Abstract
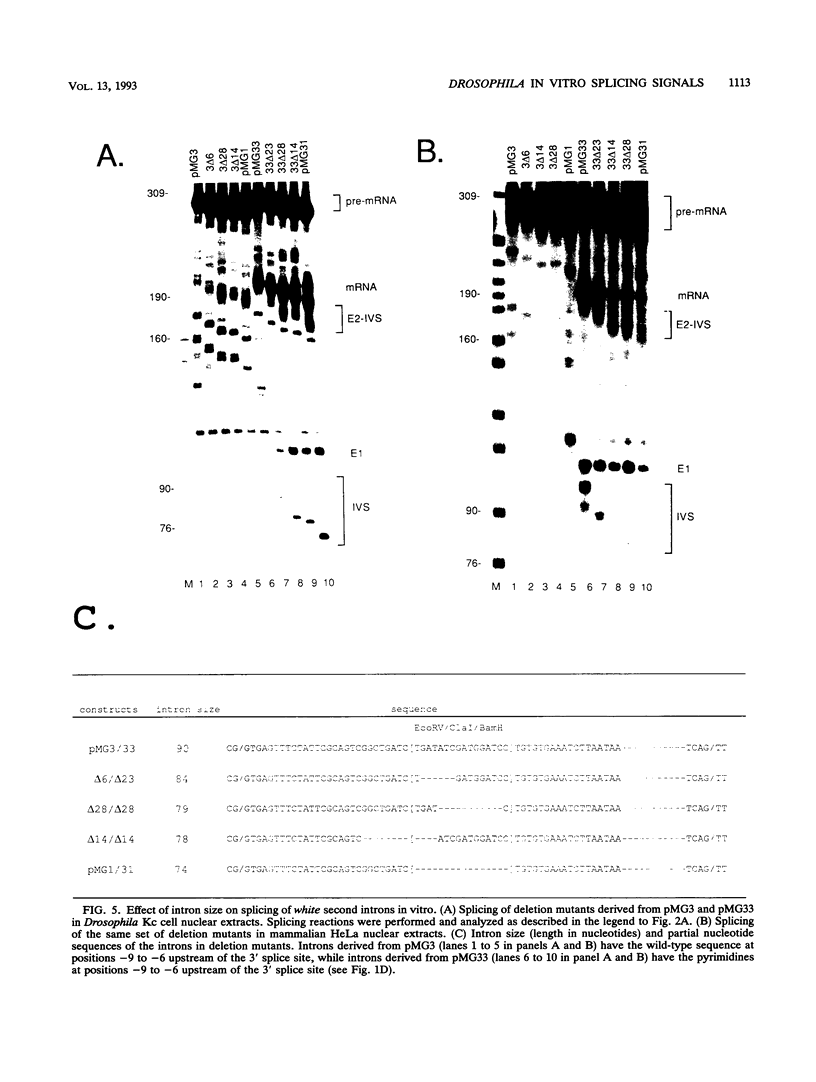
The effects of branchpoint sequence, the pyrimidine stretch, and intron size on the splicing efficiency of the Drosophila white gene second intron were examined in nuclear extracts from Drosophila and human cells. This 74-nucleotide intron is typical of many Drosophila introns in that it lacks a significant pyrimidine stretch and is below the minimum size required for splicing in human nuclear extracts. Alteration of sequences of adjacent to the 3' splice site to create a pyrimidine stretch was necessary for splicing in human, but not Drosophila, extracts. Increasing the size of this intron with insertions between the 5' splice site and the branchpoint greatly reduced the efficiency of splicing of introns longer than 79 nucleotides in Drosophila extracts but had an opposite effect in human extracts, in which introns longer than 78 nucleotides were spliced with much greater efficiency. The white-apricot copia insertion is immediately adjacent to the branchpoint normally used in the splicing of this intron, and a copia long terminal repeat insertion prevents splicing in Drosophila, but not human, extracts. However, a consensus branchpoint does not restore the splicing of introns containing the copia long terminal repeat, and alteration of the wild-type branchpoint sequence alone does not eliminate splicing. These results demonstrate species specificity of splicing signals, particularly pyrimidine stretch and size requirements, and raise the possibility that variant mechanisms not found in mammals may operate in the splicing of small introns in Drosophila and possibly other species.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G., Nevins J. R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988 Jul;7(7):2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Beggs J. D., van den Berg J., van Ooyen A., Weissmann C. Abnormal expression of chromosomal rabbit beta-globin gene in Saccharomyces cerevisiae. Nature. 1980 Feb 28;283(5750):835–840. doi: 10.1038/283835a0. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., Cline T. W. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell. 1988 Dec 23;55(6):1037–1046. doi: 10.1016/0092-8674(88)90248-6. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Judd B. H. A copy of the copia transposable element is very tightly linked to the Wa allele at the white locus of D. melanogaster. Cell. 1981 Sep;25(3):705–711. doi: 10.1016/0092-8674(81)90177-x. [DOI] [PubMed] [Google Scholar]
- Birchler J. A., Hiebert J. C. Interaction of the Enhancer of white-apricot with transposable element alleles at the white locus in Drosophila melanogaster. Genetics. 1989 May;122(1):129–138. doi: 10.1093/genetics/122.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Hiebert J. C., Rabinow L. Interaction of the mottler of white with transposable element alleles at the white locus in Drosophila melanogaster. Genes Dev. 1989 Jan;3(1):73–84. doi: 10.1101/gad.3.1.73. [DOI] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Thomas J. Cis and trans mRNA splicing in C. elegans. Trends Genet. 1988 Nov;4(11):305–308. doi: 10.1016/0168-9525(88)90107-2. [DOI] [PubMed] [Google Scholar]
- Boggs R. T., Gregor P., Idriss S., Belote J. M., McKeown M. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 1987 Aug 28;50(5):739–747. doi: 10.1016/0092-8674(87)90332-1. [DOI] [PubMed] [Google Scholar]
- Chou T. B., Zachar Z., Bingham P. M. Developmental expression of a regulatory gene is programmed at the level of splicing. EMBO J. 1987 Dec 20;6(13):4095–4104. doi: 10.1002/j.1460-2075.1987.tb02755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Ge H., Noble J., Colgan J., Manley J. L. Polyoma virus small tumor antigen pre-mRNA splicing requires cooperation between two 3' splice sites. Proc Natl Acad Sci U S A. 1990 May;87(9):3338–3342. doi: 10.1073/pnas.87.9.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W. J., Paro R. Isolation of a hybrid plasmid with homologous sequences to a transposing element of Drosophila melanogaster. Cell. 1980 Apr;19(4):897–904. doi: 10.1016/0092-8674(80)90081-1. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 1991 Sep;10(9):2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Harper D. S., Fresco L. D., Keene J. D. RNA binding specificity of a Drosophila snRNP protein that shares sequence homology with mammalian U1-A and U2-B" proteins. Nucleic Acids Res. 1992 Jul 25;20(14):3645–3650. doi: 10.1093/nar/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Johnson D., Raychaudhuri G., Beyer A. L. The Drosophila Hrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res. 1991 Jan 11;19(1):25–31. doi: 10.1093/nar/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Raychaudhuri G., Beyer A. L. The Drosophila Hrb98DE locus encodes four protein isoforms homologous to the A1 protein of mammalian heterogeneous nuclear ribonucleoprotein complexes. Mol Cell Biol. 1990 Jan;10(1):316–323. doi: 10.1128/mcb.10.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A., Rodriguez J. R., Rosbash M. A quantitative analysis of the effects of 5' junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell. 1985 Dec;43(2 Pt 1):423–430. doi: 10.1016/0092-8674(85)90172-2. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of Drosophila pre-mRNAs. Nucleic Acids Res. 1985 Jul 11;13(13):4971–4981. doi: 10.1093/nar/13.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Grabowski P. J., Padgett R. A., Sharp P. A. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985 Feb 14;313(6003):552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kurkulos M., Weinberg J. M., Pepling M. E., Mount S. M. Polyadenylylation in copia requires unusually distant upstream sequences. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3038–3042. doi: 10.1073/pnas.88.8.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Legrain P., Seraphin B., Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988 Sep;8(9):3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Bingham P. M., Rubin G. M. Physical map of the white locus of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Jan;79(2):564–568. doi: 10.1073/pnas.79.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., O'Hare K., Rubin G. M. Effects of transposable element insertions on RNA encoded by the white gene of Drosophila. Cell. 1984 Sep;38(2):471–481. doi: 10.1016/0092-8674(84)90502-6. [DOI] [PubMed] [Google Scholar]
- Levitt N., Briggs D., Gil A., Proudfoot N. J. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989 Jul;3(7):1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- Lo P. C., Mount S. M. Drosophila melanogaster genes for U1 snRNA variants and their expression during development. Nucleic Acids Res. 1990 Dec 11;18(23):6971–6979. doi: 10.1093/nar/18.23.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo R., Lo P. C., Mount S. M. Structure and expression of the Drosophila melanogaster gene for the U1 small nuclear ribonucleoprotein particle 70K protein. Mol Cell Biol. 1990 Jun;10(6):2492–2502. doi: 10.1128/mcb.10.6.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattox W., Baker B. S. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 1991 May;5(5):786–796. doi: 10.1101/gad.5.5.786. [DOI] [PubMed] [Google Scholar]
- Matunis E. L., Matunis M. J., Dreyfuss G. Characterization of the major hnRNP proteins from Drosophila melanogaster. J Cell Biol. 1992 Jan;116(2):257–269. doi: 10.1083/jcb.116.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M. J., Matunis E. L., Dreyfuss G. Isolation of hnRNP complexes from Drosophila melanogaster. J Cell Biol. 1992 Jan;116(2):245–255. doi: 10.1083/jcb.116.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Zahler A. M., Krainer A. R., Roth M. B. Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Burks C., Hertz G., Stormo G. D., White O., Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992 Aug 25;20(16):4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Green M. M., Rubin G. M. Partial revertants of the transposable element-associated suppressible allele white-apricot in Drosophila melanogaster: structures and responsiveness to genetic modifiers. Genetics. 1988 Feb;118(2):221–234. doi: 10.1093/genetics/118.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Pettersson I., Hinterberger M., Karmas A., Steitz J. A. The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro. Cell. 1983 Jun;33(2):509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Steitz J. A. Sequence of U1 RNA from Drosophila melanogaster: implications for U1 secondary structure and possible involvement in splicing. Nucleic Acids Res. 1981 Dec 11;9(23):6351–6368. doi: 10.1093/nar/9.