evidence that nuclear AUF1 may also play roles in the regulation of transcription. For example, p40AUF1 can bind and activate transcription from the complement receptor 2 gene promoter (38), and in complex with nucleolin forms the B cell-specific transcription factor LR1 (39). Furthermore, multiple AUF1 isoforms have been shown to regulate transcription of the enkephalin gene in developing brain (40), and the α-fetoprotein gene (41), which encodes an onco-developmental protein that can be re-activated during liver regeneration or cancer (42).
3.2. Structural considerations and consequences of AUF1 binding
In solution, p37AUF1 exists as a dimer (26, 31), but forms larger oligomers once bound to RNA substrates. The size of these oligomers is related to the length of the ARE target; for example, two dimers of p37AUF1 bind to form a tetrameric protein:RNA complex on the 38-nt TNFα ARE (31, 33), while the p37AUF1 complex assembled on the 70-nt ARE from c-fos mRNA is consistent with a protein hexamer (26). To date, however, the functional significance of RNA-dependent AUF1 oligomerization remains unknown.
While the “canonical” ARE substrates of AUF1 are generally considered to be largely single-stranded based on their AU-rich content, computational algorithms suggest that variants of AUF1 consensus motifs may form punctuated local secondary structures consisting of bulged and kinked stem-loops (32). Similarly, the AUF1 binding site within phosphoenolpyruvate carboxykinase mRNA includes a stem-loop structure (43). A similar structure formed by the TNFα ARE has been biochemically validated using both nuclease mapping and fluorescence-based techniques (8). However, while p37AUF1 binds the TNFα ARE substrate with nanomolar affinity, conditions that stabilize ARE folding weakened AUF1 binding. By contrast, association of the mRNA-stabilizing factor HuR was unaffected by stabilizing folding of the TNFα ARE (8). In this manner, local RNA structural potential may serve as an additional determinant of protein-binding specificity. Perhaps more importantly, alterations in local RNA structure through protein binding or other events could rapidly alter selectivity for ARE-binding proteins. Finally, AUF1 binding to RNA substrates can also modify their local structure. By monitoring the distance between the termini of RNA substrates using fluorescence resonance energy transfer (FRET), both p37AUF1 and p40AUF1 were shown to condense local RNA structures, bringing the termini closer together in solution (44, 45). Interestingly, compaction of ARE structure was abrogated by phosphorylation of p40AUF1 at Ser83 and Ser87 (45), which occurs in resting THP-1 monocytic cells and is accompanied by rapid decay of substrate mRNAs. Stimulation by phorbol esters resulted in loss of these phosphate groups, concomitant with stabilization of the TNFα and interleukin 1-β (IL-1β) transcripts (35). The direct functional significance of remodeling local RNA structure by AUF1 binding is not yet clear. However, it is likely that such modulation of local higher-order RNA structure may block or vacate nearby binding sites for other factors including other RNA-binding proteins or microRNAs (miRNAs), which may mediate antagonistic or complimentary functions on mRNA stability.
Some recent findings support the model that combinatorial influences of RNA-binding proteins and/or miRNAs can direct mRNA fates. For AREs, this possibility was envisioned early based on the overlapping nature of RNA sequences targeted by different ARE-BPs. For example, while the mRNA-destabilizing protein TTP shows a strict preference for sequences of the type UAUUUAUU (46), RNA selectivity by HuR and AUF1 appears to be more permissive, as both factors bind many RNA substrates with a generally U-rich character (32, 47). Consistent with this idea, microarray-based surveys of mRNAs co-purifying with AUF1 and HuR identified a broad range of mRNA targets that can bind both HuR and AUF1, and validation studies on selected mRNA substrates demonstrated that both bind concurrently to some mRNAs (48). Another report provides evidence that p42AUF1 and HuR compete for binding to the 3′UTR of the mRNA encoding the cap-binding translation initiation factor eIF4E (49), a potent oncoprotein that is overexpressed in many aggressive cancers (50). Similarly, AUF1 binding to c-myc mRNA enhances translation of this transcript by competing with the translational inhibitor TIA-1 for a common binding site (51). Competition and/or cooperation between AUF1 and other trans-factors are likely extensible to miRNAs as well, because some miRNAs can also associate with ARE targets (52). Considering the high degree of base pair complementarity required for miRNA targeting, protein-induced changes in local RNA structure may regulate miRNA accessibility by exposing or obstructing adjacent sequences. Precedents for such mechanisms have already been reported in other systems. For example, binding of the RNA-binding protein Dead end 1 (Dnd1) to the 3′UTR of p27 mRNA prevents targeting by miR-211 (53). Similarly, HuR can block repression of cationic amino acid transporter 1 (CAT-1) mRNA translation by miR-122 (54), although HuR can also serve as a positive regulator of miRNA action, since it enhances let-7 recruitment to c-myc mRNA (55). Together, these models predict a reciprocal relationship between RNA structure and trans-factor recruitment, where the local structural context of an ARE may dictate the relative affinity and positioning of ARE-BP and/or miRNA recruitment. In turn, local RNA conformational changes mediated by this initial binding event would then be expected to positively or negatively influence the accessibility of adjacent sites for ancillary binding factors.
3.3. The AUF1 interactome and mechanics of ARE-directed mRNA decay
Cytoplasmic turnover of most cellular mRNAs proceeds via the deadenylation-dependent mRNA decay pathway. Decay is initiated by 3′ to 5′ removal of the poly(A) tail, followed by excision of the 5′-cap and exonucleolytic degradation of the mRNA body from both 5′ and 3′ ends (reviewed in Ref. 1). Deadenylation is considered the rate limiting step of the process, since poly(A)− intermediates do not normally accumulate in vivo (4). The poly(A) tail is shortened by dedicated deadenylase enzymes, which in the cytoplasm include members of the POP2 and CCR4 protein families, and the poly(A)-specific ribonuclease (PARN) (reviewed in Ref. 56). One current model indicates that removal of the poly(A) tail makes the 3′-end of the mRNA susceptible to 3′ to 5′ degradation by the cytoplasmic exosome (57). Alternatively, deadenylated mRNAs may be rapidly degraded from the 5′-end through the activities of decapping enzymes and 5′ to 3′ exonucleases, possibly involving transport to P bodies, intracellular foci that are enriched in these factors (reviewed in Ref. 58).
AREs enhance mRNA decay by increasing mRNA deadenylation rates (4). Although AUF1 binding to ARE-containing mRNAs is most frequently associated with acceleration of mRNA decay, AUF1 itself exhibits no nucleolytic activity, nor is it competent to stimulate the decay of polysomal mRNA in its purified form (reviewed in Ref. 59). However, AUF1 was initially discovered as a protein component of a 7S cytosolic complex that enhanced polysomal c-myc mRNA turnover in cell-free mRNA decay reactions (22). Together, these observations have prompted the working model whereby AUF1 binding to ARE-containing mRNAs may function by conveying or recruiting additional trans-acting factors to the targeted mRNA substrate. The identities of AUF1-interacting proteins discovered by several groups in recent years lend some support to this model. For example, co-immunoprecipitation experiments indicated that cytoplasmic AUF1 exists in complexes with proteins including the translation initiation factor 4G (eIF4G), heat shock protein 70 (Hsp70), heat shock cognate protein 70 (Hsc70), heat shock protein 27 (Hsp27), and poly(A)-binding protein (PABP) (60–62). Each of these AUF1-binding partners has established roles in the control of mRNA decay or translation. eIF4G promotes ribosome loading by coordinating a complex of proteins that brings together the 5′-cap, 3′-poly(A) tail, and the 40S ribosomal subunit (reviewed in Ref. 63). Interestingly, several groups had previously shown that rapid decay of ARE-containing mRNAs requires ongoing translation (64–66). Hsp70, Hsc70, and Hsp27 can all function independently as ARE-binding factors, and in the case of Hsc70, can stabilize targeted transcripts in cytokine-activated hematopoietic cells (67, 68). Finally, cytoplasmic PABP forms high affinity complexes with mRNA poly(A) tails to both protect the tail and enhance translational initiation by aiding in the recruitment of eIF4G (62).
Yeast two-hybrid assays have identified p37AUF1 interactions with itself, p40AUF1, ubiquitin conjugating enzyme E2I (UBCE2I), and additional RNA-binding proteins including nuclease sensitive element binding protein 1 (NSEP-1), synaptotagmin binding, cytoplasmic RNA-interacting protein 1 (NSAP-1) and insulin-like growth factor 2 mRNA-binding protein 2 (IMP-2) (69). It is possible that UBCE2I binding is associated with control of AUF1 protein turnover, since both p37AUF1 and p40AUF1 can be polyubiquitinated in cells and degraded by the proteasome (70). Independent two-hybrid surveys have shown that AUF1 also associates with proteins αCP1 and αCP2 (71). These factors are members of the α-globin mRNA stability complex that bind a pyrimidine-rich element within the α-globin mRNA 3′UTR and contribute to the extreme stability of this transcript, although the role of AUF1 in this process remains unclear. Finally, AUF1 can interact with components of the exosome (20) and co-localizes with exosome proteins in cells (48). While this indicates an immediate and direct link between AUF1 and a very efficient 3′ to 5′ exonucleolytic machine, exosomes alone do not appear to deadenylate mRNA substrates (20), but rather, contribute to rapid degradation of the deadenylated intermediate.
Taken together, these findings suggest that AUF1 binding and oligomerization on ARE-containing mRNAs may direct rapid decay by nucleating assembly of a multi-subunit trans-acting complex, which at some point must enhance substrate mRNA deadenylation. Conceivably, this may involve direct recruitment of deadenylating activities (described above), but alternatively may involve modulating the interaction of PABP with the mRNA poly(A) tail to enhance accessibility of 3′-adenylates to these 3′ to 5′ exonucleolytic activities. The latter possibility is particularly intriguing, given the observation that the PABP is a component of an AUF1-containing cytoplasmic complex (61). Following deadenylation, it is likely that AUF1-directed recruitment of the exosome then enhances rapid degradation of the mRNA body.
