Abstract
Antigen receptor genes acquire junctional inserts upon assembly from their component, germ line-encoded V, D, and J segments. Inserts are generally of random sequence, but a small number of V-D, D-J, or V-J junctions are exceptional. In such junctions, one or two added base pairs inversely repeat the sequence of the abutting germ line DNA. (For example, a gene segment ending AG might acquire an insert beginning with the residues CT upon joining). It has been proposed that the nonrandom residues, termed "P nucleotides," are a consequence of an obligatory end-modification step in V(D)J recombination. P insertion in normal, unselected V(D)J joining products, however, has not been rigorously established. Here, we use an experimentally manipulable system, isolated from immune selection of any kind, to examine the fine structure of V(D)J junctions formed in wild-type lymphoid cells. Our results, according to statistical tests, show the following, (i) The frequency of P insertion is influenced by the DNA sequence of the joined ends. (ii) P inserts may be longer than two residues in length. (iii) P inserts are associated with coding ends only. Additionally, a systematic survey of published P nucleotide data shows no evidence for variation in P insertion as a function of genetic locus and ontogeny. Together, these analyses establish the generality of the P nucleotide pattern within inserts but do not fully support previous conjectures as to their origin and centrality in the joining reaction.
Full text
PDF
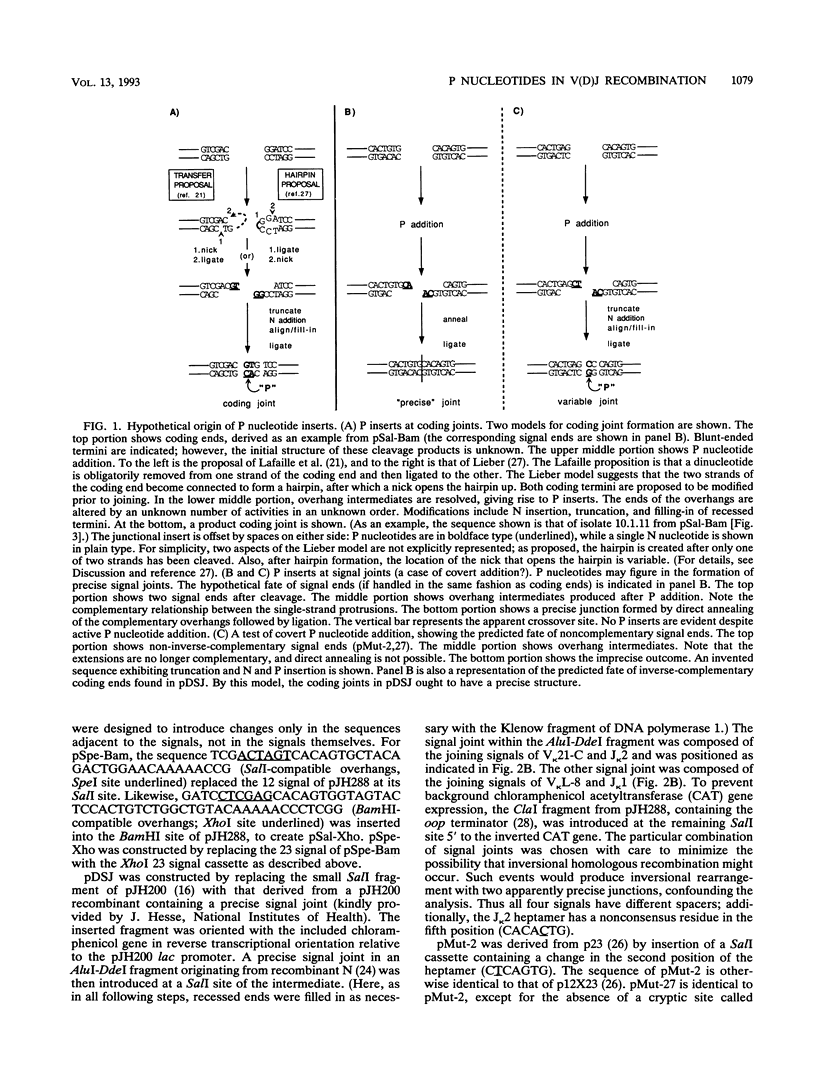
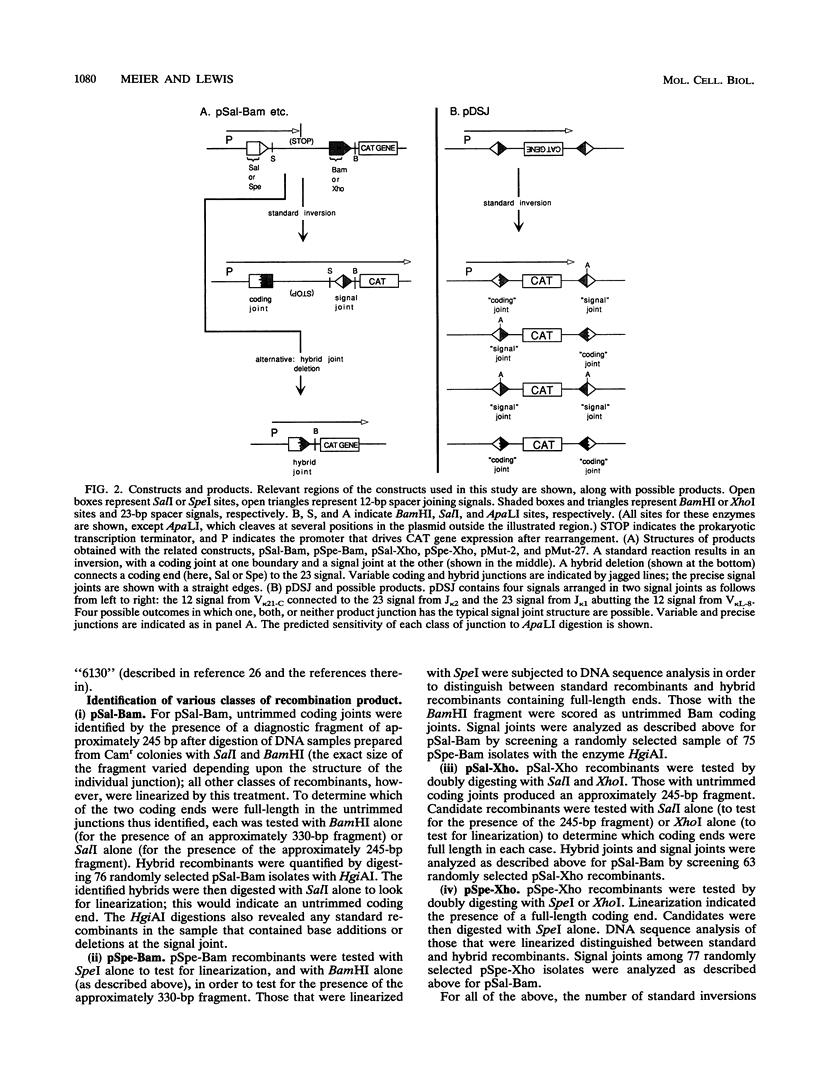
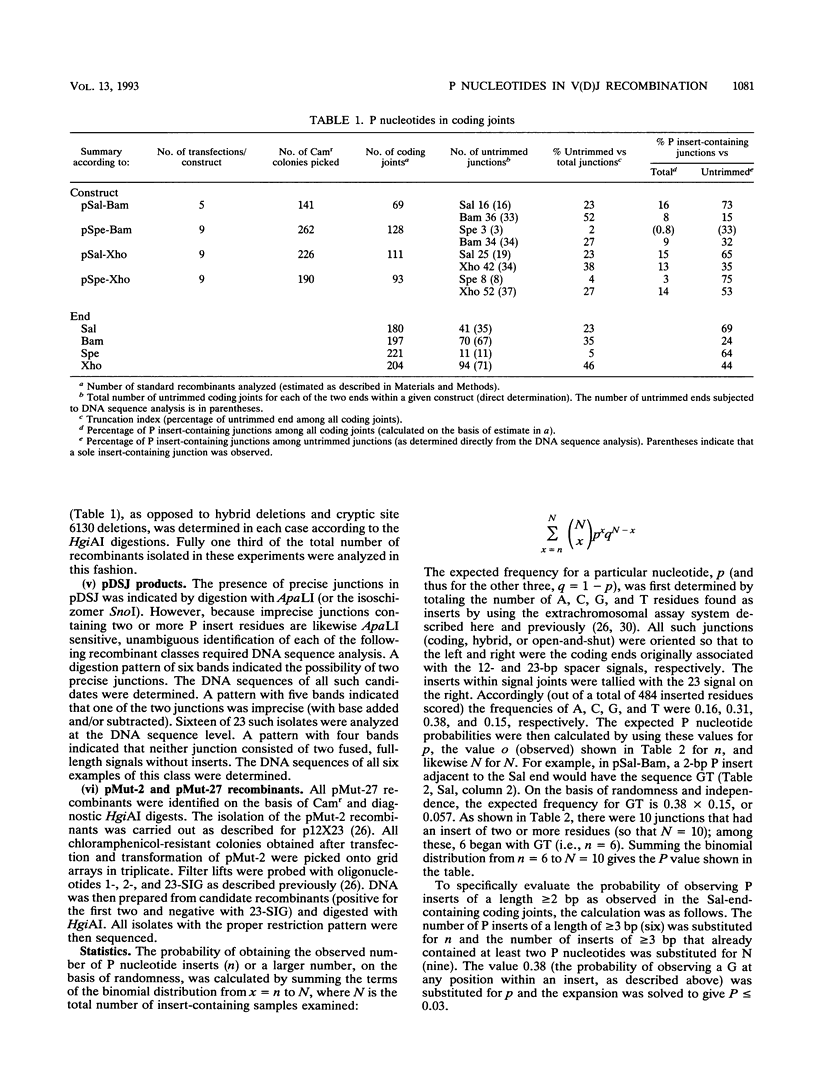





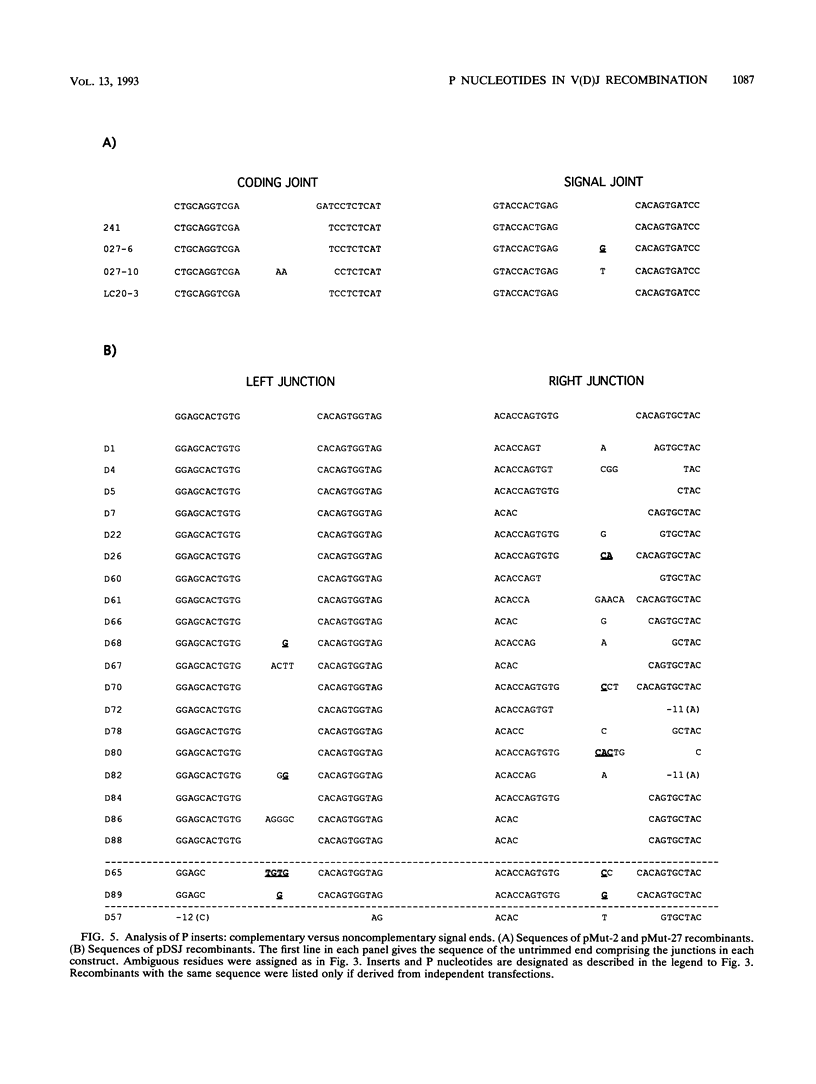




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bangs L. A., Sanz I. E., Teale J. M. Comparison of D, JH, and junctional diversity in the fetal, adult, and aged B cell repertoires. J Immunol. 1991 Mar 15;146(6):1996–2004. [PubMed] [Google Scholar]
