Abstract
Epigenetic mechanisms mediating expression of the Runt-related transcription factor Runx2 are critical for controlling its osteogenic activity during skeletal development. Here, we characterized bona fide regulatory elements within 120 kbp of the endogenous bone-related Runx2 promoter (P1) in osteoblasts by genomic DNase I footprinting and chromatin immunoprecipitations (ChIPs). We identified a ~10 kbp genomic domain spanning the P1 promoter that interacts with acetylated histones H3 and H4 reflecting an open chromatin conformation in MC3T3 osteoblasts. This large chromatin domain contains a single major DNaseI hypersensitive (DHS) region that defines a 0.4 kbp “basal core” promoter. This region encompasses two endogenous genomic protein/DNA interaction sites (i.e., footprints at Activating Protein 1 [AP1], E-box and Runx motifs). Helix-Loop-Helix (HLH)/E-box occupancy and presence of the DHS region persists in several mesenchymal cell types, but AP1 site occupancy occurs only during S phase when Runx2 expression is minimal. Point-mutation of the HLH/E box dramatically reduces basal promoter activity. Our results indicate that the Runx2 P1 promoter utilizes two stable principal protein/DNA interaction domains associated with AP1 and HLH factors. These sites function together with dynamic and developmentally responsive sites in a major DHS region to support epigenetic control of bone-specific transcription when osteoblasts transition into a quiescent or differentiated state.
Runt-related transcription factors are principal developmental regulators that control lineage commitment and cell type-specificity in diverse species. Null mutations in each of the three known runt-related transcription factor (Runx) genes in mouse causes dramatic tissue-specific phenotypes. Mutations and/or deregulation of the corresponding human genes are linked to familial diseases and genetic predispositions related to cancer and distinct abnormalities in tissue-formation (Blyth et al., 2005; Ito, 2008). While Runx proteins may functionally compensate each other, the unique phenotypes of mice with Runx null mutations are attributable to tissue- and developmental stage-specific activation of transcription. The Runx2 gene is prominently transcribed in the mesenchymal lineage to support normal development of bone and cartilage in vivo which accounts for the observed skeletal phenotypes of mice with Runx2 mutations that render mesenchymal cells Runx2 deficient (Komori et al., 1997; Otto et al., 1997; Choi et al., 2001; Lengner et al., 2002; Jeong et al., 2008; Lou et al., 2009; Zhang et al., 2009a; Liu et al., 2011).
Runx2 transcription, as the initial rate-limiting step in its expression, is exquisitely regulated by a multitude of developmental signals, regulatory promoter elements and cognate transcription factors (Lian et al., 2006; Franceschi et al., 2007; Marie, 2008; Long, 2012). The Runx2 gene is expressed from two promoters (P1 and P2), and the upstream P1 promoter supports osteoblast-specific gene transcription. Of note, polymorphisms in the P1 and P2 promoter have been correlated with bone mineral density and viral integration sites are capable of ectopic activation of Runx2 gene transcription in non-osseous cell types (Stewart et al., 2002; Doecke et al., 2006; Lee et al., 2009). The P1 promoter is autoregulated through at least seven Runx binding sites (Drissi et al., 2000, 2002b) and responds to steroid hormones (Tou et al., 2001; Drissi et al., 2002a), BMPs (Xiao et al., 2001; Tou et al., 2003; Lee et al., 2005) and WNTs (Gaur et al., 2005). The P1 promoter is controlled by several homeodomain proteins including AP1 (Drissi et al., 2002a), Dlx5 (Gaur et al., 2005), Nkx3.2 (Lengner et al., 2005), HoxA10 (Hassan et al., 2007), SP1 and Ets proteins (Zhang et al., 2009b), Hif2α (Tamiya et al., 2008), C/EBPβ (Wiper-Bergeron et al., 2007; Henriquez et al., 2011) and NF-1 related proteins (Zambotti et al., 2002). Regulation of Runx2 gene transcription by this cohort of primary DNA binding proteins occurs within the context of nucleosomal organization and higher order chromatin structure that together modulate accessibility of transcription factors to gene promoters.
The Runx2 gene, which is in throughout the osteogenic lineage, and the osteocalcin (OC) gene, which is transcriptionally activated at the maturation stage of osteoblast differentiation, together represent two versatile and intensively studied models for understanding transcriptional control during osteogenesis (Lian et al., 2004; Montecino et al., 2008). Activation of OC gene expression occurs concomitant with creation of nuclease hypersensitive sites, increased acetylation of histone H3 and H4, as well as specific binding of multiple transcription factors including Runx2 (Javed et al., 1999; Shen et al., 2002, 2003; Hassan et al., 2004). Studies using rat osteosarcoma cells and trans-differentiated mouse myoblasts have revealed SWI/SNF dependent changes in the chromatin organization of the Runx2 P1 promoter (Cruzat et al., 2009).
In this study, we established key parameters of chromatin fine-structure of the Runx2 gene promoter in mouse osteoblasts in which the Runx2 P1 promoter is naturally activated. We define histone modifications, nuclease hypersensitive sites and two genomic protein/DNA interactions that mediate transcriptional regulation of Runx2 gene expression. Our key finding is that the Runx2 P1 promoter contains two stable genomic protein-DNA interaction domains that may transcriptionally control the multiple physiological activities of Runx2 during skeletal development and bone formation in vivo.
Materials and Methods
Cell culture and synchronization
MC3T3-E1 osteoblasts were maintained in αMEM medium supplemented with 10% fetal bovine serum (FBS). C2C12 myoblasts, NIH3T3 fibroblasts, mouse embryonic fibroblasts (MEFs) and mouse Runx2 null calvarial cells were cultured as described previously (Pratap et al., 2003; Galindo et al., 2005; Lengner et al., 2005). MC3T3-E1 cells were synchronized by growth arrest through serum deprivation and released from quiescence by serum stimulation. For these experiments, cells were initially seeded in 100-mm plates at a density of 0.4 × 106 cells/plate, and grown in a subconfluent state for 72 h until the onset of exponential growth. Cells were washed three times in PBS and then cultured in serum-free medium for 48 h. Non-viable detached cells were discarded and the remaining adherent cells were stimulated to progress through the cell cycle by the administration of αMEM plus 10% FBS. After serum stimulation, cells were harvested at selected time points for nuclear run-on analysis, Northern blot analysis and DNase I digestion. Cell cycle synchrony was monitored by analyzing H4 gene expression using semi-quantitative PCR. PCR reactions were performed in a total volume of 50 μl using the Titan One tube RT-PCR system (Roche) according to the instructions of the manufacturer. Primers used for PCR analysis were: H4/N forward: 5′ TAT CGG GCT CCA GCG GTC ATG TC′ and H4/N reverse: 5′ GGA TCG AAA CGT AAG CTG GAG; GAPDH forward: 5′ TCA CCA TCT TCC AGG AGC G and GADPDH reverse: 5′ CTG CTT CAC CAC CTT CTT GA.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation analysis was performed according to the protocol provided in the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY). Histones were cross-linked to DNA by adding formaldehyde directly to culture medium to a final concentration of 1% and incubated at room temperature for 10 min. Cells were then washed twice with ice-cold PBS and resuspended in 200 μl of lysis buffer (150 mM NaCl, 50 mM Tris–HCl/pH 8.0, 1% NP-40) containing Complete protease inhibitors (Roche). Cells were incubated on ice for 20 min and sonicated for 15 sec eight times at an empirically determined power setting. Cell lysates were transferred to Eppendorf tubes and subjected to micro-centrifugation (14,000 rpm for 15 min at 4°C). Sonicated lysates were then diluted 10-fold with ChIP dilution buffer.