4. POSITIVE AND NEGATIVE CONTRIBUTIONS OF AUF1 TO NEOPLASIA
The prototypical mRNA to which AUF1 binds is one whose expression must be precisely and rapidly regulated. These mRNAs include many encoding cyclins and cyclin-dependent kinase inhibitors, regulators of apoptosis, proto-oncogenes, and cytokines (Table I). Cyclins promote the progression of the cell cycle. Conversely, cyclin-dependent kinase inhibitors arrest the cell at checkpoints to slow proliferation and maximize genome maintenance, thus minimizing the potential for cancer-stimulating mutations. Excessive promotion of the cell cycle without activation of apoptosis yields uncontrolled cellular proliferation, also known as hyperplasia. Hyperplasia can progress into dysplasia, the precursor to malignancy, as cells accumulate genetic errors through repeated rounds of premature division. Cell proliferation rates can be further enhanced by overexpression or over-activation of proto-oncogenes or lack of apoptosis activation (reviewed in Ref. 72).
Table I.
AUF1 substrate mRNAs that encode cancer-related products
| Gene symbol | Gene name | Regulation by AUF1 binding | Reference(s) |
|---|---|---|---|
| Cell Cycle Regulators | |||
| CCND1 | Cyclin D1 | destabilize mRNA | (48) |
| CDKN1A | p21 | destabilize mRNA | (48) |
| CDKN1B | p27 | decreased mRNA | (124) |
| RB1 | Retinoblastoma protein | increased protein expression associated with decreased AUF1 expression | (124) |
| CDKN2A | p16INK4a | destabilize mRNA | (147, 148) |
| Apoptosis Regulators | |||
| BAX | BCL2-associated X protein | decreased protein expression associated with increased mRNA-AUF1 association | (79) |
| BCL2 | B cell leukemia | destabilize mRNA | (83) |
| GADD45A | Growth arrest and DNA-damage-inducible α | destabilize mRNA | (149) |
| IER2 | Immediate early response 2 | stabilize mRNA | (129) |
| CASP2 | Caspase 2 | decreased protein expression associated with increased mRNA-AUF1 association | (79) |
| Metastasis Regulators | |||
| MMP9 | Matrix metaloproteinase -9 | increased mRNA with decreased AUF1 expression | (126) |
| FGF9 | Fibroblast growth factor 9 | p42AUF1 destabilizes mRNA | (106) |
| Inflammatory Mediators | |||
| GM-CSF | Granulocyte-macrophage colony-stimulating factor | destabilize mRNA | (99) |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor | stabilize mRNA | (98) |
| IL1B | Interleukin 1β | stabilize mRNA | (150) |
| IL10 | Interleukin 10 | LPS induces stabilization by p40AUF1 | (92) |
| IL6 | Interleukin 6 | destabilize and stabilize mRNA | (91) |
| NOS | human inducible nitri oxide synthase | destabilize mRNA | (151) |
| DNA Repair and Replication Regulators | |||
| FOS | c-fos | destabilize mRNA | (152) |
| TYMS | thymidylate synthase | destabilize mRNA of one allele type | (153) |
| JUND | jun D proto-oncogene | destabilize mRNA | (154) |
Cytokines are encoded by a distinct subset of highly regulated mRNAs. Although cytokines are intrinsically linked to inflammation, connections between inflammatory syndromes and the development and progression of cancer are becoming established (reviewed in Ref. 73). For example, chronic inflammation can include sustained production and secretion of many cytokines and chemokines including TNFα, interleukins-1α, -1β, -6, -8, and -18, and vascular endothelial growth factor (VEGF). Accumulation of these factors generates local microenvironments that can activate tumorigenic properties in nearby cells, including proliferation, inhibition of apoptotic signals, and angiogenesis (74). In this section, we will first overview the broad scope of AUF1-binding mRNAs and then discuss AUF1-dependent regulation of specific transcripts that encode cell cycle/apoptotic factors and cytokines, where AUF1 exhibits activities consistent with a tumor suppressor.
4.1. Insights from AUF1 target mRNAs
Shortly after AUF1 was identified, binding experiments demonstrated selective binding to AREs from a few proto-oncogene (c-myc, c-fos) and cytokine (GM-CSF) mRNAs (23, 30). However, several ribonome-wide surveys of AUF1-binding mRNAs subsequently demonstrated that AUF1 function could be intimately linked to malignancy at multiple levels (summarized in Table I). The first such study was performed by Jim Malter’s lab, which used repeated rounds of AUF1-affinity chromatography to isolate AUF1-binding mRNAs from activated peripheral blood mononuclear cells (75). Sequencing cDNA micro-libraries generated from the AUF1-binding mRNA pool revealed a plethora of early response gene targets, which comprised 33% of all mRNAs recovered. Putative AUF1 substrate mRNAs identified in this screen included those encoding plasminogen activator inhibitor, IL-8, the transcription factors JunD and BTF2, the chemokine Gro-β, and the cyclin-dependent kinase inhibitor p21. Proteins encoded by these mRNAs have diverse roles in the control of several tumorigenic phenotypes including cell proliferation, apoptosis, and cell motility. Additional AUF1 target mRNAs encoded factors involved in mRNA metabolism including the splicing regulators SC35 and hnRNP A1, and translation factors eEF-1α and the p36 subunit of eIF3. Regulated production of these factors would be expected to modulate the diversity of spliced gene products and possibly global protein synthesis.
The next large-scale screen for AUF1 substrate mRNAs was performed in Myriam Gorospe’s lab, where AUF1-bound mRNAs were recovered from HeLa cell lysates by RNP-IP and identified using cDNA arrays (48). These experiments identified over 450 potential AUF1 substrate mRNAs, including many that encode proteins closely associated with oncogenic processes. For example, the α isozyme of the catalytic subunit of protein phosphatase 1 (PPP1CA) is a potential activator of p53-induced cell cycle arrest (76). The chemokine (C-X-C motif) ligand 5 (CXCL5) is a pro-angiogenic inflammatory mediator and stimulator of cell proliferation that is up-regulated at the mRNA and/or protein level in prostate and selected other cancers (77). XRCC5 mRNA encodes a component of the DNA-dependent protein kinase required for non-homologous end-joining (NHEJ), an integral component of the cellular DNA repair machinery. Disruption of NHEJ is associated with increased cancer risk in murine models and human patients (reviewed in Ref. 78). Other AUF1-targeted mRNAs identified in this study that encode products related to cell growth and division include the translation initiation factor eIF4A2 and cyclin dependent kinase 7 (CDK7) (48).
A third ribonome-scale screen for AUF1 substrates performed by Ron Gartenhaus’ lab specifically searched for mRNAs exhibiting differential association with AUF1 in non-tumorigenic breast epithelial cells versus those transformed by the oncogene MCT-1 (79). While AUF1 expression levels did not detectably change as a result of cell transformation, there were significant differences in the mRNA subpopulations that associated with AUF1 between these cell models. For example, AUF1 binding to mRNAs encoding several cell cycle regulators (CDK1, CDK7, cyclins D1 and D2, and p21), as well as the apoptosis regulatory factor BAX, apoptotic protease caspase 2, and the translation regulatory factor eIF4EBP2 was enhanced in the transformed cell model. Western analyses showed that levels of BAX, caspase 2, and eIF4EBP2 proteins were significantly repressed in the transformed cells, consistent with a negative influence of AUF1 on the expression of each factor. Conversely, some AUF1-targeted mRNAs showed poorer binding to AUF1 in transformed versus non-tumorigenic cells. These included mRNAs encoding the pro-apoptotic and -angiogenic cytokine TWEAK, the Ras-related signal transduction mediator RAB2, and the DNA repair regulatory factor RAD51-associated protein 1 (RAD51AP1) (79). These observations that different mRNA subpopulations show increased or decreased AUF1 binding in non-tumorigenic versus MCT-1-transformed cells suggest that the mRNA selectivity of AUF1 may be modulated during cell transformation. Biochemical and cellular mechanisms that can regulate various aspects of AUF1 function are discussed in detail in sections 5, 6, and 7 (below).
4.2. Anti-tumorigenic roles of AUF1 in regulation of the cell cycle and apoptosis
The examples of section 4.1 show that AUF1-targeted mRNAs include those encoding both positive and negative regulators of neoplasia. The complexity of these gene regulatory networks thus precludes a universal definition of AUF1 as an exclusively pro- or anti-tumorigenic factor. However, in selected gene regulatory circuits this ambiguity is minimized. In this section, we describe two systems in which AUF1 function has distinctly anti-tumorigenic effects.
A pro-apoptotic role for AUF1 has been defined by its ability to suppress expression of the proto-oncogene bcl-2. Bcl-2 is an integral membrane protein that associates with the cytoplasmic surfaces of mitochondrial, nuclear, and endoplasmic reticular membranes. Bcl-2 inhibits apoptosis in many cell models, which enhances cell survival and can obstruct the efficacy of radiological and chemotherapeutic treatments for cancer (reviewed in Ref. 80). The 3′UTR of bcl-2 mRNA includes an extended AU-rich sequence that binds all four isoforms of AUF1, and deletion of this AU-rich sequence significantly stabilizes bcl-2 mRNA (81). Irradiating cells with UVC increases the cytoplasmic concentration of p45AUF1 and enhances AUF1 binding to bcl-2 mRNA, which may aid in UVC-induced apoptosis by suppressing bcl-2 mRNA levels. However, other cellular mechanisms can interfere with suppression of bcl-2 expression by AUF1. For example, CDIR, a non-coding RNA overexpressed in lung cancer, binds Hsp27 and together they sequester AUF1 away from bcl-2 mRNA (82). Formation of CDIR-AUF1 complexes can thus inhibit apoptosis by preventing AUF1 from targeting bcl-2 mRNA, resulting in increased production of Bcl-2 protein. The RNA-binding protein nucleolin can also block AUF1-dependent suppression of bcl-2 expression, but does so by competing for the AUF1 binding site in the bcl-2 mRNA 3′UTR (83).
