Abstract
Botulinum neurotoxins (BoNTs), produced by Clostridium botulinum, are a group of seven (A–G) immunologically distinct proteins and cause the paralytic disease botulism. These toxins are the most poisonous substances known to humans and are potential bioweapon agents. Therefore, it is necessary to develop highly sensitive assays for the detection of BoNTs in both clinical and environmental samples. In the current study, we have developed an enzyme-linked immunosorbent assay (ELISA)-based protein antibody microarray for the sensitive and simultaneous detection of BoNT serotypes A, B, C, D, E, and F. With engineered high-affinity antibodies, the BoNT assays have sensitivities in buffer ranging from 1.3 fM (0.2 pg/ml) to 14.7 fM (2.2 pg/ml). Using clinical and food matrices (serum and milk), the microarray is capable of detecting BoNT serotypes A to F to similar levels as in standard buffer. Cross-reactivity between assays for individual serotype was also analyzed. These simultaneous, rapid, and sensitive assays have the potential to measure botulinum toxins in a high-throughput manner in complex clinical, food, and environmental samples.
Keywords: Botulinum neurotoxin, Biodefense, ELISA, Protein microarray, Antibody, High-throughput assays
Botulinum neurotoxin (BoNT),1 produced by the bacterium Clostridium botulinum, is the most toxic substance known with an extremely low LD50 of approximately 1 ng/kg [1]. BoNT poisoning results in botulism, a deadly paralytic disease that typically occurs through the ingestion of contaminated food, infection of an open wound, or colonization of the bacteria in the gastrointestinal tract of infants [2–4]. In addition, BoNT poses a critical threat to public security due to its potent toxicity and ease of production, resulting in its listing by the Centers for Disease Control and Prevention as one of the six highest risk threat agents for bioterrorism (a class A agent) [2].
Seven immunologically distinct BoNT serotypes (A–G) have been identified, each of which is approximately 150 kDa with an N-terminal 50-kDa light chain (Lc) and a C-terminal 100-kDa heavy chain (Hc) [5]. Following exposure, BoNTs are endocytosed into presynaptic nerve cells, where the BoNT Lc functions as a zinc-dependent endoprotease and specifically cleaves SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins, leading to inhibition of acetylcholine release and resultant neuroparalysis [6].
BoNT poisoning can result in flaccid paralysis, respiratory failure, or death [7]. Even patients who survive botulism require prolonged hospitalization with mechanical ventilation, making early and accurate diagnosis of exposure to BoNT critical to the administration of appropriate and effective medical care [2,3]. Human botulism is most commonly associated with exposures to BoNT serotypes A, B, and E. BoNT/F is less often implicated in human botulism, and BoNT/G has never been associated with human botulism. Although serotypes C and D have rarely been associated with human botulism, they are typically responsible for botulism in animals such as cattle [8–11]. However, BoNT/C has been shown to induce paralysis as long-lasting as that caused by BoNT/A [12], making it a potential bioweapon threat. The standard method to detect BoNT is the mouse bioassay, or lethality test, which is highly sensitive, with a limit of detection (LOD) of 10 to 20 pg/ml (67–133 fM) for BoNT/A [2,13–15]. However, this assay is cumbersome, low-throughput, and time-consuming (1–4 days) and involves the use of large numbers of animals. Furthermore, a subsequent neutralization bioassay is required in order to confirm toxin presence and differentiate toxin serotype. These limitations highlight the increasing need to replace this assay. Recently, several alternative assays have been developed with equal or better sensitivity, including various immunoassays [14,16–26] and assays that quantify BoNT protease activity [27–33]. However, these assays were designed to detect only one or a limited number of the toxin serotypes.
Enzyme-linked immunosorbent assays (ELISAs) are exceptionally sensitive and specific methods for quantitating the concentration of proteins in complex biological fluids such as blood. As such, ELISAs are routinely used for the analysis of single-protein concentrations; however, they require large sample volumes for the detection of multiple proteins. In contrast, ELISA-based protein microarrays allow for the simultaneous analysis of up to 50 proteins in a small sample volume (~20 μl) within a single experiment, making them especially suited to high-throughput analyses [34,35]. Based on a “sandwich” ELISA antibody microarray that we described previously for the sensitive detection of BoNT/A [21], we applied high-affinity BoNT antibodies to protein microarray technology and developed sensitive assays for the simultaneous detection of BoNT serotypes A, B, C, D, E, and F. We demonstrate that the developed assays are highly sensitive in buffer, milk, and serum matrices. In addition, the microarray is simple and has the potential to analyze large numbers of clinical, food, or environmental samples for the sensitive and simultaneous detection of BoNTs.