After preclearance by centrifugation (4,000 rpm for 3 min), 2 ml of supernatant was transferred to a new tube and incubated with 8 μl antibody overnight at 4°C on a rotating wheel (at 30 rpm). Normal rabbit serum (Santa Cruz Biotechnology Inc., Santa Cruz, CA) was used as a negative control. The following antibodies were purchased from Upstate Biotechnology and recognize acetylated histones: tetra-acetylated histone H4 (“H4Ac-α1,” catalog # 06-866), penta-acetylated histone H4 (“H4Ac-α2,” catalog # 06-946), and acetylated histone H3 (“H3Ac,” catalog # 06-599). Chromatin immuno-complexes were mixed with 60 μl of protein A agarose beads (50% slurry) followed by incubation for 2 h at 4°C with gentle rotation. Beads were collected by micro-centrifugation at 3,000 rpm for 2 min at 4°C. The bead pellets were sequentially washed with 500 μl each of the following buffers: low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–Cl/pH 8.1, 150 mM NaCl), high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–Cl pH/8.1, 500 mM NaCl), and LiCl wash buffer (0.25 mM LiCl, 1% Nonidet-P40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris–Cl pH 8.1). The beads were then washed three times using 500 μl of TE buffer (Tris–HCl/pH 8.0, 1 mM EDTA/pH 8.0). The immuno-complexes were eluted with two consecutive 200 μl aliquots of freshly prepared elution buffer (1% SDS, 100 mM NaHCO3). Each aliquot was incubated with beads for 10 min with fast rotation (80 rpm) followed by 2 min microfugation at 3,000 rpm after each elution. Formaldehyde cross-links were reversed by adding 40 μl of 3M sodium acetate (pH 5.2) and incubating each sample at 65°C overnight. DNA was recovered by phenol/chloroform extractions, precipitated using ethanol and dissolved in 400 μl TE buffer. Aliquots of 5 μl DNA solution were used for quantitative PCR (ABI real time PCR machine) using SYBR Green dye fluorescence (BioRad, Hercules, CA). The specificity of the PCR products was demonstrated by the presence of one peak. To quantitate DNA amplification based on the threshold cycle number, a standard curve for each primer-pair and PCR set was generated from serial twofold dilutions of mouse genomic DNA. The slope of the standard curve for each test reaction was −3.3 ± 0.4. Multiple primer pairs were used to amplify regions across the Runx2 gene locus (see Supplementary Fig. S1).
Nuclease hypersensitivity assays using indirect end-labeling
Nuclei were isolated from 3 × 107 cells by Dounce homogenization (loose pestle) in 40 ml RSB buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2) with 0.5% (vol/vol) NP-40 and protease inhibitors (Complete, Roche). Nuclei were washed three times with RSB buffer and cell lysis was evaluated by staining with 0.4% Trypan blue 1:1 (vol/vol). DNaseI hypersensitivity at the Runx2 gene locus was analyzed as previously described (Hovhannisyan et al., 2003; Lou et al., 2009). In brief, nuclear pellets were resuspended in 3 ml RSB buffer without NP-40 and the DNA concentration was estimated by absorption at 260 nm. Alequots of 20 A260 units were digested with increasing concentrations of DNase I (0–10 U; Worthington Biochemicals, Freehold, NJ) in a 400 μl final volume of RSB buffer containing 1 mM CaCI2 for 5 min at room temperature. The reaction was stopped by adding 400 μl Stop buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 50 mM EDTA, 0.3% SDS) and incubated with 10 units of RNase ONE ribonuclease (Promega, Madison, WI) at 37°C for 1 h. The samples were extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1). Nucleic acids were precipitated with 0.7 volumes of isopropanol, washed with 70% ethanol and resuspended in 1×TE buffer.
DNaseI hypersensitive regions were defined by indirect end labeling using PCR amplified probes in the Runx2 promoter region. The purified DNA (15 μg) was subjected to a secondary digestion by PstI or AflI, then electrophoresed in 1.3% agarose gels in 1×TAE buffer, and transferred to Hybond N+ membrane (Amersham Pharmacia Biotech, Arlington Heights, IL). The specific DNA sequences were detected by hybridization with random-primed or PCR-labeled radioactive probes. Hybridization were performed at 65°C, using 5× SSC, 0.7% SDS and 5× Denhardt solution, followed by in stringent buffer conditions. The blots were analyzed by autoradiography or by using a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Genomic DNaseI footprinting using ligation-mediated (LM) PCR
LM-PCR was carried out essentially by the classical method of Mueller et al. (1997) with some modifications. Initial extension and amplification/labeling was with Vent DNA polymerase (New England Biolabs, Beverly, MA). Linker ligation was carried out with three units of T4 DNA ligase in its optimized buffer (Promega). 5′ primer labeling with γ32P ATP was carried out by T4 polynucleotide kinase (New England Biolabs). The primers used were: LM1 (−96 to −78) 5′-GAC ATG ACC GCT GGA GCC CGA TA-3′; LM8 (−31 to −7) 5′-GCT GGA GCC CGA TAG ACA GCT TTC TG-3′; LM9 (−21 to +6) 5′-GCT GGA GCC CGA TAG ACA GCT TCT GTC A-3′. The initial extension reaction was initiated with 4 μg of DNA, but after linker ligation and precipitation only half of this amount was used for the amplification step. Annealing temperatures used were LM7 =64°C; LM8 = 66°C, and LM9 =68°C. Amplification was for 20 cycles. PCR products were labeled by primer extension using 32P-labeled nested primers for two cycles, extracted by phenol/chloroform, precipitated by ethanol and analyzed on 6% denaturing polyacrylamide gels.