A second anti-tumor role for AUF1 is exemplified by its control over expression of cyclin D1. The major function of cyclin D1 is to activate the cell cycle transition between G1 and S phases. Following mitogenic stimulation, cyclin D1 forms complexes with one of a number of cyclin-dependent kinases (generally CDK4), which are in turn activated by phosphorylation. Downstream targets of cyclin D1/CDK4 include the retinoblastoma-associated protein (pRb); inactivation of pRb by cyclin D1/CDK4-directed hyperphosphorylation initiates a cascade of events leading to S phase. Cyclin D1 also functions independently of CDKs, largely by regulating the activity of selected transcription factors (reviewed in Ref. (84)). In HeLa cells, UVC-induced cell cycle arrest was accompanied by increased AUF1 binding to cyclin D1 mRNA, resulting in decreased levels of cyclin D1 mRNA and protein (48). Furthermore, depletion of AUF1 using siRNA resulted in stabilization of cyclin D1 mRNA and accumulation of cyclin D1 protein. In the non-small-cell lung cancer cell line H1299, prostaglandin A2 treatment induces expression of p45AUF1, which binds within the 3′UTR of cyclin D1 mRNA and suppresses its expression by accelerating decay of this transcript (85). These examples highlight how AUF1 interactions with elements in the cyclin D1 mRNA 3′UTR function to limit expression of cyclin D1, and hence cellular potential for cell cycle activation. Consistent with this model, many aggressive mantle cell lymphoma (MCL) tumors express 3′-truncated forms of cyclin D1 mRNA, which lack AUF1 binding sites and hence express cyclin D1 protein at very high levels (86).
4.3. AUF1 regulates expression of factors contributing to pre-cancerous chronic inflammation
Inflammation, particularly in chronic states, can promote transformation of surrounding tissues through enhanced local concentrations of growth factors, angiogenic factors, reactive oxygen species, and cytokines (73, 87). Many mRNAs encoding cytokines and other proteins contributing to both inflammation and neoplasia contain AREs and can be regulated by AUF1; such transcripts include COX-2 (88), IL-1β, IL-6, GM-CSF, iNOS, bcl-2, and c-fos (reviewed in Ref. 89 and Table I). For example, IL-6 can contribute to proliferation of myeloma cells by activating cellular signaling cascades (90). However, p37AUF1 and p42AUF1 can suppress IL-6 production by enhancing degradation of its mRNA through interactions with the IL-6 ARE sequence (91). Conversely, lipopolysaccharide-stimulated production of the anti-inflammatory factor IL-10 in THP-1 promonocytic leukemia cells requires p40AUF1 (92). Perhaps most dramatically, AUF1 knockout mice experience severe endotoxic shock due to overexpression of TNFα and IL-1β, owing to enhanced levels of mRNAs encoding these inflammatory cytokines (93). Consistent with a central role in regulating cytokine production, AUF1 protein levels are relatively high in organs rich in lymphoid (thymus) and myeloid (spleen) tissues, as well as the brain, testes, uterus, and ovaries (94).
4.4. Pro-tumorigenic roles of AUF1: over-expression induces malignancy
While the roles of AUF1 in suppressing many target mRNAs encoding cell cycle, apoptotic, and inflammatory factors (described above) are consistent with an anti-tumorigenic function for this RNA-binding protein, other data indicate that substantial increases in AUF1 levels can promote the initiation and/or progression of cancer (summarized in Table II). First, in three transgenic mice lines engineered to overexpress p37AUF1, many animals developed undifferentiated sarcomas with high vascularization and cellularity that usually progressed to a late stage resulting in animal death (95). The murine tumors explicitly showed increased expression of cyclin D1, c-myc, and c-fos, especially in cell types that normally exhibit low endogenous AUF1 levels, but decreased expression of GM-CSF and TNFα in other tissues. Endogenous AUF1 is also overexpressed in some tumors, suggesting that cellular AUF1 levels may be clinically significant. For example, AUF1 expression is elevated in human hepatocellular carcinoma samples relative to benign controls, but decreases as cells are induced to differentiate (96). Similarly, AUF1 levels were elevated in neoplastic murine lung tissues (97).
Table II.
Modification of AUF1 abundance, localization, or function in specific cancer models
| Cancer/tissue site | AUF1 phenotype in malignancy | Model | Reference |
|---|---|---|---|
| lung | increased cytoplasmic localization | urethane-induced neoplasia and butylated hydroxytoluene-induced hyperplasia in murine lung tissue | (97) |
| melanoma | restricted to the nucleus | MNT1 cells vs. normal melanocytes | (122) |
| thyroid carcinoma | elevated cytoplasmic concentrations | malignant vs. benign human tissue samples | (124) |
| liver | elevated cytoplasmic concentrations | hepatocellular carcinoma vs. benign human liver samples | (96) |
| anaplastic large cell lymphoma (ALCL) | hyperphosphorylation correlated with target mRNA stabilization and increased cell survival | nucleophosmin–anaplastic lymphoma kinase (NPM-ALK)-expressing cells | (101) |
| sarcoma-like tumors at many tissue sites | overexpression | transgenic mice overexpressing p37AUF1 | (95) |
| breast | altered mRNA binding | oncogenic transformation of breast epithelial cells | (79) |
The possibility that elevated AUF1 levels could enhance expression of selected pro-tumorigenic or pro-inflammatory factors was also noted in cultured cell-based models. For example, ectopic overexpression of AUF1 in NIH3T3 fibroblasts stabilized reporter transcripts containing AREs from several mRNAs including c-fos and GM-CSF (98). Conversely, p37AUF1 overexpression destabilized a reporter mRNA containing the ARE from GM-CSF mRNA in CHO cells (99). While these discrepancies between mRNA-stabilizing and -destabilizing roles for AUF1 were previously attributed to differences in cell type and physiological conditions (98), we believe that two other explanations also merit consideration. First, the loose RNA consensus motifs for AUF1 recognition resolved by ribonome-wide RNP-IP surveys suggests that the RNA sequence specificity for AUF1 binding may be very broad (32). As such, there is a strong likelihood that AUF1 proteins may bind to physiologically irrelevant RNA targets (or sequences therein) when the protein is expressed at supra-physiological concentrations. An observation consistent with this model is that a 3′UTR-localized U32 homopolymeric stretch cannot accelerate the decay kinetics of a stable reporter mRNA (100), even through p37AUF1 protein binds this sequence with a Kd of 4 nM, only 4-fold weaker than its affinity for the ARE from TNFα mRNA (44). Since the overall changes in cellular AUF1 levels have not always been reported in published overexpression studies, this potentially critical variable cannot be compared. An additional complication is that a fraction of cellular AUF1 protein may be sequestered in cytoplasmic granules while target mRNAs are stabilized (101). A second likely consequence of elevating cellular AUF1 levels comes from a systems biology perspective. Recent work by Myriam Gorospe’s lab has demonstrated a high degree of regulatory crosstalk controlling the relative expression levels of different ARE-BPs. In particular, AUF1 binds to elements in the 3′UTRs of mRNAs encoding HuR, KSRP, NF90, TIA-1, and TIAR (102). Furthermore, several of these factors can reciprocally bind and regulate AUF1 mRNA. As a result, the consequences of substantially elevating AUF1 expression may be amplified by downstream effects on the expression and/or activity of other ARE-BPs.
To date, published studies have provided evidence that experimental or pathological elevation of AUF1 expression may destabilize or stabilize selected mRNA targets, thus potentially giving AUF1 properties of both a pro- and anti-neoplastic factor. Although the biochemical and depletion-based studies of AUF1 function described previously are most consistent with this factor targeting mRNAs for decay, the seemingly contradictory data revealed by overexpression studies present us with new questions regarding AUF1’s role(s) in the mechanics of regulated mRNA decay. For example, does AUF1 binding trigger an mRNA decay cascade in all RNA contexts, or only for a subset thereof? While competition between AUF1 and other ARE-BPs is well documented, do AUF1 and other ARE-BPs also exert combinatorial effects on decay of mRNA substrates? Is overexpression of AUF1 sufficient to activate the protein, or are post-translational modifications required? Are there isoform-specific differences in AUF1 activity? Recent progress in addressing some of these concepts is discussed below.
5. FUNCTIONAL AND REGULATORY DISTINCTIONS AMONG AUF1 ISOFORMS
Although most studies of AUF1 function to date have considered all isoforms as an ensemble, some work has described opposing or unique functions for individual protein variants. Furthermore, the individual isoforms show unique expression profiles in response to some stimuli, many of which are linked to carcinogenesis, and specific isoforms have been associated with the decay of some mRNAs. As will be detailed later, AUF1 isoforms also have distinct subcellular localization patterns, partly owing to unique preferences for protein binding partners.
On a functional level, regulated decay of some AUF1-targeted mRNAs relies on the presence of individual isoforms or combinations thereof, which conceivably may vary in a cell type- or mRNA substrate-specific manner. As reported above, overexpression of p37AUF1, or to a lesser degree, p40AUF1, destabilized a reporter mRNA containing the GM-CSF ARE in CHO cells (99). However, in THP-1 monocytes IL-10 mRNA and protein levels are increased by p40AUF1 but not by p37AUF1 (92). Fibroblast growth factor 9 (FGF9) mRNA, which encodes an autocrine/paracrine growth factor that stimulates cell growth (103) and is oncogenic in NIH3T3 fibroblasts (104), is destabilized by the p42AUF1 isoform (105). This is a critical gene regulatory target in cancer, since uncontrolled FGF9 expression can promote cell transformation and invasion (106). Finally, while these examples indicate preferences for individual AUF1 isoforms in control of ARE-directed mRNA decay, both p40AUF1 and p45AUF1 were required to destabilize IL-3 mRNA in HT1080 human fibrosarcoma cells (13). Interestingly, no changes in IL-3 mRNA stability were noted when all isoforms were suppressed or when p42AUF1 and p45AUF1 were depleted concomitantly.