- Bogue M., Candéias S., Benoist C., Mathis D. A special repertoire of alpha:beta T cells in neonatal mice. EMBO J. 1991 Dec;10(12):3647–3654. doi: 10.1002/j.1460-2075.1991.tb04931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma M. J., Carroll A. M. The SCID mouse mutant: definition, characterization, and potential uses. Annu Rev Immunol. 1991;9:323–350. doi: 10.1146/annurev.iy.09.040191.001543. [DOI] [PubMed] [Google Scholar]
- Candéias S., Waltzinger C., Benoist C., Mathis D. The V beta 17+ T cell repertoire: skewed J beta usage after thymic selection; dissimilar CDR3s in CD4+ versus CD8+ cells. J Exp Med. 1991 Nov 1;174(5):989–1000. doi: 10.1084/jem.174.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L., Overmo C., Holmberg D. Selection against N-region diversity in immunoglobulin heavy chain variable regions during the development of pre-immune B cell repertoires. Int Immunol. 1992 May;4(5):549–553. doi: 10.1093/intimm/4.5.549. [DOI] [PubMed] [Google Scholar]
- Carroll A. M., Bosma M. J. T-lymphocyte development in scid mice is arrested shortly after the initiation of T-cell receptor delta gene recombination. Genes Dev. 1991 Aug;5(8):1357–1366. doi: 10.1101/gad.5.8.1357. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Carpenter R., Martin C. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell. 1986 Oct 24;47(2):285–296. doi: 10.1016/0092-8674(86)90451-4. [DOI] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Decker D. J., Boyle N. E., Koziol J. A., Klinman N. R. The expression of the Ig H chain repertoire in developing bone marrow B lineage cells. J Immunol. 1991 Jan 1;146(1):350–361. [PubMed] [Google Scholar]
- Feeney A. J. Junctional sequences of fetal T cell receptor beta chains have few N regions. J Exp Med. 1991 Jul 1;174(1):115–124. doi: 10.1084/jem.174.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney A. J. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med. 1990 Nov 1;172(5):1377–1390. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney A. J. Predominance of the prototypic T15 anti-phosphorylcholine junctional sequence in neonatal pre-B cells. J Immunol. 1991 Dec 15;147(12):4343–4350. [PubMed] [Google Scholar]