Electrophoretic mobility shift analysis (EMSA)
EMSAs were performed with wild type and mutant oligonucleotides that span a segment in the Runx2 P1 promoter that contains HLH and Runx2 consensus sites (Supplementary Fig. S2). Sense strand oligonucleotides were each end-labeled by incubation at a final concentration of 10 μM in 10 μl with 50 μCi of γ-P32-ATP the presence of 10 units of T4 polynucleotide kinase (New England Biolabs) for 30 min at 37°C. Samples were then boiled for 10 min in the presence of excess EDTA (10 mM final concentration) to inactivate the kinase. Unlabelled anti-sense strand oligonucleotides (10 μl of a 30 μM DNA solution) was then added to each reaction followed by boiling for 10 min to denature DNA strands. The reaction was then gradually allowed to cool to room temperature to allow annealing of sense and antisense strands. Double stranded oligos were then purified over a G-25 Sephadex column to remove unincorporated nucleotides.
DNA binding assays were performed by incubating 10 fmoles (50 nM) of radio-labeled double stranded oligonucleotide (~80,000 cpm) with 5 μg of nuclear proteins from MC3T3 cells and 1 μg of poly(dI-dC)*poly(dI-dC) DNA. Nuclear protein preparations were prepared from MC3T3-E1 cells that were cultured to 90% confluence and harvested in ice-cold PBS buffer by scraping. Protein concentrations were determined by the Bradford method (Pierce Chemical Co., Rockford, IL). To visualize specifically binding of HLH proteins to the labeled probe, binding of Runx2 to an overlapping motif in the probe was suppressed in some experiments by inclusion of a 100-fold excess of unlabeled Runx2 consensus oligonucleotide (see Supplementary Fig. S2). Complexes were visualized after separation in a 4.5% 40:1 acrylamide:bis-acrylamide gel in the presence of 0.5× TBE buffer followed by autoradiography.
Results
Molecular mechanisms that control the activity of the Runx2 gene promoter have previously been examined using transgenic or reporter gene knock-in mice, transiently transfected promoter/reporter gene constructs, chromatin immune-precipitation (ChIP) assays and in vitro protein/DNA interactions with cell lysates. These studies have revealed an intricate spectrum of important regulatory transactions at the two promoters (P1 and P2) of the Runx2 gene that control its developmental and tissue-specific expression (Fig. 1). Here, we are complementing these findings by establishing a novel model for protein/DNA interactions at the bone-related Runx2 P1 promoter that integrates data on the prevalence of histone modifications characteristic of open chromatin (e.g., H3 and H4 acetylation), the location of nuclease hypersensitive sites and the actual location of key protein/DNA interaction events as detected by genomic DNaseI footprinting.
Fig 1.
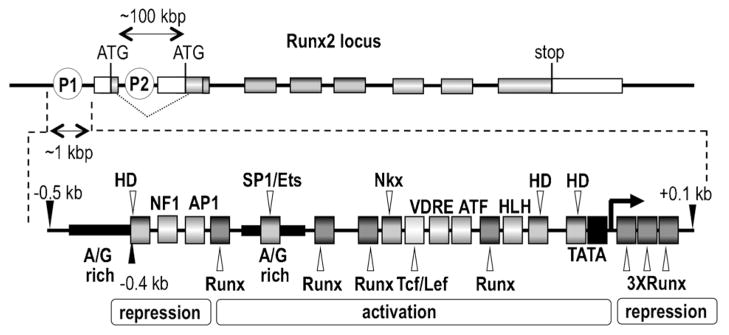
Genomic organization of the Runx2 locus. The top portion shows the two promoters (P1 and P2) of the Runx2 gene that generate distinct mRNAs encoding proteins with different N-termini due to alternative splicing and different translational start codons. The P1 mRNA encodes the bone-related p57/TypeII/MASNS Runx2 protein isoform and the P2 mRNA encodes the p56/TypeI/MRIPV Runx2 protein isoform. The diagram shows only the relative location of structural and regulatory elements in the Runx2 locus and is not drawn to scale. (e.g., distance between the P1 and P2 promoters is around 100 kbp, while the break-out of the P1 promoter spans only about 1 kbp). The bottom portion shows the relative location of selected regulatory DNA motifs and cognate transcription factors that have been identified in previous studies (Drissi et al., 2000, 2002a,b; Gaur et al., 2005; Lee et al., 2005; Lengner et al., 2005; Hassan et al., 2007; Zhang et al., 2009b).
The P1 promoter of the Runx2 locus selectively associates with acetylated histones H3 and H4 in mesenchymal cells
We examined the long-range chromatin organization of the Runx2 locus in mesenchymal cells using ChIP-qPCR assays with antibodies recognizing acetylated histones H3 or H4 and primers that amplify a series of segments across a ~100 kbp region of the Runx2 gene (−45 to +77 kbp relative to the upstream bone-related P1 promoter; Fig. 2). In MC3T3 osteoblasts that actively express Runx2 P1 mRNA, highest levels of acetylated H3 and H4 histones are present between −4 and +8 kbp of the P1 promoter (Fig. 2). Sequences between +65 and +77 kpb are also associated with acetylated histones H3 and H4 (data not shown). Histone acetylation also occurs within the same region of the P1 promoter in NIH3T3 fibroblasts and C2C12 myoblasts, albeit that the overall enrichment of this modification is lower in non-osteoblastic cell types (data not shown and Supplementary Figs. S3 and S4). The enrichment of acetylated histones H3 and H4 in the Runx2 P1 gene promoter supports the active expression of Runx2 P1 mRNAs in osteoblasts and defines a broad ~10–12 kbp chromatin region that participates in transcriptional control.
Fig 2.
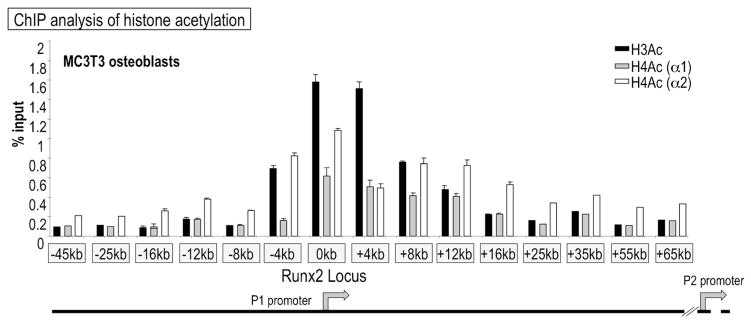
The Runx2 P1 promoter associates with acetylated histones H3 and H4 in osteoblasts. The graph presents data from chromatin immunoprecipitation analysis with antibodies recognizing acetylated histones H3 and H4. Genomic primers amplify the P1 promoter and the first intron of the Runx2 locus in MC3T3 osteoblasts. The antibodies recognize multiple acetylated forms of histones H3 and H4 (i.e., antibodies are not lysine specific) that generally mark “open” chromatin.