Several mechanisms may account for AUF1 isoform-specific influences on mRNA stability. First, the different isoforms may recruit unique ancillary factors that lead to changes in localization or function (69). Second, AUF1 isoforms can have different binding affinities for ARE-containing mRNAs. For example, p37AUF1 has an approximately 4-fold greater affinity for the TNFα ARE sequence than p40AUF1 does (33, 45). As such, at equivalent protein concentrations p37AUF1 might saturate the ARE with oligomeric complexes, but p40AUF1 only exhibit a single dimer binding event. Variations in the oligomeric status of the major AUF1-RNA species could conceivably alter the function of the complex by differential recruitment of ancillary factors (see above) or by unique consequences on local RNA structure (44). This concept may explain why both AUF1 overexpression and knockdown were observed to stabilize IL-6 mRNA (91). Under low AUF1 concentrations (ie: basal levels or siRNA knockdown), AUF1 complexes on the IL-6 ARE may have been limited to protein dimers. Conversely, when AUF1 expression was dramatically increased (ie: by ectopic overexpression), AUF1 tetramers or larger complexes may form on the IL-6 ARE, which could exert stabilizing, rather than destabilizing influences on the mRNA. Finally, the individual isoforms of AUF1 may be subject to unique combinations of post-translational modifications that could impact both subcellular localization and the functional consequences of mRNA binding.
Consistent with isoform-dependent functional consequences of AUF1 binding, the expression and/or activity of the individual isoforms can also be differentially regulated. For example, testosterone and dihydrotestosterone induce changes in the expression of individual AUF1 isoforms in a tissue and sex-specific fashion in mouse models (107). For example, p37AUF1 is the predominant AUF1 isoform in submaxillary glands from male mice, while these glands from female mice express principally p40AUF1 and p45AUF1. However, these patterns are reversed when male mice are deprived of androgens (by castration), or female mice are treated with testosterone. By contrast, the distributions of AUF1 isoforms in kidney are not influenced by androgens in either model (107). In several other experimental systems, levels of p45AUF1 are specifically induced. For example, estradiol treatment increased p45AUF1 expression in ovine uterus, leading to increased protein binding to estrogen receptor α mRNA and stabilization of this transcript (108). Similarly, p45AUF1 was specifically induced in H1299 non-small-cell lung carcinoma cells treated with prostaglandin A2, and was linked to cell cycle arrest through enhanced degradation of cyclin D1 mRNA (85). In the developing rat cerebellum, AUF1 mRNA is globally down-regulated very quickly at the end of embryonic development and after birth, but the p45AUF1 protein is progressively up-regulated at later stages of development (109). At present there are no details available regarding the molecular mechanisms responsible for altered levels and/or activity of AUF1 isoforms, but theoretically these may include regulated pre-mRNA splicing or isoform-specific control of AUF1 mRNA decay, protein turnover, post-translational modifications, or association with ancillary factors.
6. SUBCELLULAR LOCALIZATION OF AUF1
AUF1 was independently identified as heterogeneous nuclear ribonucleoprotein D (hnRNP D) based on its enrichment in nuclear RNA fractions and association with telomeric repeat sequences (110). However, concomitant with the cloning of the first AUF1 cDNA in the early 1990’s, Brewer and colleagues demonstrated that different isoforms of AUF1 displayed variable subcellular distributions (23). In most cell types, the two largest two isoforms (p42AUF1 and p45AUF1) are mainly nuclear, while p37AUF1 and p40AUF1 are typically distributed in both nuclear and cytoplasmic compartments (35, 111, 112).
Nucleocytoplasmic shuttling is a common property of many RNA-binding proteins (reviewed in Ref. 113), and modulating the subcellular distribution of specific factors can have pathological consequences. Among the ARE-BPs, the prototypical example for this model is HuR, as cytoplasmic accumulation of this mRNA-stabilizing factor is associated with malignancy (reviewed in Ref. 114). While AUF1 can also shuttle between nuclear and cytoplasmic compartments, its subcellular distribution is highly isoform-dependent and can be modulated by cellular signaling pathways and other extrinsic stimuli. For example, during heat shock or prolonged inhibition of the proteasome, cytoplasmic AUF1 is transported to the nucleus or perinuclear region concomitant with stabilization of AUF1 target mRNAs (60). There is also a shift in the cytoplasmic isoforms during granulopoiesis (115). In skeletal muscle, chronic contractile activity increases cytoplasmic levels of p37AUF1, p40AUF1, and p45AUF1, resulting in enhanced decay of mRNAs that encode regulators of mitochondrial biogenesis (116). Considering accumulating data indicating distinct roles for AUF1 in nuclear versus cytoplasmic cell compartments (described in section 3.1), we submit that the subcellular distribution of AUF1 isoforms may constitute a critical factor in the regulated expression of many genes.
6.1. AUF1 translocation mechanisms
Nuclear import of all AUF1 isoforms is mediated by a common 19-amino acid domain at the extreme C-terminus that can bind transportin 1 (117). However, the biochemical rationale for isoform-specific differences in AUF1 subcellular distribution remains contentious. For example, an alternative nuclear import model suggests that insertion of the exon 7-encoded domain inhibits nuclear import of p42AUF1 and p45AUF1, implying that their accumulation in the nucleus may require transport as part of a larger AUF1-containing protein complex (118). If so, then additional mechanisms must contribute to preferential retention of these AUF1 isoforms in the nucleus. One such mechanism may involve interactions between the exon 7-encoded domains of p42AUF1 and p45AUF1 with nuclear scaffold attachment factor-β (111). Another possibility is that a subpopulation of p37AUF1 and p40AUF1 is sequestered in the cytoplasm by association with 14-3-3σ. The binding site for this 14-3-3 family member on AUF1 overlaps and occludes the AUF1 nuclear localization signal, but is interrupted by the exon 7-encoded domain, which precludes its association with p42AUF1 and p45AUF1 (119). Finally, some evidence suggests that AUF1 isoforms may differentially associate with specific subpopulations of nuclear RNA. In HeLa cells, nuclear p37AUF1 and p40AUF1 are freely diffusible in mRNA-protein complexes (nmRNPs) (120). These complexes contain the mRNA export factor REF, but neither pre-mRNAs nor non-shuttling proteins, and form late in the mRNA maturation pathway in anticipation of cytoplasmic export. By contrast, the larger isoforms of p42AUF1 and p45AUF1 preferentially associate with hnRNP complexes in the nucleoplasm (120) which are not ready for export, but rather are instrumental in pre-mRNA processing (121). By this model, p37AUF1 and p40AUF1 may be selectively exported to the cytoplasm in conjunction with mRNA cargoes. Taken together, these emerging data indicate that the subcellular distribution of AUF1 proteins may be a combined function of isoform-dependent transport across the nuclear envelope, and location-specific sequestration of individual protein variants.
6.2. AUF1 relocalization in malignant cells
AUF1-dependent control of mRNA decay is most consistently associated with cytoplasmic AUF1 proteins. As such, perturbation of cytoplasmic AUF1 levels would be expected to impact the ability of these proteins to regulate expression of targeted mRNAs, which could lead to profound changes in cellular physiology. Consistent with this model, the subcellular localization of AUF1 proteins is misregulated in diverse cancers. For example, cytoplasmic AUF1 levels increased in urethane-induced neoplasia and butylated hydroxytoluene-induced compensatory hyperplasia in murine lung tissue, and proliferation of neoplastic epithelial cell lines corresponded with increased cytoplasmic AUF1 concentrations when compared to non-tumorigenic counterparts (97). By contrast, in MNT1 melanoma cells AUF1 is restricted to the nucleus, while normal melanocytes express some AUF1 proteins in the cytoplasm which are competent to bind ARE-containing mRNAs (122). In this model, retention of AUF1 in melanoma cell nuclei is coordinated with a 10-fold stabilization of IL-10 mRNA. The IL-10 protein is commonly overexpressed in malignancies and may minimize host tumor rejection (reviewed in Ref. 123). However, perhaps the most comprehensive study to date examining AUF1 localization in cancer was published in 2009 by Cuong Hoang-Vu’s group. They surveyed 55 patient specimens to show that AUF1 accumulates to higher levels in the cytoplasm of thyroid carcinoma tissues versus their benign counterparts, and noted even higher cytoplasmic AUF1 levels in carcinoma cells undergoing mitosis (124). Decreasing AUF1 levels using siRNA substantially retarded proliferation of thyroid carcinoma cell lines, concomitant with increased expression of cell cycle inhibitory factors p21, p27, and Rb1, each encoded by an AUF1-binding mRNA. These findings support a pro-tumorigenic role for cytoplasmic AUF1 in this context, since enhanced levels of AUF1 in the cytoplasm can suppress expression of cell cycle inhibitory proteins important for limiting proliferation.
6.3. Cellular regulation of AUF1 localization
Emerging findings from several labs have shown that a variety of cellular signaling pathways can modulate the subcellular distribution of AUF1 isoforms, often coincident with altered expression of cancer-related gene products. Activation of mitogen-activated protein kinase (MAPK) pathways is commonly associated with stabilization of ARE-containing mRNAs, and is accompanied by increased cytoplasmic accumulation of both HuR and AUF1 (125). Interestingly, HuR accumulates more rapidly in the cytoplasm than AUF1, raising the possibility that preferential enrichment of HuR could be responsible for stabilizing ARE substrate mRNAs early following MAPK activation, but afterwards must compete with AUF1 for many of its mRNA targets. A very recent study showed that p42AUF1 relocalized from the nucleus to the cytoplasm in human fibroblast cells treated with the inflammatory mediator leukotriene B4 by a mechanism involving Ras/c-Raf/ERK signaling and the nuclear export receptor CRM1 (88). Enhanced cytoplasmic accumulation of p42AUF1 resulted in stabilization of COX-2 mRNA, leading to enhanced expression of COX-2 protein. COX-2 activity is closely associated with promotion of several tumorigenic phenotypes, particularly cell proliferation, apoptotic resistance, metastasis, and angiogenesis. Accordingly, overexpression of COX-2 is associated with many cancers (reviewed in Ref. 89).