- Ferrier P., Covey L. R., Li S. C., Suh H., Malynn B. A., Blackwell T. K., Morrow M. A., Alt F. W. Normal recombination substrate VH to DJH rearrangements in pre-B cell lines from scid mice. J Exp Med. 1990 Jun 1;171(6):1909–1918. doi: 10.1084/jem.171.6.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Förster I., Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO J. 1990 Jul;9(7):2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K., Yamagishi H. Lack of feedback inhibition of V kappa gene rearrangement by productively rearranged alleles. J Exp Med. 1991 Feb 1;173(2):409–415. doi: 10.1084/jem.173.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse J. E., Lieber M. R., Gellert M., Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987 Jun 19;49(6):775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- Kallenbach S., Doyen N., Fanton d'Andon M., Rougeon F. Three lymphoid-specific factors account for all junctional diversity characteristic of somatic assembly of T-cell receptor and immunoglobulin genes. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2799–2803. doi: 10.1073/pnas.89.7.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienker L. J., Kuziel W. A., Garni-Wagner B. A., Kumar V., Tucker P. W. T cell receptor gamma and delta gene rearrangements in scid thymocytes. Similarity to those in normal thymocytes. J Immunol. 1991 Dec 15;147(12):4351–4359. [PubMed] [Google Scholar]
- Lafaille J. J., DeCloux A., Bonneville M., Takagaki Y., Tonegawa S. Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell. 1989 Dec 1;59(5):859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- Lewis S. M., Hesse J. E. Cutting and closing without recombination in V(D)J joining. EMBO J. 1991 Dec;10(12):3631–3639. doi: 10.1002/j.1460-2075.1991.tb04929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. M., Hesse J. E., Mizuuchi K., Gellert M. Novel strand exchanges in V(D)J recombination. Cell. 1988 Dec 23;55(6):1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- Lewis S., Gellert M. The mechanism of antigen receptor gene assembly. Cell. 1989 Nov 17;59(4):585–588. doi: 10.1016/0092-8674(89)90002-0. [DOI] [PubMed] [Google Scholar]