Nuclease hypersensitivity of the “basal core” P1 promoter of Runx2
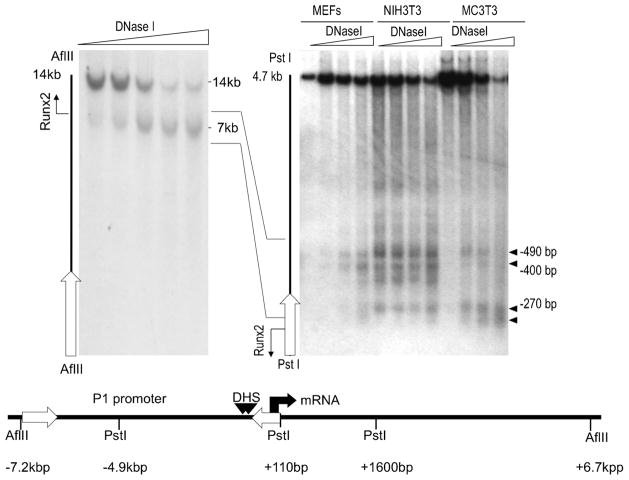
We examined whether increased acetylation of histones H3 and H4 at the Runx2 P1 promoter correlates with increased changes in chromatin accessibility as reflected by DNaseI nuclease sensitivity in MC3T3 osteoblasts and other mesenchymal cell types (Fig. 3). We observed one major nuclease hypersensitive region which is located near the transcriptional start site of the P1 promoter (Fig. 3). At higher resolution, this hypersensitive region can be resolved into several distinct nuclease hypersensitive sites using indirect end-labeling with probes closer to the transcription start site (Fig. 3). The nuclease hypersensitive sites are located within a 0.4 kbp region of the Runx2 P1 promoter which is associated with increased levels of acetylated histones H3 and H4 (see Fig. 2). Because this 0.4 kbp encompasses all key regulatory elements that have been identified to date (see Fig. 1), these data define a “basal core” promoter for bone-specific expression of Runx2.
Fig 3.
The initial 0.4 kbp of the Runx2 P1 promoter exhibits DNaseI hypersensitivity. DNaseI hypersensitive sites in the Runx2 promoter were detected by indirect end-labeling of chromatin fragments. Chromatin was digested in situ using empirically determined amounts of DNaseI (sloped triangle). Purified DNA was digested with the indicated restriction enzymes and subjected to Southern blot analysis with radio-labeled DNA probes (open arrows) that hybridize with one end of each restriction fragment. Restriction digestion will yield a full length DNA fragment that resisted DNaseI digestion (e.g., AflII/14 kbp, PstI/4.7 kbp), as well as one or more sub-fragments that were pre-cleaved with DNaseI in situ (closed horizontal triangles). The single 7 kbp band observed for the AflII digestion indicates that there is only one DNaseI hypersensitive region between −7.2 and +6.7 kbp. This region resolves into several distinct DNaseI sub-sites within the first 0.4 kbp upstream from the transcriptional start site of the P1 promoter (right part, PstI digestion). Similar results were obtained with different mesenchymal cell types (e.g., fibroblasts and osteoblasts).
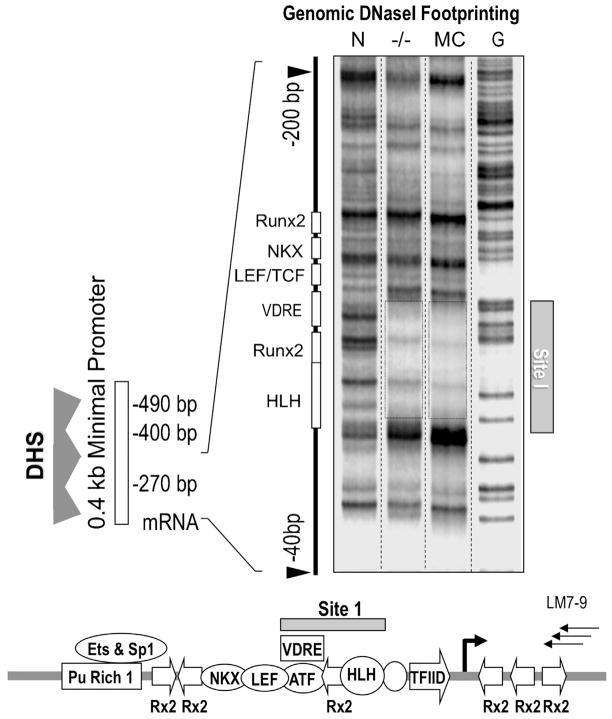
Two major sites of in vivo protein/DNA interaction at the P1 promoter
The functional organization of the Runx2 P1 “basal core” promoter was further analyzed for transcription factor occupancy at single nucleotide resolution using DNaseI protection assays combined with ligation-mediated PCR to detect genomic DNaseI footprints in intact cells. We initially used primers that are optimized for the examination of protein/DNA interactions on the anti-sense strand within the first 0.0 to − 0.3 kbp relative to the transcriptional start site of P1 mRNAs. These primers detect one major nuclease protected region (DNaseI footprint: Site 1; −93/−65; Fig. 4 and Supplementary Fig. S5). This genomic protein/DNA interaction domain exhibits occupancy in lineage-committed MC3T3 osteoblasts that actively transcribe Runx2, as well as in Runx2 null osteoprogenitor cells that are not yet committed to the osteoblastic phenotype (Fig. 4). Using primers that amplify distal sequences of the Runx2 P1 promoter (−0.6 to −0.3 kbp), we detect a second genomic footprint between nt −417 to −377 (Site 2) on both the sense and anti-sense strand in MC3T3 cells (Fig. 5 and Supplementary Fig. S5). Sites 1 and 2 represent the only genomic DNaseI footprints that were routinely detected in several mesenchymal cell types (Figs. 4 and 5), indicating that many functional protein/DNA interactions at the Runx2 promoter (see Fig. 1) are not sufficiently stable to be detected by DNaseI protection analysis.
Fig 4.
Identification of Site 1 in the Runx2P1 promoter by genomic DNaseI protection analysis. LM-PCR mediated DNaseI footprinting of intact nuclei reveals a stable genomic protein/DNA interaction domain, Site 1, in the proximal region of the P1 promoter in Runx2 null calvarial cells (−/−) and MC3T3 osteoblasts (MC). The lane on the left shows the DNaseI digestion pattern of naked DNA (N) and the right lane shows a ladder of DNA cleaved at G residues. The diagram at the bottom shows the location of Site 1 in relation to known transcription factor motifs in the Runx2 P1 promoter and the three nested LM-PCR primers used for the analysis (LM7-9).