In addition to MAPK pathways, several other cellular signaling systems have been shown to influence the subcellular localization of AUF1. For example, nitric oxide exposure increased AUF1 cytoplasmic concentrations leading to an AUF1-dependent decrease in expression of matrix metalloproteinase-9 (MMP-9) mRNA (126). MMP-9 is a gelatinase that can penetrate the basement membrane to facilitate angiogenesis, and generally contributes to tumor growth and metastasis (127). Conversely, prostaglandin-E2 treatment decreased cytoplasmic AUF1 concentrations without affecting global AUF1 protein levels in cultured human endometrial stromal cells, leading to induction of FGF9 mRNA (105), an AUF1-targeted transcript that is misregulated in a variety of cancers (section 5).
Finally, the subcellular distribution of AUF1 can be influenced by hormonal signals. In breast epithelial cells, lactogenic hormone promotes nuclear retention of AUF1 concomitant with changes in gene expression patterns that restrict proliferation and promote differentiation of breast epithelium (128). By contrast, in ovariectomized rats, estradiol promoted transient accumulation of p40AUF1 in the cytoplasm of uterine cells, which was coupled to induction of AUF1-targeted mRNAs encoding the A20 binding inhibitor of nuclear factor (NF)-κB activation-2 (ABIN2) and immediate early response-2 (Ier2/pip92/Chx1) proteins (129). ABIN2 is a potential activator of A20, an inhibitor of NF-κB, and functions to inhibit activation of inflammatory genes (130). ABIN2 also interacts with the anti-inflammatory receptor tyrosine kinase Tie2 which is necessary for angiogenesis and blood vessel maintenance (131) and regulates apoptosis (132).
Together, these findings support the hypothesis that modulation of cytoplasmic AUF1 concentrations can influence the stability of some mRNAs that encode cancer-related products. While nuclear retention of AUF1 largely promotes stabilization of AUF1-targeted transcripts, shifting abnormally large concentrations of AUF1 to the cytoplasm may also stabilize some mRNAs. Furthermore, the consequences of AUF1 relocalization may be profoundly impacted by coordinated changes in other RBPs. For example, AUF1 and HuR can compete for many mRNA targets (48) and present very similar tissue distributions (94), suggesting that the relative cytoplasmic concentrations of these normally antagonistic factors may be just as important as their individual levels on the net stability of their substrate mRNAs.
7. AUF1 MODIFICATIONS
In the 1993 study by the Brewer lab reporting the cloning of the first AUF1 cDNA, AUF1 immunoprecipitations from 32P-labeled K562 cells revealed that the various AUF1 isoforms could exist as phosphoproteins (23). In addition, many examples have been reported where the decay kinetics of specific ARE-containing mRNAs is regulated by the activation or suppression of cellular signaling events. Among the most extensively studied of these regulatory pathways are the MAPK (including p38MAPK, ERK, and c-Jun N-terminal kinase (JNK)) and phosphatidylinositol 3-kinase (PI3-K) cascades, which modulate the stability of many ARE-containing transcripts that encode factors involved in both inflammation and tumorigenesis (reviewed in Ref. 89). Together, these observations contributed to an early hypothesis that regulation of AUF1 localization and/or function could be mediated by post-translational modifications, particularly phosphorylation. Many subsequent studies that have added support to this model are described above, including prostaglandin A2-induced decay of cyclin D1 mRNA in lung cancer cells (section 4.2), which is accompanied by activation of the ERK pathway (133), and ERK-dependent accumulation of p42AUF1 in the cytoplasm of human fibroblasts (section 6.3).
Although phosphorylation of AUF1 shows promise as a potential mechanism for regulating the activity of these proteins in post-transcriptional control of gene expression, emerging data suggest that other covalent modifications of AUF1 may also influence its abundance and/or function. First, many RNA-binding proteins, particularly among the hnRNP family, can be methylated on arginine residues within conserved RGG motifs (134). Recently, high throughput proteomic screening for methylated proteins revealed that an arginine located near the most C-terminal RGG motif of all AUF1 isoforms (Arg345 in p45AUF1) could be dimethylated (135), although the function of this modification remains unknown. Among the ARE-BPs, arginine methylation has been demonstrated on HuR at Arg217 (136), and may be coupled to cytoplasmic accumulation of the protein (96). A second alternative post-translational modification to consider may be modification by poly-(ADP-ribose). In Drosophila, poly-(ADP-ribosyl)ation of the mRNA processing factor Squid inhibits its binding to RNA substrates and regulates Squid-dependent pre-mRNA splicing events (137). Although Squid is the Drosophila orthologue of AUF1 (11), it is not known whether AUF1 can be similarly modified. Finally, both p37AUF1 and p40AUF1 can be polyubiquitinated in cells (70), which likely contributes to post-translational control of their cytoplasmic concentrations. However, interference with the ubiquitin-proteasome pathway by chemical inhibition of the proteasome, inactivation of the ubiquitin-activating enzyme E1, or overexpression of a deubiquitinating protein all blocked ARE-directed mRNA decay (60, 138), suggesting that a functional link may exist between the mRNA and protein degradation machinery.
These examples highlight a variety of possible mechanisms that may link cellular signaling pathways with post-transcriptional control of gene expression through AUF1. Conceivably, such covalent modifications of AUF1 could regulate its function(s) in many ways, including modulation of subcellular localization, abundance, affinity for specific mRNAs, the local structure of bound RNA substrates, and interactions with ancillary binding partners, among other possible mechanisms. Furthermore, the complexity of these regulatory switches is compounded by the likelihood that at least some of these modifications may be specific for one or a subset of AUF1 isoforms.
7.1. Identified AUF1 phosphorylation events
The best characterized post-translational modifications of AUF1 are the phosphorylation of polysomal p40AUF1 at Ser83 and Ser87. Since these residues reside within the exon 2-encoded domain, they are excluded from the p37AUF1 and p42AUF1 isoforms. Activation of THP-1 monocytic cells with 12-O-tetradecanoylphorbol-13-acetate (TPA) leads to rapid dephosphorylation at these sites, concomitant with stabilization of AUF1-bound mRNAs encoding TNFα and IL-1β (35). Purified p40AUF1 can be stoichiometrically phosphorylated in vitro at Ser83 and Ser87 by glycogen synthase kinase-3β (GSK-3β) and protein kinase A (PKA), respectively (45). Interestingly, these modifications did not significantly alter the abundance or subcellular distribution of p40AUF1 in THP-1 cells (35), nor the affinity of this protein for ARE substrates in vitro; however, they did strongly influence the conformation of bound RNA substrates. While unphosphorylated p40AUF1 compacts the local structure of associated RNAs, phosphorylation of p40AUF1 on Ser83 and Ser87 induces an extended conformation on RNAs assembled into ribonucleoprotein complexes (45). It remains unknown how maintaining AREs in an elongated conformation may contribute to rapid mRNA decay, but it is conceivable that this structural arrangement provides unique opportunities to recruit ancillary factors required to direct the mRNA substrate for degradation.
A high throughput proteomic screen for phosphoproteins provided orthogonal support for AUF1 phosphorylation events on Ser83 and Ser87 (139). However, this survey also identified other Ser residues within the exon 2-encoded domain (contained within p40AUF1 and p45AUF1) close to Ser83 and Ser87 that may also be phosphorylated in cells. In addition, this report revealed AUF1 phosphorylation at a distinct site located within C-terminal RRM (Ser190 of p45AUF1). While the functional significance or kinases responsible for these modifications are currently unknown, an intriguing possibility is that at least one of these sites may be responsible for recognition by the 14-3-3σ protein. As described previously (section 6.1), 14-3-3σ can bind p37AUF1 and p40AUF1, and may contribute to cytoplasmic retention of these isoforms. However, recognition by the 14-3-3 proteins normally involves Ser- or Thr-phosphorylated sites (140). This relationship has been well described for TTP and butyrate response factor (BRF1), two other mRNA-destabilizing ARE-BPs. Phosphorylation of TTP is coordinated with mRNA stabilization, since phosphorylation-null TTP mutants show decreased association with 14-3-3 factors, increased TTP recruitment to cytoplasmic stress granules, and constitutive mRNA-destabilizing activity (141). Similarly, BRF1 protein phosphorylation enhances association with 14-3-3 factors and stabilization of mRNA substrates (142). As such, it seems plausible that AUF1 phosphorylation may also regulate 14-3-3 association and resultant influences on subcellular distribution (119), in addition to its documented role in modulating the local structure of bound RNA substrates.
7.2. Hyperphosphorylation of AUF1
The variety of potential phosphorylation sites on AUF1 (described above) raise the possibility that these proteins may be modified at multiple locations. However, some evidence suggests that AUF1 proteins may in fact be hyperphosphorylated, and that high phosphate:protein stoichiometries may elicit distinct functional consequences, most notably coupled to protein localization. First, fractionation across 2-dimensional gels demonstrated several acidic forms of p42AUF1 and p45AUF1 extracted from nuclear membranes, consistent with the distribution of these proteins across multiple phosphorylated states (120). However, these AUF1 isoforms resolved as single species in 2D-fractionated nucleoplasm, suggesting that enhanced phosphorylation was coupled to nuclear membrane localization. In a second study, nucleophosmin anaplastic lymphoma kinase (NPM-ALK), an oncogenic tyrosine kinase chimera key to lymphomagenesis in anaplastic large cell lymphoma, was found to interact directly with AUF1, with preference for the p45AUF1 isoform (101). Active NPM-ALK induced multiple anionic shifts in AUF1 on a 2D Western blot which were recognized by an anti-Tyr antibody, indicating that AUF1 phosphorylation is increased by NPM-ALK activity. NPM-ALK is largely found in the nucleolus from which AUF1 is excluded but can also co-localize with AUF1 in large cytoplasmic foci (non-P body granules). Notably, expression of active ALK stabilized several AUF1-associated mRNAs including c-myc and cyclin D1. These data led to the hypothesis that different levels of AUF1 phosphorylation may influence the protein in diverse ways. For example, some phosphorylation of AUF1 may be required for mRNA-binding activity, but hyperphosphorylation of AUF1 is associated with inactivation by sequestration (101).