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi R. N., McKeown M., Burtis K. C., Belote J. M., Baker B. S. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988 Apr 22;53(2):229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989 Oct;3(10):1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Lin R. J., Cheng S. C., Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985 Aug;42(1):335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Ge H., Chaudhuri M., Manley J. L. Factor interactions with the simian virus 40 early pre-mRNA influence branch site selection and alternative splicing. Mol Cell Biol. 1989 May;9(5):2007–2017. doi: 10.1128/mcb.9.5.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble J. C., Pan Z. Q., Prives C., Manley J. L. Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell. 1987 Jul 17;50(2):227–236. doi: 10.1016/0092-8674(87)90218-2. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. In vitro splicing of simian virus 40 early pre mRNA. Nucleic Acids Res. 1986 Feb 11;14(3):1219–1235. doi: 10.1093/nar/14.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Murphy C., Levis R., Rubin G. M. DNA sequence of the white locus of Drosophila melanogaster. J Mol Biol. 1984 Dec 15;180(3):437–455. doi: 10.1016/0022-2836(84)90021-4. [DOI] [PubMed] [Google Scholar]
- Ogg S. C., Anderson P., Wickens M. P. Splicing of a C. elegans myosin pre-mRNA in a human nuclear extract. Nucleic Acids Res. 1990 Jan 11;18(1):143–149. doi: 10.1093/nar/18.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5' splice junction uncouples recognition, cleavage, and ligation. Cell. 1985 May;41(1):107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Peng X. B., Mount S. M. Characterization of enhancer-of-white-apricot in Drosophila melanogaster. Genetics. 1990 Dec;126(4):1061–1069. doi: 10.1093/genetics/126.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling M., Mount S. M. Sequence of a cDNA from the Drosophila melanogaster white gene. Nucleic Acids Res. 1990 Mar 25;18(6):1633–1633. doi: 10.1093/nar/18.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V., Bröckl C. Transcription of the Drosophila white locus and some of its mutants. EMBO J. 1984 Mar;3(3):563–568. doi: 10.1002/j.1460-2075.1984.tb01847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinow L., Birchler J. A. A dosage-sensitive modifier of retrotransposon-induced alleles of the Drosophila white locus. EMBO J. 1989 Mar;8(3):879–889. doi: 10.1002/j.1460-2075.1989.tb03449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Rio D. C. Accurate and efficient pre-mRNA splicing in Drosophila cell-free extracts. Proc Natl Acad Sci U S A. 1988 May;85(9):2904–2908. doi: 10.1073/pnas.85.9.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Safer B. 2B or not 2B: regulation of the catalytic utilization of eIF-2. Cell. 1983 May;33(1):7–8. doi: 10.1016/0092-8674(83)90326-4. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Mutational analysis of the interactions between U1 small nuclear RNA and pre-mRNA of yeast. Gene. 1989 Oct 15;82(1):145–151. doi: 10.1016/0378-1119(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Tseng J. C., Zollman S., Chain A. C., Laski F. A. Splicing of the Drosophila P element ORF2-ORF3 intron is inhibited in a human cell extract. Mech Dev. 1991 Aug;35(1):65–72. doi: 10.1016/0925-4773(91)90042-5. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Herrero J. J., Filipowicz W. Nuclear pre-mRNA processing in plants: distinct modes of 3'-splice-site selection in plants and animals. Mol Cell Biol. 1988 May;8(5):2042–2051. doi: 10.1128/mcb.8.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zachar Z., Chou T. B., Bingham P. M. Evidence that a regulatory gene autoregulates splicing of its transcript. EMBO J. 1987 Dec 20;6(13):4105–4111. doi: 10.1002/j.1460-2075.1987.tb02756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z., Davison D., Garza D., Bingham P. M. A detailed developmental and structural study of the transcriptional effects of insertion of the Copia transposon into the white locus of Drosophila melanogaster. Genetics. 1985 Nov;111(3):495–515. doi: 10.1093/genetics/111.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991 Jan;10(1):207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]