Materials and methods
Antigens, antibodies, and other reagents
All BoNT pure holotoxins were purchased from Metabiologics (Madison, WI, USA). It should be noted that subtype A1 was used for BoNT/A and that the Metabiologics BoNT/D is a D–C mosaic toxin. Supplemental Table 1 in the supplementary material lists the antibodies tested in developing the microarray. Antibodies were purchased from Gallus Immunotech (Cary, NC, USA), R&D Systems (Minneapolis, MN, USA), List Biological Laboratories (Campbell, CA, USA), Metabiologics, Abcam (Cambridge, MA, USA), Meridian Life Science (Memphis, TN, USA), and HyTest (Turku, Finland) or were obtained using yeast display technologies for molecular evolution and selection of antibodies in the laboratory of James D. Marks at the University of California, San Francisco [36–40]. To produce the high-affinity antibodies, human immunoglobulin G (IgG) monoclonal antibodies (mAbs) were first isolated from a single-chain variable fragment (scFv) library displayed on yeast that was generated from either a human volunteer or mice immunized with pentavalent BoNT toxoid. Higher affinity antibodies were generated from the lead antibodies using molecular evolution or affinity maturation techniques [38,39]. Once a selected scFv was affinity matured, it was converted to a full-length IgG as described previously [39]. The epitopes that the engineered antibodies bind have been defined [36,38,41–43] and are indicated in Supplemental Table 1.
Antibodies used for detection were biotinylated using the EZ-Link NHS–LC–LC Biotin Kit from Pierce (Rockford, IL, USA) following the manufacturer’s protocol. The biotinyltyramide was obtained from PerkinElmer (Waltham, MA, USA). The horseradish peroxidase (HRP)–streptavidin and cyanine dye 3 (Cy3)–streptavidin conjugates were obtained from Jackson ImmunoResearch (West Grove, PA, USA). Milk was 2% milk fat and was purchased locally. Human blood serum was obtained from a healthy anonymous female donor (Golden West Biologicals, Temecula, CA, USA). Aminopropylsilane-coated glass slides stamped with a hydrophobic barrier to generate 16 wells (2 × 8) on each single slide were purchased from Thermo Scientific (Portsmouth, NH, USA).
Production of antibody microarrays
Antibody microarrays were generated as described previously [21]. Briefly, antibodies used for capture were adjusted to concentrations of 0.8 to 1.0 mg/ml in phosphate-buffered saline (PBS, pH 7.2) and printed on the aminopropylsilane-coated slides using either a Microgrid II robot (Genomic Solutions, Ann Arbor, MI, USA) or a GeSiM NanoPlotter 2.1 (Quantum Analytics, Foster City, CA, USA). Antibodies to each toxin serotype were printed in each of the 16 wells on each slide to make identical arrays, with each antibody printed as five replicate spots in a consecutive row. Spots were printed approximately 400 μm apart and with approximately 1 nl of antibody per spot. Quality of the printing was evaluated by red reflect scanning using ScanArray ExpressHT (PerkinElmer). Printed slides were incubated in a humid chamber for at least 2 to 3 h, and a hydrophobic barrier pen was used on the stamped barrier to enhance the hydrophobicity. The slides were air-dried for 1 h and then blocked using 1% casein in PBS (Bio-Rad, Hercules, CA, USA) for 1 h and stored dry at 4 or −20 °C.
