Abstract
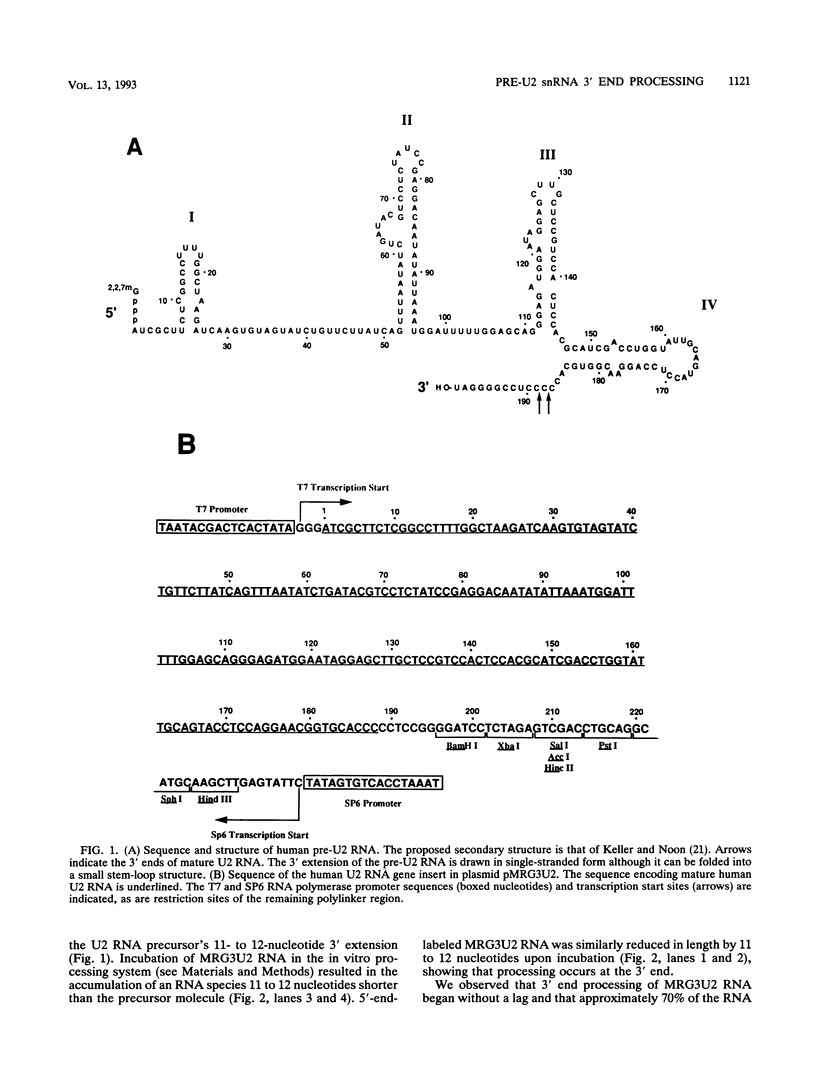
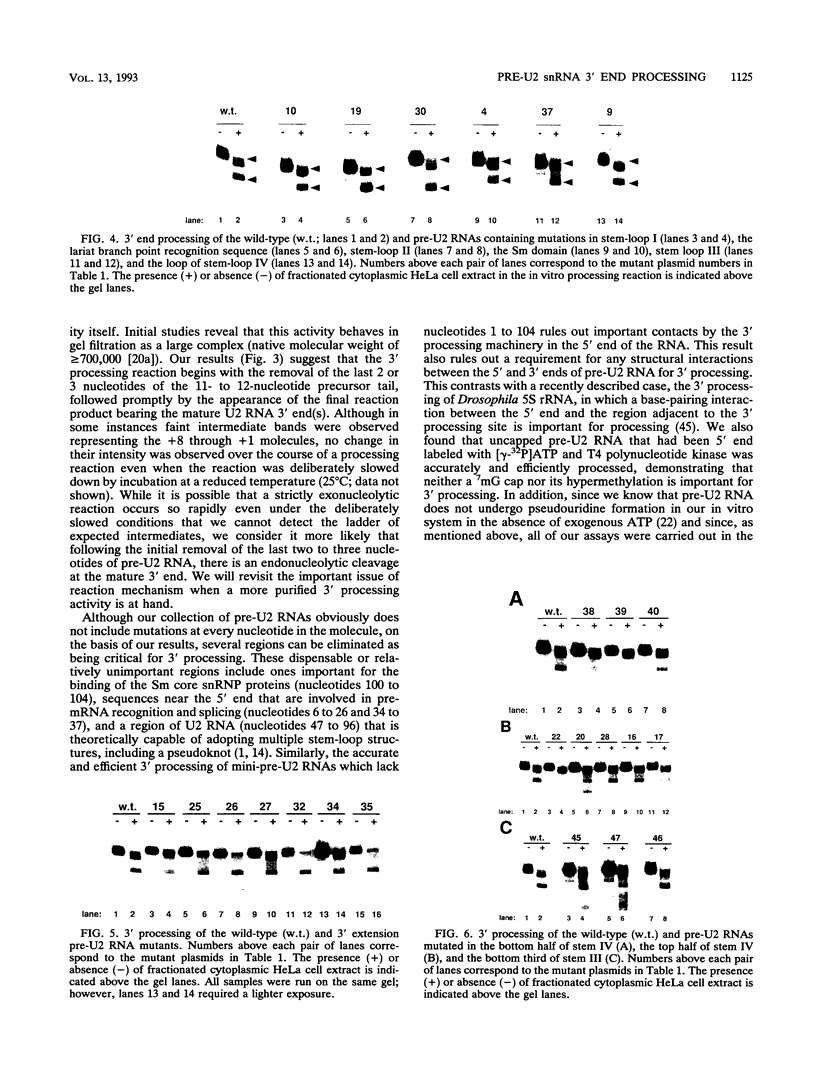
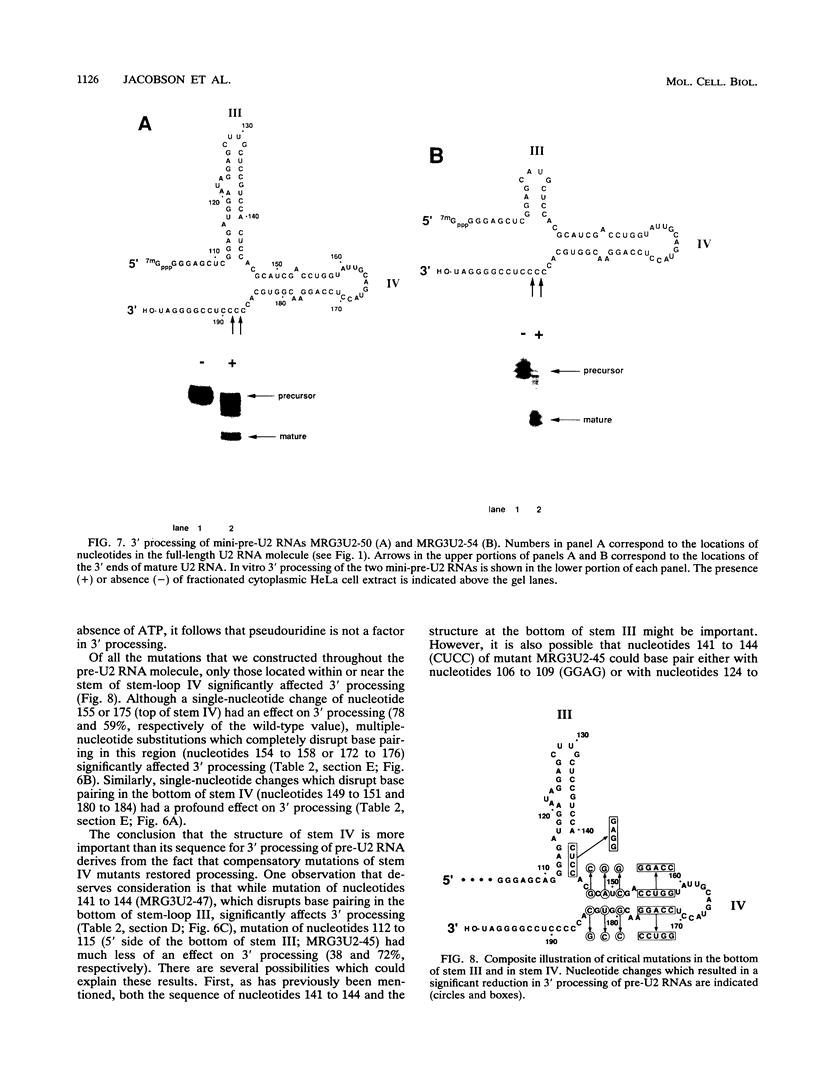
Mature U2 small nuclear RNA is generated by the removal of 11 to 12 nucleotides from the 3' end of the primary transcript. This pre-U2 RNA processing reaction takes place in the cytoplasm. In this study, the sequences and/or structures of pre-U2 RNA that are important for 3' processing have been examined in an in vitro system. The 7-methylguanosine cap, stem-loops I and II, the lariat branch site recognition sequence, the conserved Sm domain, and several other regions throughout the 5' end of U2 RNA have no apparent role in the 3' processing reaction. In fact, deletion of the entire first 104 nucleotides resulted in mini-pre-U2 RNAs which were efficiently processed. Similarly, deletion of the top two-thirds of stem-loop III or mutation of nucleotides in the loop of stem-loop IV had little effect on 3' processing. Most surprisingly, the precursor's 11- to 12-nucleotide 3' extension itself was of relatively little importance, since this sequence could be replaced with completely different sequences with only a minor effect on the 3' processing reaction. In contrast, we have defined a critical structure consisting of the bottom of stem III and the stem of stem-loop IV that is essential for 3' processing of pre-U2 RNA. Compensatory mutations which restore base pairing in this region resulted in normal 3' processing. Thus, although the U2 RNA processing activity recognizes the bottom of stem III and stem IV, the sequence of this critical region is much less important than its structure. These results, together with the surprising observation that the reaction is relatively indifferent to the sequence of the 11- to 12-nucleotide 3' extension itself, point to a 3' processing reaction of pre-U2 RNA that has sequence and structure requirements significantly different from those previously identified for pre-mRNA 3' processing.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ares M., Jr, Igel A. H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990 Dec;4(12A):2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- Bentley R. C., Keene J. D. Recognition of U1 and U2 small nuclear RNAs can be altered by a 5-amino-acid segment in the U2 small nuclear ribonucleoprotein particle (snRNP) B" protein and through interactions with U2 snRNP-A' protein. Mol Cell Biol. 1991 Apr;11(4):1829–1839. doi: 10.1128/mcb.11.