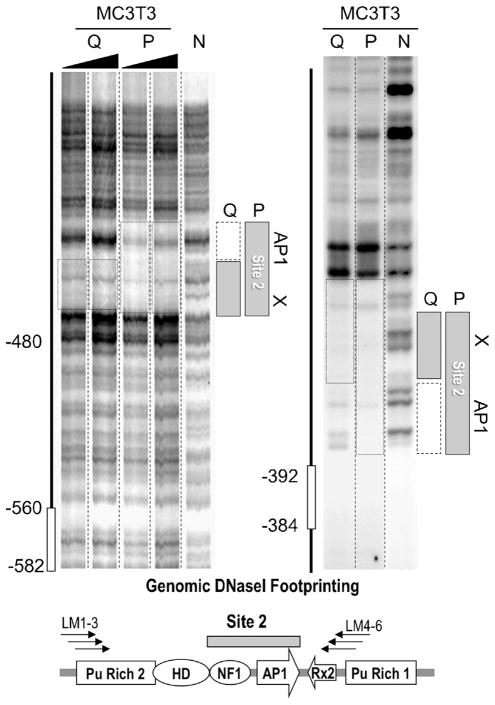
Fig 5.
Proliferation-related genomic protein/DNA interactions at Site 2 in the Runx2 P1 promoter. Genomic LM-PCR assisted DNaseI protection analysis indicates that there is a second stable genomic protein/DNA interaction domain, Site 2, in the distal region of the Runx2 P1 promoter. The size of the DNaseI footprint in quiescent (Q) MC3T3 osteoblasts is reduced compared to proliferating (P) cells, indicating selective loss of protein/DNA interactions. The left and right parts show DNaseI digestion patterns for, respectively, the sense-strand (using primers LM1-3) and the anti-sense strand (using primers LM4-6). The left part shows two replicates each for Q and P samples. The left lane in both left and right parts shows the DNaseI cleavage patterns of naked DNA (N). The diagram at the bottom shows relevant transcription factor consensus motifs.
The general region of the P1 promoter (nt −124/+1) that encompasses Site 1 is remarkably conserved among vertebrate Runx1, Runx2, and Runx3 genes (data not shown). This observation suggests that the first 0.1 kbp around the TATA-box and transcriptional start sites of the different P1 promoters of Runx genes in general are key component of a “basal core” promoter. Interestingly, sequences of Site 1 itself (−93/−65) are uniquely well-conserved among vertebrate Runx2 genes (data not shown). The Runx2-specific phylogenetic conservation of Site 1 sequences suggests that this in vivo genomic protein/DNA interaction domain may have evolved to meet physiological demands for Runx2 gene transcription during skeletal development.
Selective modifications in genomic occupancy of the P1 promoter during the cessation of osteoblast proliferation
We addressed whether changes in Runx2 gene transcription rates are accompanied by modifications in genomic protein/DNA interactions. In quiescent MC3T3 osteoblasts, endogenous Runx2 gene transcription is clearly detectable by nuclear run-on analysis, but occurs at a low basal rate in actively proliferating MC3T3 cells (Supplementary Fig. S6); for comparison, histone H4 gene transcription is enhanced in proliferating cells, while GAPDH gene transcription does not appreciably change. Occupancy at Sites 1 and 2 is observed in actively proliferating MC3T3 osteoblasts, but protein/DNA interactions at Site 2 are selectively modified in serum-deprived quiescent cells (Fig. 5), while occupancy at Site 1 is unaltered in either proliferating or quiescent cells (Supplementary Fig. S7).
Site 2 contains a well-established binding site for transcription factor AP1 (Drissi et al., 2002a; Franceschi et al., 2007) and a consensus motif for a DNA binding activity related to Nuclear Factor 1 (NF1; Zambotti et al., 2002; Marie, 2008). In quiescent cells, the interactions at the AP1 element are selectively lost while binding events at the adjacent NF1-like motif are retained (Fig. 5). The AP1 site is located within a promoter region that attenuates Runx2 gene transcription (see Fig. 1), indicating that the cognate AP1 protein may have repressive properties. Loss of these AP1 related interactions in quiescent cells correlates with increased transcription of the Runx2 gene (Supplementary Fig. S6). Thus, our data suggest that enhanced Runx2 P1 transcription in quiescent cells may ensue, because a repressive AP1 protein vacates its recognition motif in Site 2.
Site 1 interacts with a novel Helix-Loop-Helix transcription factor
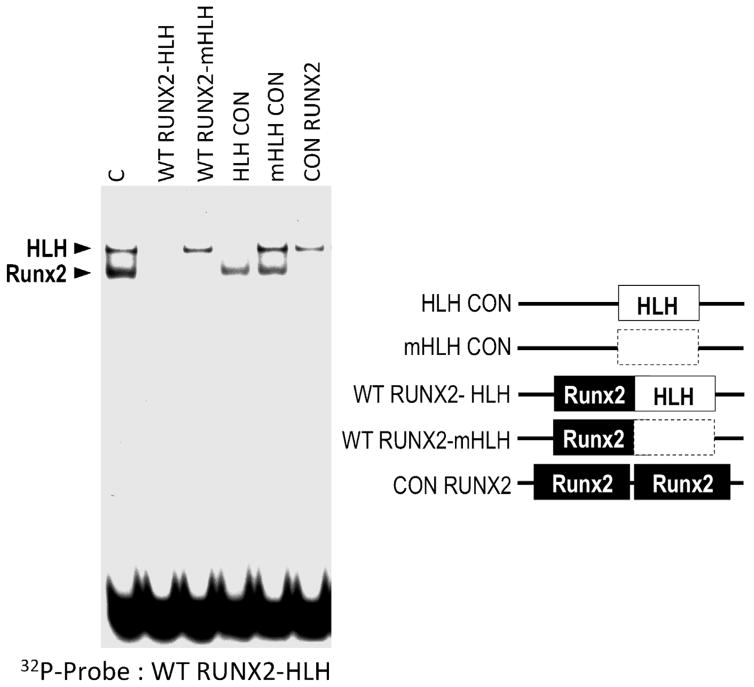
The distal part of Site 1 encompasses a (1,25)-dihydroxyvitamin D3 responsive element (VDRE; 5′AGTACT GTG AGGTCA), which binds the cognate steroid hormone receptor, and an overlapping motif for constitutive ATF-related proteins (5′TGAGGTCA; Supplementary Fig. S5). Mutation of this composite element abolishes (1,25)-dihydroxyvitamin D3 enhanced transcription, but has marginal effects on basal promoter activity (Drissi et al., 2002a; Franceschi et al., 2007). The proximal part of Site 1 contains an established recognition motif for Runx2 and a c-Myc/c-Max-related E-box consensus motif that may bind a Helix-Loop-Helix (HLH) related factor (Drissi et al., 2000; Marie, 2008). We examined in vitro protein/DNA interactions at Site 1 using EMSAs with osteoblast-derived nuclear proteins and oligonucleotides containing mutations in either the Runx2 or HLH motif in the −82/−56 promoter region (Fig. 6 and Supplementary Fig. S2). The radio-labeled −82/−56 probe detects two protein/DNA complexes that selectively compete with the unlabeled competitor oligonucleotides. The Runx2 and HLH complexes competed with all DNA fragments containing their respective wild type but not mutated recognition motifs. Strikingly, the HLH complex also competes with a related wild type HLH consensus element, but not the corresponding mutant HLH sequence (Fig. 6). Furthermore, the HLH complex on the Runx2 promoter selectively cross-competes with an HLH complex on the osteocalcin promoter (data not shown). Taken together, the data establish that the proximal part of Site 1 binds both Runx2 and HLH proteins.