ELISA microarray assays
ELISA microarray assays were processed as described previously [21,44]. Stored microarray slides were brought to room temperature and rinsed with PBS-T buffer (PBS containing 0.05% Tween 20). In general, all toxins were diluted using 0.1% casein solution in PBS, and milk and serum samples were diluted 1:4 in 0.1% casein/PBS solution. Here 20 μl of diluted antigens (toxins) or detection antibodies was used for incubation with each well of the microarray. Diluted antigen was added to microarray wells and gently mixed with an orbital shaker in a humid sealed chamber for 12 to 16 h. Three washes were performed after each incubation step with PBS-T. Following the wash step, the slides were incubated with the corresponding detection antibody mix with gentle shaking for 2 h. The signal was amplified using the biotinyltyramide amplification system [45,46]. Briefly, the slides were incubated with 1 μg/ml HRP-conjugated streptavidin for 30 min, followed by incubation with biotinyltyramide for 10 min; both incubations were done without shaking. Finally, the slides were incubated for 1 h with 1 μg/ml Cy3-conjugated streptavidin with gentle mixing in the dark, followed by a quick rinse with ultrapure water and then drying. A ScanArray 3000 (PerkinElmer) or an LS Reloaded microarray scanner (Tecan, Männedorf, Switzerland) was used for detection of Cy3 fluorescence.
Data analysis
Signals from the ELISA microarray assays were quantified by ScanArray Express software (PerkinElmer). The mean signal values were calculated from five replicates and were background-corrected. Standard curves were generated using in-house statistics software ProMAT (Protein Microarray Analysis Tool) [47]. The LOD in buffer, milk, and serum was calculated as the concentration of analyte necessary to generate a signal greater than 3 times the standard deviation above the background signal. Recoveries were calculated by dividing the expected concentration (i.e., actual concentration of antigen spiked into matrix) by the calculated concentration.
Results and discussion
Development of antibody microarray for sensitive detections of BoNTs
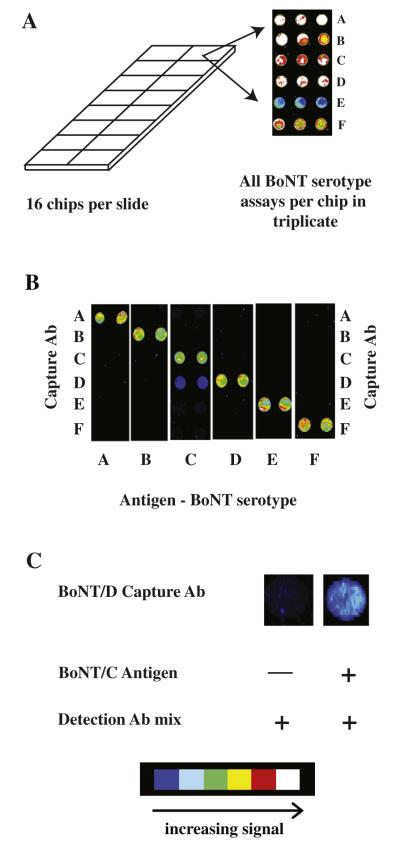
A schematic illustration of the sandwich antibody microarray that is based on our previous microarray assay of BoNT/A [21] is described in Fig. 1. This approach uses a sensitive enzymatic signal enhancement based on tyramide signal amplification in which HRP catalyzes the conversion of tyramide to reactive free radicals that bind covalently to nearby proteins, creating multiple localized biotin sites for signal amplification. Because the availability of high-affinity antibodies is a key factor in developing highly sensitive and selective immunoassays, including antibody microarrays, we screened a number of anti-BoNT antibodies from multiple sources, including commercially available and high-affinity monoclonal antibodies produced by the Marks laboratory of the University of California, San Francisco. The Marks antibodies have been employed for the detection of BoNT/A and BoNT/E in several assay formats, including protein microarrays and activity-based assays [16,21,26,48–51]. Among the 57 antibodies screened, 19 are commercial antibodies and 38 are from the Marks laboratory. Detailed information regarding antibodies used is listed in Supplemental Table 1.
Fig.1.