- Lewis S., Gifford A., Baltimore D. DNA elements are asymmetrically joined during the site-specific recombination of kappa immunoglobulin genes. Science. 1985 May 10;228(4700):677–685. doi: 10.1126/science.3158075. [DOI] [PubMed] [Google Scholar]
- Lewis S., Gifford A., Baltimore D. Joining of V kappa to J kappa gene segments in a retroviral vector introduced into lymphoid cells. 1984 Mar 29-Apr 4Nature. 308(5958):425–428. doi: 10.1038/308425a0. [DOI] [PubMed] [Google Scholar]
- Lieber M. R., Hesse J. E., Lewis S., Bosma G. C., Rosenberg N., Mizuuchi K., Bosma M. J., Gellert M. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988 Oct 7;55(1):7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- Lieber M. R., Hesse J. E., Mizuuchi K., Gellert M. Developmental stage specificity of the lymphoid V(D)J recombination activity. Genes Dev. 1987 Oct;1(8):751–761. doi: 10.1101/gad.1.8.751. [DOI] [PubMed] [Google Scholar]
- Lieber M. R., Hesse J. E., Mizuuchi K., Gellert M. Lymphoid V(D)J recombination: nucleotide insertion at signal joints as well as coding joints. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8588–8592. doi: 10.1073/pnas.85.22.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M. R. Site-specific recombination in the immune system. FASEB J. 1991 Nov;5(14):2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- McCormack W. T., Tjoelker L. W., Carlson L. M., Petryniak B., Barth C. F., Humphries E. H., Thompson C. B. Chicken IgL gene rearrangement involves deletion of a circular episome and addition of single nonrandom nucleotides to both coding segments. Cell. 1989 Mar 10;56(5):785–791. doi: 10.1016/0092-8674(89)90683-1. [DOI] [PubMed] [Google Scholar]