Fig 6.
Definition of a Helix-Loop-Helix/E-box related recognition motif in Site I. Protein/DNA interactions at Site 1 were examined in vitro by EMSAs using a radio-labeled DNA binding site probe (WTRUNX2-HLH) spanning both a Runx2 and a HLH consensus motif and a panel of unlabeled competitor oligonucleotides (see Supplementary Fig. S2). Control binding reaction with nuclear proteins from MC3T3 cells generates two complexes that self-compete with the unlabeled WT RUNX2-HLH oligonucleotide. The upper complex also competes with oligonucleotides containing wildtype (HLH CON), but not mutant HLH (mHLH CON and WTRUNX2-mHLH) consensus motifs, and is mediated by an HLH/E-box related DNA binding activity. The lower complex competes with oligonucleotides containing Runx2 binding motifs.
The HLH motif in Site 1 is critical for basal transcription of Runx2 P1 gene transcription
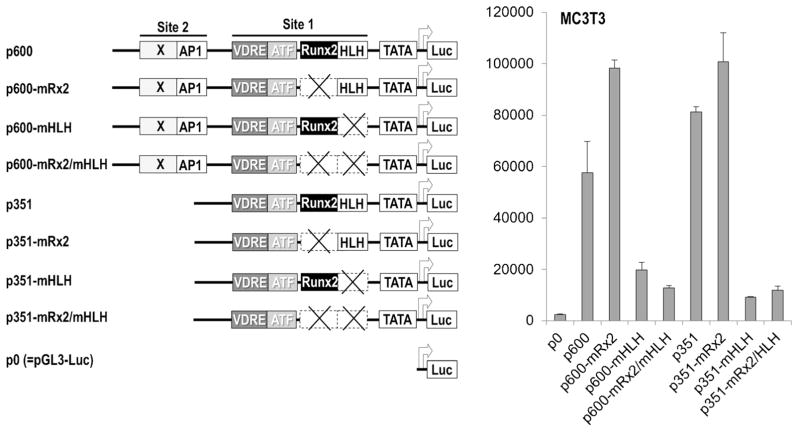
The functional contributions of the Runx2 and HLH motif in Site 1 to Runx2 P1 promoter activity were assessed using Luciferase reporter assays (Fig. 7 and Supplementary Fig. S8). Deletion of the promoter region containing Site 1 enhances Luc reporter activity by up to twofold in MC3T3 osteoblasts, NIH3T3 fibroblasts and ROS17/2.8 osteosarcoma cells (comparing p600-Luc and p351-Luc), consistent with transcriptional repression mediated by Site 1. Mutation of the Runx2 motif in Site 1 also increases Luc reporter activity by up to twofold for either the 600 or 351 bp promoters, corroborating the auto-suppression of Runx2 P1 gene transcription that we previously established (Drissi et al., 2000). In contrast, mutation of the HLH motif in MC3T3 osteoblasts results in a striking reduction of Runx2 promoter activity, independent of the length of the promoter or mutation of the adjacent Runx2 motif (Fig. 7). Very similar results were obtained with HLH mutations examined in NIH3T3 fibroblasts and ROS17/2.8 osteosarcoma cells (Supplementary Fig. S8). We conclude that the HLH motif is essential for basal transcription of the Runx2 P1 gene promoter.
Fig 7.
The HLH motifin Site 1 is essential for basal Runx2 P1 promoter activity. Expression of Luciferase (Luc) reporter genes was examined upon transient transfection in MC3T3 osteoblasts. Reporter genes were fused to two distinct Runx2 P1 promoter segments (p600, nt −600/−16; p351, nt −351/−16) and compared with a promoter-less construct (p0). The contribution of Runx2 and HLH motifs was determined by examining reporter genes with the respective binding site mutations in both the p600 and p351 constructs.
Discussion
The biological effects of Runx2 during skeletal development and dosage-insufficiency of Runx2 that causes craniofacial abnormalities are both directly related to the bone tissue-specific expression of Runx2. In this study, we have identified a ~10 kbp region of the Runx2 locus that preferentially associates with acetylated histones H3 and H4 as detected by real-time qPCR data using a large panel of primers. The presence of these chromatin marks is consistent with recent ChIP-Seq data obtained in MC3T3 cells and normal bone marrow stromal cells showing that the P1 promoter has a transcriptionally active histone code signature (unpublished observations). The Runx2 P1 promoter contains a 0.4 kbp DNaseI hypersensitive “basal core” regulatory region containing two genomic protein/DNA interaction domains, Sites 1 and 2. Sites 1 and 2 each modulate Runx2 P1 promoter activity, but they have functionally distinct roles. Site 2, which contains an AP1 motif that is vacated in quiescent osteoblasts, appears to suppress Runx2 gene transcription in immature proliferating osteoblasts. Site 1, which contains a HLH motif, is essential for establishing basal Runx2 P1 promoter activity.
The proximal part of Site 1 interacts with a novel HLH protein that is very important for basal Runx2 P1 gene transcription, because mutation of this HLH site yields the strongest quantitative effect on P1 promoter activity. A similar HLH protein binds to the osteocalcin promoter, which shares many transcription factors with the Runx2 P1 promoter, suggesting it is a key protein with an important coordinating regulatory function. We have therefore tested a large panel of candidate HLH proteins with known functions in skeletal development for effects on Runx2 gene transcription (e.g., Twist1, Twist2, c-Myc). However, none of these factors were rate-limiting in transient transfection assays (Zhang et al., 2008, 2009c). The identity of the relevant HLH protein will need to await DNA affinity purification.
One main finding of this study is that there are only two clearly detectable DNaseI footprints in the 0.4 kbp “basal core” promoter, even though there are many functionally validated Runx2 promoter binding proteins. Nevertheless, a number of known functional protein/DNA interactions cannot be detected by DNaseI protection analysis. Known recognition motifs could remain undetected due to the instability of protein/DNA interactions relative to the kinetics of nuclease digestion. Binding events can also be obscured by partitioning on redundant recognition motifs within the same promoter (e.g., Runx2) or on allelic promoters. Also, biological heterogeneity within cell populations can generate different binding states at individual promoters. Detection of Sites 1 and 2 on the Runx2 P1 promoter indicates that these genomic protein binding domains are very robust and present in the majority of cells.