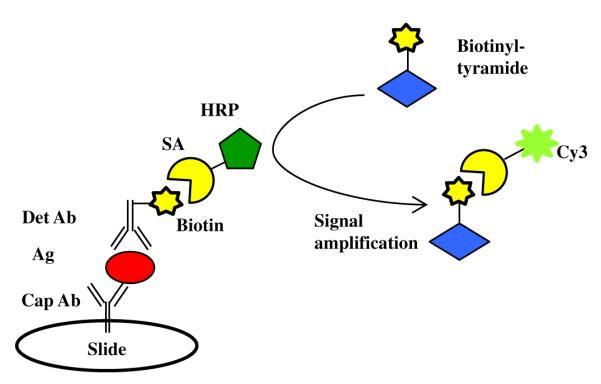
Schematic illustration of ELISA antibody microarray assay with biotinyltyramide amplification. The target antigen is captured via a surface-immobilized antibody (capture antibody). A second antibody (detection antibody) labeled with biotin, specific for a different epitope on the same antigen, is used for detection. Cap Ab, capture antibody; Ag, antigen; Det Ab, detection antibody; SA, streptavidin; HRP, horseradish peroxidase; Cy3, cyanine dye 3.
To screen the large number of antibodies for potential antibody combinations (capture and detection antibodies), smaller serotype-specific arrays were initially developed and used. For instance, to determine the optimal pair of BoNT/A antibodies, a BoNT/A-specific array was generated consisting of all possible BoNT/A capture antibodies in a single chip. A BoNT-specific array was made for each serotype, and then each specific array was incubated with corresponding holotoxin in a 4-fold dilution series ranging from 5000 down to 1.2 pg/ml and probed with all possible relevant detection antibodies. As expected, antibodies with high binding affinities typically performed better in the microarray (data not shown).
The optimal antibody pairs for the combined BoNTs/A to F microarray consisted of the following: BoNT/A, AR4 (capture) and RAZ1 (detection); BoNT/B, B2 (capture) and 1B10.1 (detection); BoNT/C, C1 (capture) and 1C1 (detection); BoNT/D, 8DC1 (capture) and 8DC2 (detection); BoNT/E, 3E6.2 (capture) and 3E4.1 (detection); and BoNT/F, 6F8 (capture) and 6F5 (detection). Among the antibody pairs, some pairs recognize nonoverlapping epitopes within the same domain, such as AR4 and RAZ1, both of which recognize the BoNT/A heavy chain (Supplemental Table 1). Other pairs recognize epitopes in different domains of BoNTs such as the 6F8 and 6F5 pair that interact with the BoNT/F heavy chain and translocation domain, respectively.
The combined BoNTs/A to F microarray was optimized for improved signal-to-noise ratio for each array by varying the concentration of detection antibody. To perform these tests, an antigen concentration was chosen within the upper third of the linear range of the standard curve and detection antibody concentrations of 6.25, 12.5, 25, 50, and 100 ng/ml were tested. The signal-to-noise ratios were found to be maximized at detection antibody concentrations of 25 ng/ml (RAZ1, 1C1, and 8DC2) and 50 ng/ml (1B10.1, 3E4.1, and 6F5) (Fig. 2).
Fig.2.
Optimization of detection antibody concentrations. Signal-to-noise ratios for each BoNT serotype over a range of detection antibody concentrations are shown. Values are shown as the percentage of the maximal signal-to-noise ratio for each assay.
Analyses of potential cross-reactivity between assays
Immunoassays can suffer from nonspecific interactions between an antibody and other components in the assay (i.e., antigen and/or another antibody). This is especially true when proteins with a high degree of similarity are assayed together such as the BoNT proteins, which have a protein sequence similarity of more than 50%. Therefore, to determine the specificity of each serotype assay, we undertook a systematic study to determine potential cross-reactivity between the assays of the BoNT antibody microarray. Similar to Gonzalez et al. [44], we tested for interactions among (i) capture antibody and antigen, (ii) capture antibody and detection antibody, and (iii) antigen and detection antibody.