- McVay L. D., Carding S. R., Bottomly K., Hayday A. C. Regulated expression and structure of T cell receptor gamma/delta transcripts in human thymic ontogeny. EMBO J. 1991 Jan;10(1):83–91. doi: 10.1002/j.1460-2075.1991.tb07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C., Even J., Jarvis J. M., Gonzalez-Fernandez A., Gherardi E. Non-random features of the repertoire expressed by the members of one V kappa gene family and of the V-J recombination. Eur J Immunol. 1992 Jun;22(6):1627–1634. doi: 10.1002/eji.1830220642. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Heteroduplex substrates for bacteriophage lambda site-specific recombination: cleavage and strand transfer products. EMBO J. 1989 Nov;8(11):3523–3533. doi: 10.1002/j.1460-2075.1989.tb08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock W. J., Dennis E. S., Gerlach W. L., Sachs M. M., Schwartz D. Insertion and excision of Ds controlling elements in maize. Cold Spring Harb Symp Quant Biol. 1984;49:347–354. doi: 10.1101/sqb.1984.049.01.041. [DOI] [PubMed] [Google Scholar]
- Reynaud C. A., Anquez V., Weill J. C. The chicken D locus and its contribution to the immunoglobulin heavy chain repertoire. Eur J Immunol. 1991 Nov;21(11):2661–2670. doi: 10.1002/eji.1830211104. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Chang X. B., Wilson J. H. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol Cell Biol. 1989 Jul;9(7):3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Menetski J. P., Nakajima P. B., Bosma M. J., Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992 Sep 18;70(6):983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Nakajima P. B., Menetski J. P., Bosma M. J., Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992 Apr 3;69(1):41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- Schuler W., Ruetsch N. R., Amsler M., Bosma M. J. Coding joint formation of endogenous T cell receptor genes in lymphoid cells from scid mice: unusual P-nucleotide additions in VJ-coding joints. Eur J Immunol. 1991 Mar;21(3):589–596. doi: 10.1002/eji.1830210309. [DOI] [PubMed] [Google Scholar]