Further support for this model was observed in a hyperparathyroidism model, where stabilization of parathyroid hormone (PTH) mRNA results in overexpression of PTH and increased cellular proliferation (143, 144). Similar to the examples above, 2D gel electrophoresis revealed enhanced levels of multiple acidic forms of AUF1 consistent with hyperparathyroidism-induced hyperphosphorylation of the protein (143). Calcimimetic treatment, which induces allosteric modifications of the calcium-sensing receptor, resulted in destabilization of PTH mRNA, decreased parathyroid cell proliferation, reversal of AUF1 modifications, and total reversal of the effects of hyperparathyroidism on these systems. Together, these examples suggest that hyperphosphorylation constitutes an additional mechanism for regulating the ability of AUF1 to direct decay of substrate mRNAs, likely based on protein sequestration.
7.3. AUF1 modifications induced by isomerization
A final post-translational modification identified in AUF1 isoforms is mediated by the peptidyl proline cis/trans isomerase Pin1, which co-immunoprecipitates with all four AUF1 isoforms in stimulated eosinophils (145). However, only the p40AUF1 and p45AUF1 isoforms have a Pin1 recognition site (Ser83-Pro84) as Pin1 recognizes phos-Ser-Pro or phos-Thr-Pro (146). Therefore binding to p37AUF1 and p42AUF1 may be indirect, possibly mediated by protein-protein interactions within AUF1 heterodimers. Phospho-Pin1-dependent isomerization of basally phosphorylated AUF1 inactivated the RNA-binding activity of AUF1 (145). By contrast, inhibition of Pin1 activity increased association of AUF1 with GM-CSF mRNA and destabilized this transcript. Irreversible inhibition of Pin1 by juglone led to the loss of all detectable cytoplasmic p40AUF1, p42AUF1, and p45AUF1 after 2 hours as a result of AUF1 degradation (145). Although frequently overlooked, isomerization may thus represent a novel mechanism for regulating AUF1 function following (hyper)phosphorylation by relocalizing or degrading the protein.
8. SUMMARY AND FURTHER QUESTIONS
Although historically we and others have tried to classify AUF1 as either an mRNA stabilizer or destabilizer, the growing body of evidence produced by many laboratories on this issue indicates that such clear-cut distinctions underestimate the complexity of this system. It is more likely that the activity of AUF1 is not a simple “on-off” switch, but rather can be regulated at multiple levels including isoform expression, subcellular localization, and post-translational modifications. Shifts in the expression and biochemical characteristics of AUF1 have been identified in multiple cancers including leukemias, lymphomas, and solid tumors with corresponding changes in AUF1 activities. The consequences of altered AUF1 functionality in these syndromes can be severe, since the principal mRNA targets of AUF1 include ARE-containing mRNAs encoding cell cycle regulators, proto-oncogenes and cytokines. Complicating matters further, dramatic induction or repression of AUF1 activity both yield severe pathological outcomes, supporting the hypothesis that AUF1 levels and function must both be tightly regulated to maintain normal cellular homeostasis.
However, the complexity of this system also provides some exciting opportunities for future research. For example, while many of the signal transduction pathways hyperactivated in malignancies are linked to perturbation of AUF1 function, it is imperative that the post-translational or gene regulatory modifications of AUF1 or its binding partners mediated by these signals be unambiguously identified. This information will be essential for downstream characterization of the biochemical consequences of these modifications, as well as their impact on the subcellular localization of AUF1 and the fate of AUF1-targeted RNA substrates. A second key area is to distinguish the functions of individual AUF1 isoforms. The hazards of dramatically overexpressing AUF1 have been described in this document; a more promising strategy will likely be to suppress endogenous AUF1 and rescue individual isoforms to endogenous expression levels. Regardless, important question to be answered include: Are different AUF1 isoforms selective for different subsets of mRNA substrates? Do they differentially interface with cellular signaling systems? Do they have selective nuclear roles, and are these independent, or can they complement cytoplasmic functions? Finally, it will be exciting to determine whether the expression and/or activity of specific AUF1 isoforms can be modulated by exogenous agents, and to see if such agents have utility in restoring the activity of post-transcriptional gene regulatory networks disrupted in neoplasia.
Acknowledgments
We would like to thank Dr. Toni Antalis and Dr. Feyruz Rassool for their encouragement and critique of this manuscript. Research in the Wilson lab contributing to this manuscript was funded by NIH R01 CA102428 (to G.M.W.). B.E.Z. is supported in part by NIH T32 GM066706.
Abbreviations
- ARE
AU-rich element
- ARE-BP
ARE-binding protein
- AUF1
AU-rich element RNA- binding protein 1
- EMSA
electrophoretic mobility shift assay
- ERK
extracellular signal-regulated kinase
- FRET
fluorescence resonance energy transfer
- hnRNP
heterogeneous nuclear ribonucleoprotein
- MAPK
mitogen activated protein kinase
- miR(miRNA)
microRNA
- NHEJ
non-homologous end joining
- RNP-IP
ribonucleoprotein immunoprecipitation
- RRM
RNA recognition motif
- TPA
12-O-tetradecanoylphorbol-13-acetate
- UTR
untranslated region
References
- 1.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 3.Fan J, Yang X, Wang W, Wood WHI, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci USA. 2002;99:10611–10606. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CYA, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 5.Bakheet T, Williams BRG, Khabar KSA. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 7.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fialcowitz EJ, Brewer BY, Keenan BP, Wilson GM. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J Biol Chem. 2005;280:22406–22417. doi: 10.1074/jbc.M500618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-acting factors. Biochem Soc Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 11.Wilson GM, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acids Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 12.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuadrado A, Navarro-Yubero C, Furneaux H, Kinter J, Sonderegger P, Munoz A. HuD binds to three AU-rich sequences in the 3′-UTR of neuroserpin mRNA and promotes the accumulation of neuroserpin mRNA and protein. Nucleic Acids Res. 2002;30:2202–2211. doi: 10.1093/nar/30.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckel-Mitchener A, Miera A, Keller R, Perrone-Bizzozero NI. Poly(A) tail length-dependent stabilization of GAP-43 mRNA by the RNA-binding protein HuD. J Biol Chem. 2002;277:27996–28002. doi: 10.1074/jbc.M201982200. [DOI] [PubMed] [Google Scholar]
- 16.Pascale A, Amadio M, Scapagnini G, Lanni C, Racchi M, Provenzani A, Govoni S, Alkon DL, Quattrone A. Neuronal ELAV proteins enhance mRNA stability by a PKCα-dependent pathway. Proc Natl Acad Sci USA. 2005;102:12065–12070. doi: 10.1073/pnas.0504702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: Binding of tristetraprolin-related zinc-finger proteins to AU-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 20.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJM, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 21.Gherzi R, Lee KY, Briata P, Wegmüller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner BJ, Long L, Rao PN, Pettenati MJ, Brewer G. Localization and physical mapping of genes encoding the A+U-rich element RNA-binding protein AUF1 to human chromosomes 4 and X. Genomics. 1996;34:219–222. doi: 10.1006/geno.1996.0269. [DOI] [PubMed] [Google Scholar]
- 25.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-RNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 26.DeMaria CT, Sun Y, Long L, Wagner BJ, Brewer G. Structural determinants in AUF1 required for high affinity binding to A+U-rich elements. J Biol Chem. 1997;272:27635–27643. doi: 10.1074/jbc.272.44.27635. [DOI] [PubMed] [Google Scholar]
- 27.Nagai K, Oubridge C, Ito N, Avis J, Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 28.Nagata T, Kurihara Y, Matsuda G, Saeki JI, Kohno T, Yanagida Y, Ishikawa F, Uesugi S, Katahira M. Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0. J Mol Biol. 1999;287:221–237. doi: 10.1006/jmbi.1999.2616. [DOI] [PubMed] [Google Scholar]
- 29.Katahira M, Miyanoiri Y, Enokizono Y, Matsuda G, Nagata T, Ishikawa F, Uesugi S. Structure of the C-terminal RNA-binding domain of hnRNP D0 (AUF1), its interactions with RNA and DNA, and change in backbone dynamics upon complex formation with DNA. J Mol Biol. 2001;311:973–988. doi: 10.1006/jmbi.2001.4862. [DOI] [PubMed] [Google Scholar]
- 30.DeMaria CT, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 31.Wilson GM, Sun Y, Lu H, Brewer G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J Biol Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- 32.Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson GM, Sutphen K, Chuang K, Brewer G. Folding of A+U-rich RNA elements modulates AUF1 binding: Potential roles in regulation of mRNA turnover. J Biol Chem. 2001;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- 34.Bisswanger H. Enzyme kinetics. Wiley-VCH; Weinheim, Germany: 2002. pp. 5–9. [Google Scholar]
- 35.Wilson GM, Lu J, Sutphen K, Sun Y, Huynh Y, Brewer G. Regulation of A+U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J Biol Chem. 2003;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- 36.Eversole A, Maizels N. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol Cell Biol. 2000;20:5425–5432. doi: 10.1128/mcb.20.15.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogenous nuclear ribonucleoprotein D. J Biol Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 38.Tolnay M, Baranyi L, Tsokos GC. Heterogeneous nuclear ribonucleoprotein D0 contains transactivator and DNA-binding domains. Biochem J. 2000;348:151–158. [PMC free article] [PubMed] [Google Scholar]