The critical test indicating whether the microarray assay will be useful for identification and discrimination of BoNT serotypes with unknown suspected BoNT-contaminated sample examines potential nonspecific interactions between the capture antibody and either the antigen or the detection antibodies. These potential nonspecific interactions were analyzed by spiking individual BoNT antigens into standard casein–PBS buffer and testing on microarrays containing all six capture antibodies and using a complete mix of the detection antibodies. To perform this test, a high concentration (1250 pg/ml) of the toxin was used. The test demonstrated that the BoNT protein microarray is capable of clearly identifying and distinguishing all six BoNT serotypes (Fig. 3). There was a minor cross-reactivity (<10%) observed between the BoNT/D capture antibody and BoNT/C antigen and/or a detection antibody that does not interfere with the ability of the assay to distinguish between serotypes C and D (Fig. 3B). To determine whether BoNT/C antigen or a detection antibody is responsible for the nonspecific interaction with BoNT/D capture antibody, an assay without BoNT/C antigen and with only the mix of BoNT detection antibodies was performed. Fig. 3C clearly shows that the signal results from a cross-reaction between the BoNT/D capture antibody and BoNT/C toxin and is not due to interactions between the BoNT/D capture antibody and a BoNT detection antibody. This possibility is supported by the high degree (65%) of protein sequence similarity between BoNT/C and mosaic BoNT/D–C. It has also been found recently that the antibodies generated against BoNT/D were raised to a mosaic strain of BoNT/D–C [52]. The D–C mosaic strain consists of BoNT/D amino acid sequence in the light chain, but with amino acid sequence similar to BoNT/C in the heavy chain. For whatever reason, it is likely that the BoNT/D capture antibody recognizes a conserved epitope, likely within the heavy chain, shared between BoNT/C and BoNT/D. Nevertheless, the observed signal from the BoNT/D capture antibody and BoNT/C antigen was minor (<10% of the BoNT/C assay) and, as stated and shown (Fig. 3B), does not have an impact on the assay’s ability to distinguish the BoNT serotypes C and D.
Fig.3.
Specificity of BoNT microarray assays. (A) Schematic illustration of a slide containing 16 identical chips. The inset depicts a fluorescent scan image of a single representative chip from a slide consisting of an array printed with all six BoNT capture antibodies in triplicate. The chip was incubated with all six toxins, each at a concentration of 1250 pg/ml, and then with a complete detection antibody mix. (B) Identical BoNT microarray chips were printed with capture antibodies to all six BoNT toxins. Each chip was incubated with one toxin antigen and then with a complete detection antibody mix. (C) The left panel shows that the BoNT/C detection antibody does not bind to the BoNT/D capture antibody when BoNT/C antigen is omitted from the assay. This indicates that the nonspecific interaction, seen in the right panel, is likely caused by interactions between BoNT/C antigen with BoNT/D capture antibody. The scale at the bottom of the figure indicates increasing fluorescence intensity from blue through white. (For interpretation of the reference to color in this figure legend, the reader is referred to the Web version of this article.)
We also tested whether any of the detection antibodies nonspecifically interacts with a different antigen in the assay. This type of nonspecific interaction does not affect the accuracy or specificity of the assay because the signal intensity remains proportional to the targeted antigen concentration in both the standards and the biological samples. Therefore, this type of nonspecific assay interaction does not compromise the ability of the BoNT microarray to specify or distinguish the BoNT serotypes. The test was performed by assaying a mixture of all antigens on chips printed with all capture antibodies using single-detection antibodies. In this evaluation, the only new cross-reactivity detected was between the BoNT/F detection antibody, 6F5, and BoNT/A and BoNT/E antigens (Fig. 4). Cross-reactivity with 6F5 (also referred to as 4E17.2) was not totally unexpected because it has been shown previously that 6F5 recognizes a translocation domain that is conserved in several of the BoNT proteins. Indeed, 6F5 has been shown to bind BoNTs/A, E, and F with affinities of 1.4, 4.8, and 2.1 nM, respectively [42]. As shown in Fig. 3B, tests for nonspecific interaction between 6F5 and any of the capture antibodies did not produce detectable signal, indicating that the nonspecific interaction between 6F5 and the BoNT/A and BoNT/E assays appears to be, as expected, between the detection antibody and the toxin antigens. Despite the cross-reactivity detected between 6F5 and the BoNT/A and BoNT/E assays, 6F5 demonstrated the best sensitivity and specificity for BoNT/F detection out of at least 10 high-affinity BoNT/F antibodies tested. The 6F5 antibody is also reported to bind BoNT/B with an affinity of 27 nM [42]; however, we do not detect any cross-reactivity between BoNT/B and detection antibody 6F5. Finally, a cross-activity was also observed between the BoNT/C antigen and BoNT/D detection antibody (8DC2), and it is again likely that 8DC2 recognizes an epitope in BoNT/D that is similar with a sequence in BoNT/C, which is similar to the cross-activity observed with the BoNT/D capture antibody, 8DC1. As stated above, these nonspecific assay interactions between the detection antibody and antigen do not compromise the ability of the BoNT microarray to specify or distinguish between serotypes.