The AP1 element in the proximal portion of the genomic DNaseI footprint at Site 2 binds heterodimers related to the c-Fos/c-Jun family (Drissi et al., 2000; Wiper-Bergeron et al., 2007). The current work and our prior studies revealed that mutation or deletion of the AP-1 site enhances basal Runx2 gene transcription, albeit that these effects are quantitatively modest (<2-fold). This AP1 site will bind in vitro whichever c-Fos/c-Jun related protein is most prevalent in nuclear extracts prepared from cells at a given stage of osteoblast differentiation (McCabe et al., 1995; Drissi et al., 2002a). Transfection experiments indicate that AP1 can activate or suppress bone-specific promoters depending on the specific c-Fos/c-Jun combination (McCabe et al., 1995). The latter findings suggest that the AP1 site in the Runx2 P1 promoter may be similarly bifunctional at different developmental stages. EMSA assays established that AP1/DNA interactions in the Runx2 P1 promoter are responsive to (1,25)-dihydroxyvitamin D3 (Drissi et al., 2002a), providing a plausible mechanism for switching the function of AP1 depending on physiological bone anabolic requirements.
Site 2 is similar to the CE2 domain that was identified by other investigators (Zambotti et al., 2002) using in vitro DNaseI protection assays using crude nuclear protein extracts from NIH3T3 fibroblasts and 3T3L1 pre-adipocytes. These investigators proposed that the proximal AP1 site interacts with FosB to stimulate Runx2 P1 promoter activity, and that the distal NF1 motif binds a unique NF1-specific isoform (NF1-A) that suppresses transcription in non-osteoblastic cells. These interpretations are somewhat incongruent with our observations that this NF1 motif remains stably occupied in quiescent osteoblasts, that genomic occupancy of the AP1 site correlates with transcriptional suppression, and that the NF1 and AP1 sites are rather promiscuous and interact with multiple isoforms in many cell types. Regardless of interpretative differences, the identity of Site 1/CE1 using either in vitro assays with nuclear extracts (Zambotti et al., 2002) or genomic occupancy with intact nuclei (this work), provides strong experimental evidence for the functional importance of Site 2.
Site 1 is a complex multi-partite protein/DNA interaction site that encompasses a negative (1,25)-dihydroxyvitamin D3 responsive element (VDRE) that overlaps with an ATF motif, as well as a Runx2 motif (one of seven empirically validated Runx2 sites in the P1 promoter) and an HLH consensus motif (Drissi et al., 2000, 2002b). As with the AP1 site in Site 2, mutation of the VDRE/ATF element in Site 1 has only marginal effects on basal Runx2 P1 gene transcription (Drissi et al., 2002a), but abolishes suppression by (1,25)-dihydroxyvitamin D3. Mutation of the Runx2 motif increases transcription by twofold, suggesting that this Runx2 motif is important for auto-regulation of Runx2 gene transcription (Drissi et al., 2000). Taken together, at least three elements contained within Sites 1 and 2 are directly or indirectly linked to (1,25)-dihydroxyvitamin D3 signaling. Compounding twofold transcriptional effects could theoretically amount up to one order of magnitude (~8 fold), while even modest percentage changes in Runx2 levels may cause dosage insufficiency and craniofacial abnormalities in mice that resemble those of cleidocranial dysplasia. We conclude that transcriptional control of Runx2 is intricate and dynamic, and that genomic Sites 1 and 2 represent critical basal core components that support induction of Runx2 gene transcription in osteoblasts.
Acknowledgments
This work was funded by NIH grant R01 AR039588 to GSS and JBL, R01 AR049069 to AvW, and R37 DE012528 to JBL. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Additional supporting information may be found in the online version of this article.
Literature Cited
- Blyth K, Cameron ER, Neil JC. The runx genes: Gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Choi J-Y, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein GS. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruzat F, Henriquez B, Villagra A, Hepp M, Lian JB, van Wijnen AJ, Stein JL, Imbalzano AN, Stein GS, Montecino MA. SWI/SNF-independent nuclease hypersensitivity and increased histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry. 2009;48:7287–7295. doi: 10.1021/bi9004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doecke JD, Day CJ, Stephens AS, Carter SL, van DA, Kotowicz MA, Nicholson GC, Morrison NA. Association of functionally different RUNX2 P2 promoter alleles with BMD. J Bone Miner Res. 2006;21:265–273. doi: 10.1359/JBMR.051013. [DOI] [PubMed] [Google Scholar]
- Drissi H, Luc Q, Shakoori R, Chuva de Sousa Lopes S, Choi J-Y, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Drissi H, Pouliot A, Koolloos C, Stein JL, Lian JB, Stein GS, van Wijnen AJ. 1,25(OH)2Vitamin D3 suppresses the bone-related Runx2/Cbfa1 gene promoter. Exp Cell Res. 2002a;274:323–333. doi: 10.1006/excr.2002.5474. [DOI] [PubMed] [Google Scholar]
- Drissi H, Pouliot A, Stein JL, van Wijnen AJ, Stein GS, Lian JB. Identification of novel protein/DNA interactions within the promoter of the bone-related transcription factor Runx2/Cbfa1. J Cell Biochem. 2002b;86:403–412. doi: 10.1002/jcb.10238. [DOI] [PubMed] [Google Scholar]
- Franceschi RT, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Ann N Y Acad Sci. 2007;1116:196–207. doi: 10.1196/annals.1402.081. [DOI] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1 related anti-proliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PVN, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating RUNX2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: Temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez B, Hepp M, Merino P, Sepulveda H, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. C/EBPbeta binds the P1 promoter of the Runx2 gene and up-regulates Runx2 transcription in osteoblastic cells. J Cell Physiol. 2011;226:3043–3052. doi: 10.1002/jcp.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan H, Cho B, Mitra P, Montecino M, Stein GS, van Wijnen AJ, Stein JL. Maintenance of open chromatin and selective genomic occupancy at the cell-cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Mol Cell Biol. 2003;23:1460–1469. doi: 10.1128/MCB.23.4.1460-1469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. RUNX genes in development and cancer: Regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- Javed A, Gutierrez S, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D responsive transcription and contribute to chromatin organization. Mol Cell Biol. 1999;19:7491–7500. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JH, Jin JS, Kim HN, Kang SM, Liu JC, Lengner CJ, Otto F, Mundlos S, Stein JL, van Wijnen AJ, Lian JB, Stein GS, Choi JY. Expression of Runx2 transcription factor in non-skeletal tissues, sperm and brain. J Cell Physiol. 