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens W., Scherly D., Beijer R. P., Jansen E. J., Dathan N. A., Mattaj I. W., van Venrooij W. J. A weak interaction between the U2A' protein and U2 snRNA helps to stabilize their complex with the U2B" protein. Nucleic Acids Res. 1991 Feb 11;19(3):455–460. doi: 10.1093/nar/19.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet J. P., Meyer L. M., Pederson T. Small nuclear RNA U2 is base-paired to heterogeneous nuclear RNA. Science. 1982 Jul 30;217(4558):456–458. doi: 10.1126/science.6178162. [DOI] [PubMed] [Google Scholar]
- Calvet J. P., Pederson T. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell. 1981 Nov;26(3 Pt 1):363–370. doi: 10.1016/0092-8674(81)90205-1. [DOI] [PubMed] [Google Scholar]
- Datta B., Weiner A. M. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991 Aug 29;352(6338):821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M., Lienhard S., Parisot R. F. Intracellular transport of microinjected 5S and small nuclear RNAs. Nature. 1982 Feb 18;295(5850):572–577. doi: 10.1038/295572a0. [DOI] [PubMed] [Google Scholar]
- Eliceiri G. L., Sayavedra M. S. Small RNAs in the nucleus and cytoplasm of HeLa cells. Biochem Biophys Res Commun. 1976 Sep 20;72(2):507–512. doi: 10.1016/s0006-291x(76)80070-8. [DOI] [PubMed] [Google Scholar]
- Eliceiri G. L. Short-lived, small RNAs in the cytoplasm of HeLa cells. Cell. 1974 Sep;3(1):11–14. doi: 10.1016/0092-8674(74)90031-2. [DOI] [PubMed] [Google Scholar]
- Fischer U., Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990 Aug 17;249(4970):786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hamm J., Darzynkiewicz E., Tahara S. M., Mattaj I. W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990 Aug 10;62(3):569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Mattaj I. W. Functional analysis of mutant Xenopus U2 snRNAs. Cell. 1989 Oct 6;59(1):159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- Hausner T. P., Giglio L. M., Weiner A. M. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990 Dec;4(12A):2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3' end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985 Jul;4(7):1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Igel A. H., Ares M., Jr Internal sequences that distinguish yeast from metazoan U2 snRNA are unnecessary for pre-mRNA splicing. Nature. 1988 Aug 4;334(6181):450–453. doi: 10.1038/334450a0. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of Drosophila pre-mRNAs. Nucleic Acids Res. 1985 Jul 11;13(13):4971–4981. doi: 10.1093/nar/13.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt A. M., Patton J. R., Pederson T. U2 small nuclear RNP assembly in vitro. Nucleic Acids Res. 1989 Jun 26;17(12):4817–4828. doi: 10.1093/nar/17.12.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt A. M., Pederson T. Accurate and efficient 3' processing of U2 small nuclear RNA precursor in a fractionated cytoplasmic extract. Mol Cell Biol. 1987 Sep;7(9):3131–3137. doi: 10.1128/mcb.7.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt A. M., Pederson T. RNA processing and ribonucleoprotein assembly studied in vivo by RNA transfection. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1283–1287. doi: 10.1073/pnas.87.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G. R., Maser R. L., Calvet J. P., Pederson T. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8575–8579. doi: 10.1073/pnas.83.22.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G. R., Pederson T. Transcription boundaries of U1 small nuclear RNA. Mol Cell Biol. 1985 Sep;5(9):2332–2340. doi: 10.1128/mcb.5.9.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G. R., Pederson T. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev. 1988 Feb;2(2):196–204. doi: 10.1101/gad.2.2.196. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Steitz J. A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E. Cyclic 2',3'-phosphates and nontemplated nucleotides at the 3' end of spliceosomal U6 small nuclear RNA's. Science. 1992 Jan 17;255(5042):327–330. doi: 10.1126/science.1549778. [DOI] [PubMed] [Google Scholar]
- Madore S. J., Wieben E. D., Kunkel G. R., Pederson T. Precursors of U4 small nuclear RNA. J Cell Biol. 1984 Sep;99(3):1140–1144. doi: 10.1083/jcb.99.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madore S. J., Wieben E. D., Pederson T. Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA-protein complexes. J Biol Chem. 1984 Feb 10;259(3):1929–1933. [PubMed] [Google Scholar]
- Madore S. J., Wieben E. D., Pederson T. Intracellular site of U1 small nuclear RNA processing and ribonucleoprotein assembly. J Cell Biol. 1984 Jan;98(1):188–192. doi: 10.1083/jcb.98.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F. Histone 3' ends: essential and regulatory functions. Gene Expr. 1992;2(2):93–97. [PMC free article] [PubMed] [Google Scholar]