Fig.4.
Cross-reactivity of antigens and detection antibodies. Graphic illustration of all data obtained from an analysis of the six antigens assayed at a concentration of 1250 pg/ml using microarray chips that contain all six capture antibodies and probed with individual detection antibodies is shown.
A final test of the assays was undertaken in which a single antigen or detection antibody is removed from the assay mixture, an “N – 1” dropout assay [44]. If there is no interference in an assay, then the removal of antigen or detection antibody is expected to decrease the assay signal to the level of the background signal. In the first test, the antigen dropout assay, six unique antigen mixes were prepared, each with a different target antigen missing. Only one antigen dropout assay, BoNT/D, showed cross-reactivity with another assay with a cross-reactivity of less than 10% (Fig. 5, red column). This is consistent with our previous result (Fig. 3) demonstrating that the BoNT/D capture antibody interacts with BoNT/C toxin with subsequent detection by the BoNT/D detection antibody. The second test, the detection antibody dropout assay, uses a mixture of antigens and detection antibodies with one target detection antibody missing. In these tests, no additional cross-reactivities were detected beyond what was already identified in earlier experiments, including increased background signal from (i) BoNT/A when BoNT/A detection antibody (RAZ1) was dropped out, (ii) BoNT/C and BoNT/D when BoNT/C and BoNT/D detection antibodies (1C1 and 8DC2, respectively) were dropped out, and (iii) BoNT/E when BoNT/E detection antibody (3E4.1) was dropped out (Fig. 5). The increased signals for numbers (i) and (iii) were likely caused by the interactions between BoNT/F detection antibody 6F5 and BoNT/A and BoNT/E antigens, whereas number (ii) was probably caused by the cross-recognitions between BoNT/C and BoNT/D antigens and detection antibodies, as described in previous sections. Finally, no background signal was detected in chips incubated without antigen (blank) but with detection antibody confirming that there is no cross-reactivity between the capture and detection antibodies.
Fig.5.
Evaluation of cross-reactivity analyzed by systematically removing single assay reagents. The “All Ag mix” shows the signal produced when the complete antigen and detection antibody mixes are incubated to the microarray chip. The “N – 1 Ag” column shows the signal produced when only the indicated antigen for the listed assay is omitted from the antigen mix before incubating with the complete detection antibody mix. The “N – 1 detAb” column shows the signal produced when the complete antigen mix is incubated on the chip but the indicated detection antibody is omitted from the assay. Finally, the “Blank” column shows the results of chips that were incubated with blank buffer containing no antigens and then incubated with the complete detection antibody mix. (For interpretation of the reference to color in the text description of this figure, the reader is referred to the Web version of this article.)