- 39.Dempsey LA, Hanakahi LA, Maizels N. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J Biol Chem. 1998;273:29224–29229. doi: 10.1074/jbc.273.44.29224. [DOI] [PubMed] [Google Scholar]
- 40.Dobi A, Szemes M, Lee C, Palkovits M, Lim F, Gyorgy A, Mahan MA, Agoston DV. AUF1 Is expressed in the developing brain, binds to AT-rich double-stranded DNA, and regulates enkephalin gene expression. J Biol Chem. 2006;281:28889–28900. doi: 10.1074/jbc.M511858200. [DOI] [PubMed] [Google Scholar]
- 41.Jiao R, He QY, Chen H, Hua Z, Jiao Q, Chiu JF. AUF1-like protein binds specifically to DAS cis-acting element that regulates mouse α-fetoprotein gene expression. J Cell Biochem. 2006;98:1257–1270. doi: 10.1002/jcb.20843. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Egan JO, Chiu JF. Regulation and activities of α-fetoprotein. Crit Rev Eukaryot Gene Expr. 1997;7:11–41. doi: 10.1615/critreveukargeneexpr.v7.i1-2.20. [DOI] [PubMed] [Google Scholar]
- 43.Hajarnis S, Schroeder JM, Curthoys NP. 3′-Untranslated region of phosphoenolpyruvate carboxykinase mRNA contains multiple instability elements that bind AUF1. J Biol Chem. 2005;280:28272–28280. doi: 10.1074/jbc.M501204200. [DOI] [PubMed] [Google Scholar]
- 44.Wilson GM, Sutphen K, Moutafis M, Sinha S, Brewer G. Structural remodeling of an A+U-rich RNA element by cation or AUF1 binding. J Biol Chem. 2001;276:38400–38409. doi: 10.1074/jbc.M106509200. [DOI] [PubMed] [Google Scholar]
- 45.Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ylsa RM, Charles S, Brewer G. Phosphorylation of p40AUF1 regulates binding to A+U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J Biol Chem. 2003;278:33039–33048. doi: 10.1074/jbc.M305775200. [DOI] [PubMed] [Google Scholar]
- 46.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: Conformational changes coupled to the bipartite nature of AU-rich mRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 47.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Topisirovic I, Siddiqui N, Orolicki S, Skrabanek LA, Tremblay M, Hoang T, Borden KLB. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol. 2009;29:1152–1162. doi: 10.1128/MCB.01532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 51.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 52.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JAF, Slanchev K, le Sage C, Nagel R, Voorhoeve M, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 54.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harbor Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 55.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 57.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 59.Wilson GM, Brewer G. Regulation of mRNA stability by AUF1. In: Sandberg K, Molroney SE, editors. RNA Binding Proteins: New Concepts in Gene Regulation. Vol. 16. Kluwer Academic Publishers; Norwell, MA: 2002. pp. 101–117. [Google Scholar]
- 60.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteosome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 61.Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12:883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinsimer KS, Gratacos FM, Knapinska AM, Lu J, Krause CD, Wierzbowski AV, Maher LR, Scrudato S, Rivera YM, Gupta S, et al. Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Mol Cell Biol. 2008;28:5223–5237. doi: 10.1128/MCB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 64.Savant-Bhonsale S, Cleveland DW. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 65.Winstall E, Gamache M, Raymond V. Rapid mRNA degradation mediated by the c-fos 3′ AU-rich element and that mediated by the granulocyte-macrophage colony-stimulating factor 3′ AU-rich element occur through similar polysome-associated mechanisms. Mol Cell Biol. 1995;15:3796–3804. doi: 10.1128/mcb.15.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curatola AM, Nadal MS, Schneider RJ. Rapid degradation of AU-rich element (ARE) mRNAs is activated by ribosome transit and blocked by secondary structure at any position 5′ to the ARE. Mol Cell Biol. 1995;15:6331–6340. doi: 10.1128/mcb.15.11.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson GM, Sutphen K, Bolikal S, Chuang K, Brewer G. Thermodynamics and kinetics of Hsp70 association with A+U-rich mRNA-destabilizing sequences. J Biol Chem. 2001;276:44450–44456. doi: 10.1074/jbc.M108521200. [DOI] [PubMed] [Google Scholar]
- 68.Matsui H, Asou H, Inaba T. Cytokines direct the regulation of Bim mRNA stability by heat-shock cognate protein 70. Mol Cell. 2007;25:99–112. doi: 10.1016/j.molcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 69.Moraes KC, Quaresma AJ, Maehnss K, Kobarg J. Identification and characterization of proteins that selectively interact with isoforms of the mRNA binding protein AUF1 (hnRNP D) Biol Chem. 2003;384:25–37. doi: 10.1515/BC.2003.004. [DOI] [PubMed] [Google Scholar]
- 70.Laroia G, Schneider RJ. Alternate exon insertion controls selective ubiquitination and degradation of different AUF1 protein isoforms. Nucleic Acids Res. 2002;30:3052–3058. doi: 10.1093/nar/gkf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kiledjian M, DeMaria CT, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weber GF. Molecular Mechanisms of Cancer. Springer; Dordrecht, The Netherlands: 2007. [Google Scholar]
- 73.Marx J. Inflammation and cancer: the link grows stronger. Science. 2004;306:966–968. doi: 10.1126/science.306.5698.966. [DOI] [PubMed] [Google Scholar]
- 74.Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G. Inflammation and cancer: how hot is the link? Biochem Pharmacol. 2006;72:1605–1621. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 75.Bhattacharya S, Giordano T, Brewer G, Malter JS. Identification of AUF-1 ligands reveals vast diversity of early response gene mRNAs. Nucleic Acids Res. 1999;27:1464–1472. doi: 10.1093/nar/27.6.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Castro ME, Ferrer I, Cascón A, Guijarro MV, Lleonart M, Ramón y Cajal S, Leal JF, Robledo M, Carnero A. PPP1CA contributes to the senescence program induced by oncogenic Ras. Carcinogenesis. 2008;29:491–499. doi: 10.1093/carcin/bgm246. [DOI] [PubMed] [Google Scholar]
- 77.Begley LA, Kasina S, Mehra R, Adsule S, Admon AJ, Lonigro RJ, Chinnaiyan AM, Macoska JA. CXCL5 promotes prostate cancer progression. Neoplasia. 2008;10:444–454. doi: 10.1593/neo.07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thacker J, Zdzienicka MZ. The XRCC genes: expanding roles in DNA double-strand break repair. DNA Repair. 2008;3:1081–1090. doi: 10.1016/j.dnarep.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 79.Mazan-Mamczarz K, Hagner PR, Dai B, Wood WH, Zhang Y, Becker KG, Liu Z, Gartenhaus RB. Identification of transformation-related pathways in a breast epithelial cell model using a ribonomics approach. Cancer Res. 2008;68:7730–7735. doi: 10.1158/0008-5472.CAN-08-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 81.Lapucci A, Donnini M, Papucci L, Witort E, Tempestini A, Bevilacqua A, Niolin A, Brewer G, Schiavone N, Capaccioli S. AUF1 is a bcl-2 A+U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J Biol Chem. 2002;277:16139–16146. doi: 10.1074/jbc.M201377200. [DOI] [PubMed] [Google Scholar]
- 82.Shchors K, Yehiely F, Kular R, Kotlo KU, Brewer G, Deiss LP. Cell death inhibiting RNA (CDIR) derived from a 3′-untranslated region binds AUF1 and Heat Shock Protein 27. J Biol Chem. 2002;277:47061–47072. doi: 10.1074/jbc.M202272200. [DOI] [PubMed] [Google Scholar]
- 83.Ishimaru D, Zuraw L, Ramalingam S, Sengupta TK, Bandyopadhyay S, Reuben A, Fernandes DJ, Spicer EK. Mechanism of regulation of Bcl-2 mRNA by nucleolin and A+U rich element binding factor 1 (AUF1) J Biol Chem. 2010;285:27182–27191. doi: 10.1074/jbc.M109.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gautschi O, Ratschiller D, Gugger M, Betticher DC, Heighway J. Cyclin D1 in non-small cell lung cancer: a key driver of malignant transformation. Lung Cancer. 2007;55:1–14. doi: 10.1016/j.lungcan.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 85.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M. Down-regulation of cyclin D1 expression by prostaglandin A2 is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol. 2000;20:7903–7913. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, Liu H, Rosenwald A, Muller-Hermelink HK, Ott G, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599–4606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan B, Wang H, Rabbani ZN, Zhao Y, Li W, Yuan Y, Li F, Dewhirst MW, Li CY. Tumor necrosis factor-α is a potent endogenous mutagen that promotes cellular transformation. Cancer Res. 2006;66:11565–11570. doi: 10.1158/0008-5472.CAN-06-2540. [DOI] [PubMed] [Google Scholar]
- 88.Zhai B, Yang H, Mancini A, He Q, Antoniou J, Di Battista JA. Leukotriene B4 BLT receptor signaling regulates the level and stability of cyclooxygenase-2 (COX-2) mRNA through restricted activation of Ras/Raf/ERK/p42AUF1 pathway. J Biol Chem. 2010;285:23568–23580. doi: 10.1074/jbc.M110.107623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khabar KSA. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ishikawa H, Tsuyama N, Abroun S, Lui S, Otsuyama KI, Zheng X, Kawano MM. Interleukin-6, CD45 and the src-kinases in myeloma cell proliferation. Leukemia Lymphoma. 2003;44:1477–1481. doi: 10.3109/10428190309178767. [DOI] [PubMed] [Google Scholar]
- 91.Paschoud S, Dogar AM, Kuntz C, Grisoni-Neupert B, Richman L, Kuhn LC. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol Cell Biol. 2006;26:8228–8241. doi: 10.1128/MCB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sarkar S, Sinsimer KS, Foster RL, Brewer G, Pestka S. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J Interferon Cytokine Res. 2008;28:679–691. doi: 10.1089/jir.2008.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- 95.Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- 96.Vázquez-Chantada M, Fernández-Ramos D, Embade N, Martínez-Lopez N, Varela-Rey M, Woodhoo A, Luka Z, Wagner C, Anglim PP, Finnel RH, et al. HuR/Methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol Carcinog. 2000;28:76–83. [PubMed] [Google Scholar]