- Maser R. L., Calvet J. P. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5' external transcribed spacer of pre-ribosomal-RNA. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6523–6527. doi: 10.1073/pnas.86.17.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters D. S., Fabrizio P., Abelson J. In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev. 1989 Dec;3(12B):2124–2136. doi: 10.1101/gad.3.12b.2124. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman de Vegvar H. E., Dahlberg J. E. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol. 1990 Jul;10(7):3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J. R., Pederson T. The Mr 70,000 protein of the U1 small nuclear ribonucleoprotein particle binds to the 5' stem-loop of U1 RNA and interacts with Sm domain proteins. Proc Natl Acad Sci U S A. 1988 Feb;85(3):747–751. doi: 10.1073/pnas.85.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Chromatin structure and the cell cycle. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2224–2228. doi: 10.1073/pnas.69.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penman S. RNA metabolism in the HeLa cell nucleus. J Mol Biol. 1966 May;17(1):117–130. doi: 10.1016/s0022-2836(66)80098-0. [DOI] [PubMed] [Google Scholar]
- Preiser P. R., Levinger L. F. Drosophila 5 S RNA processing requires the 1-118 base pair and additional sequence proximal to the processing site. J Biol Chem. 1991 Dec 15;266(35):23602–23605. [PubMed] [Google Scholar]
- Reddy R., Henning D., Das G., Harless M., Wright D. The capped U6 small nuclear RNA is transcribed by RNA polymerase III. J Biol Chem. 1987 Jan 5;262(1):75–81. [PubMed] [Google Scholar]
- Rohleder A., Wieben E. Solid-phase processing of U2 snRNA precursors. Biochemistry. 1986 Oct 7;25(20):5910–5914. doi: 10.1021/bi00368a011. [DOI] [PubMed] [Google Scholar]
- Roy S., Parkin N. T., Rosen C., Itovitch J., Sonenberg N. Structural requirements for trans activation of human immunodeficiency virus type 1 long terminal repeat-directed gene expression by tat: importance of base pairing, loop sequence, and bulges in the tat-responsive sequence. J Virol. 1990 Mar;64(3):1402–1406. doi: 10.1128/jvi.64.3.1402-1406.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster E. O., Guthrie C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988 Oct 7;55(1):41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- Simmons D. T., Loeber G., Tegtmeyer P. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J Virol. 1990 May;64(5):1973–1983. doi: 10.1128/jvi.64.5.1973-1983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T., Watanabe-Nagasu N., Okada N., Ohshima Y. Molecular cloning and characterization of a gene for rat U2 small nuclear RNA. J Mol Biol. 1983 Aug 15;168(3):579–594. doi: 10.1016/s0022-2836(83)80303-9. [DOI] [PubMed] [Google Scholar]