2008;217:511–517. doi: 10.1002/jcp.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao Y-H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Lee MH, Kim YJ, Yoon WJ, Kim JI, Kim BG, Hwang YS, Wozney JM, Chi XZ, Bae SC, Choi KY, Cho JY, Choi JY, Ryoo HM. Dlx5 specifically regulates Runx2-II expression by binding to homeodomain response elements in the Runx2 distal promoter. J Biol Chem. 2005;280:35579–35587. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Koh JM, Hwang JY, Choi KY, Lee SH, Park EK, Kim TH, Han BG, Kim GS, Kim SY, Lee JY. Association of a RUNX2 promoter polymorphism with bone mineral density in postmenopausal Korean women. Calcif Tissue Int. 2009;84:439–445. doi: 10.1007/s00223-009-9246-6. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Drissi H, Choi J-Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Activation of the bone related Runx2/Cbfa1 promoter in mesenchymal condensations and developing chondrocytes of the axial skeleton. Mech Dev. 2002;114:167–170. doi: 10.1016/s0925-4773(02)00050-3. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Hassan MQ, Serra RW, Lepper C, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Nkx3.2 mediated repression of RUNX2 promotes chondrogenic differentiation. J Biol Chem. 2005;280:15872–15879. doi: 10.1074/jbc.M411144200. [DOI] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: Role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- Liu JC, Lengner CJ, Gaur T, Lou Y, Hussain S, Jones MD, Borodic B, Colby JL, Steinman HA, van Wijnen AJ, Stein JL, Jones SN, Stein GS, Lian JB. Runx2 protein expression utilizes the Runx2 P1 promoter to establish osteoprogenitor cell number for normal bone formation. J Biol Chem. 2011;286:30057–30070. doi: 10.1074/jbc.M111.241505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F. Building strong bones: Molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- Lou Y, Javed A, Hussain S, Colby J, Frederick D, Pratap J, Xie R, Gaur T, van Wijnen AJ, Jones SN, Stein GS, Lian JB, Stein JL. A Runx2 threshold for the cleidocranial dysplasia phenotype. Hum Mol Genet. 2009;18:556–568. doi: 10.1093/hmg/ddn383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- McCabe LR, Kockx M, Lian J, Stein J, Stein G. Selective expression of fos- and jun-related genes during osteoblast proliferation and differentiation. Exp Cell Res. 1995;218:255–262. doi: 10.1006/excr.1995.1154. [DOI] [PubMed] [Google Scholar]
- Montecino M, Stein GS, Stein JL, Lian JB, van Wijnen AJ, Carvallo L, Marcellini S, Cruzat F, Arriagada G. Vitamin D control of gene expression: Temporal and spatial parameters for organization of the regulatory machinery. Crit Rev Eukaryot Gene Expr. 2008;18:163–172. doi: 10.1615/critreveukargeneexpr.v18.i2.50. [DOI] [PubMed] [Google Scholar]
- Mueller P, Garrity P, Wold B. Ligation-mediated PCR for genomic sequencing and footprinting. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, editors. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 1997. pp. 15.5.1–15.5.26. [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, Beddington RSP, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi J-Y, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cell growth regulatory role of Runx2 during proliferative expansion of pre-osteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- Shen J, Montecino MA, Lian JB, Stein GS, van Wijnen AJ, Stein JL. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D dependent, bone tissue-specific transcription. J Biol Chem. 2002;277:20284–20292. doi: 10.1074/jbc.M112440200. [DOI] [PubMed] [Google Scholar]
- Shen J, Hovhannisyan H, Lian JB, Montecino MA, Stein GS, Stein JL, van Wijnen AJ. Transcriptional induction of the osteocalcin gene during osteoblast differentiation involves acetylation of histones h3 and h4. Mol Endocrinol. 2003;17:743–756. doi: 10.1210/me.2002-0122. [DOI] [PubMed] [Google Scholar]
- Stewart M, Mackay N, Cameron ER, Neil JC. The common retroviral insertion locus Dsi1 maps 30 kilobases upstream of the P1 promoter of the murine Runx3/Cbfa3/Aml2 gene. J Virol. 2002;76:4364–4369. doi: 10.1128/JVI.76.9.4364-4369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiya H, Ikeda T, Jeong JH, Saito T, Yano F, Jung YK, Ohba S, Kawaguchi H, Chung UI, Choi JY. Analysis of the Runx2 promoter in osseous and non-osseous cells and identification of HIF2A as a potent transcription activator. Gene. 2008;416:53–60. doi: 10.1016/j.gene.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Tou L, Quibria N, Alexander JM. Regulation of human cbfa1 gene transcription in osteoblasts by selective estrogen receptor modulators (SERMs) Mol Cell Endocrinol. 2001;183:71–79. doi: 10.1016/s0303-7207(01)00594-9. [DOI] [PubMed] [Google Scholar]
- Tou L, Quibria N, Alexander JM. Transcriptional regulation of the human Runx2/Cbfa1 gene promoter by bone morphogenetic protein-7. Mol Cell Endocrinol. 2003;205:121–129. doi: 10.1016/s0303-7207(03)00151-5. [DOI] [PubMed] [Google Scholar]
- Wiper-Bergeron N, St-Louis C, Lee JM. CCAAT/Enhancer binding protein beta abrogates retinoic acid-induced osteoblast differentiation via repression of Runx2 transcription. Mol Endocrinol. 2007;21:2124–2135. doi: 10.1210/me.2006-0452. [DOI] [PubMed] [Google Scholar]
- Xiao ZS, Liu SG, Hinson TK, Quarles LD. Characterization of the upstream mouse Cbfa1/Runx2 promoter. J Cell Biochem. 2001;82:647–659. doi: 10.1002/jcb.1192. [DOI] [PubMed] [Google Scholar]
- Zambotti A, Makhluf H, Shen J, Ducy P. Characterization of an osteoblast-specific enhancer element in the CBFA1 gene. J Biol Chem. 2002;277:41497–41506. doi: 10.1074/jbc.M204271200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hassan MQ, Li ZY, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Intricate gene regulatory networks of Helix-Loop-Helix (HLH) proteins support regulation of bone-tissue related genes during osteoblast differentiation. J Cell Biochem. 2008;105:487–496. doi: 10.1002/jcb.21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xiao Z, Luo J, He N, Mahlios J, Quarles LD. Dose-dependent effects of Runx2 on bone development. J Bone Miner Res. 2009a;24:1889–1904. doi: 10.1359/JBMR.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hassan MQ, Xie RL, Hawse JR, Spelsberg TC, Montecino M, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Co-stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats) J Biol Chem. 2009b;284:3125–3135. doi: 10.1074/jbc.M807466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The Notch-responsive transcription factor Hes-1 attenuates osteocalcin promoter activity in osteoblastic cells. J Cell Biochem. 2009c;108:651–659. doi: 10.1002/jcb.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]