- 98.Xu N, Chen CYA, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sarkar B, Xi Q, He C, Schneider RJ. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol Cell Biol. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello D. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 102.Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Naruo K, Seko C, Kuroshima K, Matsutani E, Sasada R, Kondo T, Kurokawa T. Novel secretory heparin-binding factors from human glioma cells (glia-activating factors) involved in glial cell growth. Purification and biological properties. J Biol Chem. 1993;268:2857–2864. [PubMed] [Google Scholar]
- 104.Miyamoto M, Naruo K, Seko C, Matsumoto S, Kondo T, Kurokawa T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Mol Cell Biol. 1993;13:4251–4259. doi: 10.1128/mcb.13.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen T-M, Hsu C-H, Tsai S-J, Sun HS. AUF1 p42 isoform selectively controls both steady-state and PGE2-induced FGF9 mRNA decay. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq717. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hendrix ND, Wu R, Kuick R, Schwartz DR, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 107.Sheflin LG, Spaulding SW. Testosterone and dihydrotestosterone regulate AUF1 isoforms in a tissue-specific fashion in the mouse. Am J Physiol Endocrinol Metab. 2000;278:E50–E57. doi: 10.1152/ajpendo.2000.278.1.E50. [DOI] [PubMed] [Google Scholar]
- 108.Ing NH, Massuto DA, Jaeger LA. Estradiol up-regulates AUF1p45 binding to stabilizing regions within the 3′-untranslated region of estrogen receptor α mRNA. J Biol Chem. 2008;283:1764–1772. doi: 10.1074/jbc.M704745200. [DOI] [PubMed] [Google Scholar]
- 109.Hambardzumyan D, Sergent-Tanguy S, Thinard R, Bonnamain V, Masip M, Fabre A, Boudin H, Neveu I, Naveilhan P. AUF1 and Hu proteins in the developing rat brain: implication in the proliferation and differentiation of neural progenitors. J Neurosci Res. 2009;87:1296–1309. doi: 10.1002/jnr.21957. [DOI] [PubMed] [Google Scholar]
- 110.Ishikawa F, Matunis MJ, Dreyfuss G, Cech TR. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arao Y, Kuriyama R, Kayama F, Kato S. A nuclear matrix-associated factor, SAF-β, interacts with specific isoforms of AUF1/hnRNP D. Arch Biochem Biophys. 2000;380:228–236. doi: 10.1006/abbi.2000.1938. [DOI] [PubMed] [Google Scholar]
- 112.Inoue A, Arao Y, Omori A, Ichinose S, Nishio K, Yamamoto N, Kinoshita Y, Mita S. Identification of S1 proteins B2, C1 and D1 as AUF1 isoforms and their major role as heterogeneous nuclear ribonucleoprotein proteins. Biochem J. 2003;372:775–785. doi: 10.1042/BJ20021719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shyu AB, Willkinson M. The double lives of shuttling mRNA binding proteins. Cell. 2000;102:135–138. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- 114.Yuan Z, Sanders AJ, Ye L, Jiang WG. HuR, a key post-transcriptional regulator, and its implication in progression of breast cancer. Histol Histopathol. 2010;25:1331–1340. doi: 10.14670/HH-25.1331. [DOI] [PubMed] [Google Scholar]
- 115.Steinman RA. mRNA stability control: a clandestine force in normal and malignant hematopoiesis. Leukemia. 2007;21:1158–1171. doi: 10.1038/sj.leu.2404656. [DOI] [PubMed] [Google Scholar]
- 116.Lai RY, Ljubicic V, D’souza D, Hood DA. Effect of chronic contractile activity on mRNA stability in skeletal muscle. Am J Physiol Cell Physiol. 2010;299:C155–C163. doi: 10.1152/ajpcell.00523.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suzuki M, Iijima M, Nishimura A, Tomozoe Y, Kamei D, Yamada M. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 2005;272:3975–3987. doi: 10.1111/j.1742-4658.2005.04820.x. [DOI] [PubMed] [Google Scholar]
- 118.Sarkar B, Lu J, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- 119.He C, Schneider RJ. 14-3-3σ is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mili S, Shu HJ, Zhao Y, Piñol-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: Candidate intermediates in formation and export of mRNA. Mol Cell Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Piñol-Roma S. hnRNP proteins and the nuclear export of mRNA. Semin Cell Dev Biol. 1997;8:57–63. doi: 10.1006/scdb.1996.0122. [DOI] [PubMed] [Google Scholar]
- 122.Brewer G, Saccani S, Sarkar S, Lewis A, Pestka S. Increased interleukin-10 mRNA stability in melanoma cells is associated with decreased levels of A+U-rich element binding factor AUF1. J Interferon Cytokine Res. 2003;23:553–564. doi: 10.1089/107999003322485053. [DOI] [PubMed] [Google Scholar]
- 123.Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Trojanowicz B, Brodauf L, Sekulla C, Rainer F, Henning D, Hoang-Vu C. The role of AUF1 in thyroid carcinoma progression. Endocr Relat Cancer. 2009;16:857–871. doi: 10.1677/ERC-08-0234. [DOI] [PubMed] [Google Scholar]
- 125.David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE-binding proteins, HuR and p37AUF1. RNA. 2007;13:1453–1468. doi: 10.1261/rna.501707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu W, Rosenberg GA, Liu KJ. AUF-1 mediates inhibition by nitric oxide of lipopolysaccharide-induced matrix metalloproteinase-9 expression in cultured astrocytes. J Neurosci Res. 2006;84:360–369. doi: 10.1002/jnr.20895. [DOI] [PubMed] [Google Scholar]
- 127.Deryugina E, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 128.Nagaoka K, Tanaka T, Imakawa K, Sakai S. Involvement of RNA binding proteins AUF1 in mammary gland differentiation. Exp Cell Res. 2007;313:2937–2945. doi: 10.1016/j.yexcr.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 129.Arao Y, Kikuchi A, Kishida M, Yonekura M, Inoue A, Yasuda S, Wada S, Ikeda K, Kayama F. Stability of A+U-rich element binding factor 1 (AUF1)-binding messenger ribonucleic acid correlates with the subcellular relocalization of AUF1 in the rat uterus upon estrogen treatment. Mol Endocrinol. 2004;18:2255–2267. doi: 10.1210/me.2004-0103. [DOI] [PubMed] [Google Scholar]
- 130.VanHuffel S, Delaei F, Heyninck K, DeValck D. Identification of a novel A20-binding inhibitor of nuclear factor-κ B activation termed ABIN-2. J Biol Chem. 2001;276:30216–30223. doi: 10.1074/jbc.M100048200. [DOI] [PubMed] [Google Scholar]
- 131.Hughes DP, Marron MB, Brindle NP. The antiinflammatory endothelial tyrosine kinase Tie2 interacts with a novel nuclear factor-κ B inhibitor ABIN-2. Circ Res. 2003;92:630–636. doi: 10.1161/01.RES.0000063422.38690.DC. [DOI] [PubMed] [Google Scholar]
- 132.Tadros A, Hughes DP, Dunmore BJ, Brindle NP. ABIN-2 protects endothelial cells from death and has a role in the antiapoptotic effect of angiopoietin-1. Blood. 2003;102:4407–4409. doi: 10.1182/blood-2003-05-1602. [DOI] [PubMed] [Google Scholar]
- 133.Yang X, Wang W, Fan J, Lal A, Yang D, Cheng H, Gorospe M. Prostaglandin A2-mediated stabilization of p21 mRNA through an ERK-dependent pathway requiring the RNA-binding protein HuR. J Biol Chem. 2004;279:49298–49306. doi: 10.1074/jbc.M407535200. [DOI] [PubMed] [Google Scholar]
- 134.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 136.Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. J Biol Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 137.Ji Y, Tulin AV. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 2009;37:3501–3513. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Laroia G, Sarkar B, Schneider RJ. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc Natl Acad Sci USA. 2002;99:1842–1846. doi: 10.1073/pnas.042575699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 140.Aitken A, Baxter H, Dubois T, Clokie S, Mackie S, Mitchell K, Peden A, Zemlickova E. 14-3-3 Proteins in cell regulation. Biochem Soc Trans. 2002;30:351–360. doi: 10.1042/bst0300351. [DOI] [PubMed] [Google Scholar]
- 141.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Benjamin D, Schmidlin M, Min L, Gross B, Moroni C. BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol Cell Biol. 2006;26:9497–9507. doi: 10.1128/MCB.01099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Levi R, Ben-Dov I, Lavi-Moshayoff V, Dinur M, Martin D, Naveh-Many T, Silver J. Increased parathyroid hormone gene expression in secondary hyperparathyroidism of experimental uremia is reversed by calcimimetics: Correlation with posttranslational modifications of the trans acting factor AUF1. J Am Soc Nephol. 2006;17:107–112. doi: 10.1681/ASN.2005070679. [DOI] [PubMed] [Google Scholar]
- 144.Silver J, Kilav R, Naveh-Many T. Mechanisms of secondary hyperparathyroidism. Am J Physiol Renal Physiol. 2002;283:F367–F376. doi: 10.1152/ajprenal.00061.2002. [DOI] [PubMed] [Google Scholar]
- 145.Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol. 2005;6:1280–1287. doi: 10.1038/ni1266. [DOI] [PubMed] [Google Scholar]
- 146.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1883–1884. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 147.Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol. 2010;30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang W, Martindale JL, Yang X, Chrest FJ, Gorospe M. Increased stability of the p16 mRNA with replicative senescence. EMBO reports. 2005;6:158–164. doi: 10.1038/sj.embor.7400346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45α by genotoxic stress. Mol Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 150.Sirenko O, Böcker U, Morris JS, Haskill JS, Watson JM. IL-1β transcript stability in monocytes is linked to cytoskeletal reorganization and the availability of mRNA degradation factors. Immunol Cell Biol. 2002;80:328–339. doi: 10.1046/j.1440-1711.2002.01085.x. [DOI] [PubMed] [Google Scholar]
- 151.Pautz A, Linker K, Altenhöfer S, Heil S, Schmidt N, Art J, Knauer S, Stauber R, Sadri N, Pont A, et al. Similar regulation of human inducible nitric-oxide synthase expression by different isoforms of the RNA-binding protein AUF1. J Biol Chem. 2009;284:2755–2766. doi: 10.1074/jbc.M809314200. [DOI] [PubMed] [Google Scholar]
- 152.Loflin P, Chen CYA, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destablization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pullman R, Abdelmohsen K, Lal A, Martindale JL, Ladner RD, Gorospe M. Differential stability of thymidylate synthase 3′-untranslated region polymorphic variants regulated by AUF1. J Biol Chem. 2006;281:23465–23463. doi: 10.1074/jbc.M600282200. [DOI] [PubMed] [Google Scholar]
- 154.Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′-untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