- Van Arsdell S. W., Weiner A. M. Human genes for U2 small nuclear RNA are tandemly repeated. Mol Cell Biol. 1984 Mar;4(3):492–499. doi: 10.1128/mcb.4.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Keller W. The biochemistry of 3'-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- Westin G., Lund E., Murphy J. T., Pettersson U., Dahlberg J. E. Human U2 and U1 RNA genes use similar transcription signals. EMBO J. 1984 Dec 20;3(13):3295–3301. doi: 10.1002/j.1460-2075.1984.tb02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G., Zabielski J., Hammarström K., Monstein H. J., Bark C., Pettersson U. Clustered genes for human U2 RNA. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3811–3815. doi: 10.1073/pnas.81.12.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieben E. D., Madore S. J., Pederson T. Protein binding sites are conserved in U1 small nuclear RNA from insects and mammals. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1217–1220. doi: 10.1073/pnas.80.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieben E. D., Nenninger J. M., Pederson T. Ribonucleoprotein organization of eukaryotic RNA. XXXII. U2 small nuclear RNA precursors and their accurate 3' processing in vitro as ribonucleoprotein particles. J Mol Biol. 1985 May 5;183(1):69–78. doi: 10.1016/0022-2836(85)90281-5. [DOI] [PubMed] [Google Scholar]
- Wu J. A., Manley J. L. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991 Aug 29;352(6338):818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Yang H., Moss M. L., Lund E., Dahlberg J. E. Nuclear processing of the 3'-terminal nucleotides of pre-U1 RNA in Xenopus laevis oocytes. Mol Cell Biol. 1992 Apr;12(4):1553–1560. doi: 10.1128/mcb.12.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo C. Y., Ares M., Jr, Weiner A. M. Sequences required for 3' end formation of human U2 small nuclear RNA. Cell. 1985 Aug;42(1):193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- Zavanelli M. I., Ares M., Jr Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev. 1991 Dec;5(12B):2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]