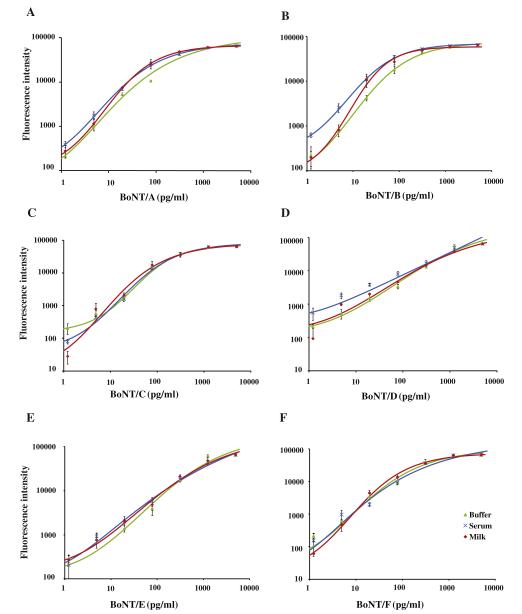
Simultaneous detection of BoNTs/A to F holotoxins in buffer, milk, and serum
The optimized assays were combined into a single BoNTs/A to F multiplexed microarray. Using BoNTs/A to F holotoxins and the optimized detection antibody concentrations, calibration curves were obtained in buffer (Fig. 6). In addition, to measure the combined BoNT microarray in more complex sample matrices, we spiked various concentrations of the BoNT holotoxins directly into milk or blood serum. The calibration curves for the six BoNT holotoxins in buffer, milk, and serum are shown in Fig. 6. A majority of the standard curves in buffer, milk, and serum had a goodness of fit R2 value of at least 0.98 (Table 1). The LODs in buffer ranged from 1.33 fM (0.2 pg/ml) for BoNT/E to 14.7 fM (2.2 pg/ml) for BoNT/C and are listed, along with the LODs in milk or serum, in Table 1. Recovery studies were performed by using individual BoNT holotoxins spiked into standard buffer, milk, and serum, respectively, to assess the accuracy of the assays. Two concentrations were tested ranging from the low end (20 pg/ml) to the high end (313 pg/ml) of the standard curve. The fluorescence signals of the spiked samples were used for concentration predication by the standard curves. Toxin recovery varied from 84% to 116% in samples spiked with the various BoNT toxins (Table 1).
Fig.6.
Standard curves for the simultaneous detection of the BoNT serotypes in buffer, milk, and serum using an ELISA protein microarray. Mixtures of BoNT serotypes A, B, C, D, E, and F were serially diluted in PBS (diamonds), serum (triangles), or milk (squares) followed by protein microarray detection. Calibration curves are shown for BoNT/A (A), BoNT/B (B), BoNT/C (C), BoNT/D (D), BoNT/E (E), and BoNT/F (F). Error bars refer to the standard deviations of five microarray spots.
Table 1.
Assay characteristics and statistics for the optimized detection of BoNT serotypes A, B, C, D, E, and F measured in buffer, milk, or serum
| Assay | Capture Ab | Detection Ab | Matrix | LOD (pg/ml) | R 2 | % Recoverya | % Recoveryb |
|---|---|---|---|---|---|---|---|
| BoNT/A | AR4 | RAZ1 | Buffer | 0.8 | 0.989 | 98.4 | 109.5 |
| Milk | 0.8 | 0.993 | 93.9 | 116.3 | |||
| Serum | 0.9 | 0.996 | 101.2 | 98.5 | |||
| BoNT/B | B2 | 1B10.1 | Buffer | 0.5 | 0.996 | 99.3 | 85.5 |
| Milk | 0.8 | 0.987 | 101.9 | 108.5 | |||
| Serum | 0.8 | 0.995 | 95.8 | 97.8 | |||
| BoNT/C | C1 | 1C1 | Buffer | 2.2 | 0.989 | 90.5 | 86.3 |
| Milk | 2.7 | 0.978 | 93.2 | 84.5 | |||
| Serum | 3.5 | 0.992 | 90.1 | 98.2 | |||
| BoNT/D | 8DC1 | 8DC2 | Buffer | 0.9 | 0.984 | 89.5 | 100.9 |
| Milk | 1.1 | 0.990 | 98.4 | 97.2 | |||
| Serum | 5.1 | 0.946 | 94.8 | 97.5 | |||
| BoNT/E | 3E6.2 | 3E4.1 | Buffer | 0.2 | 0.980 | 95.2 | 93.0 |
| Milk | 0.2 | 0.978 | 92.3 | 96.7 | |||
| Serum | 0.5 | 0.981 | 97.4 | 90.2 | |||
| BoNT/F | 6F8 | 6F5 | Buffer | 1.4 | 0.991 | 93.9 | 99.2 |
| Milk | 2.2 | 0.972 | 101.2 | 95.2 | |||
| Serum | 1.7 | 0.920 | 101.1 | 98.0 |
Percentage recovery of antigen added at a toxin spike level concentration of 313 pg/ml.
Percentage recovery of antigen added at a toxin spike level concentration of 20 pg/ml.
The detection limits reported here for the BoNT ELISA protein microarray are lower than for the mouse bioassay and among the lowest reported for assays measuring all six BoNT serotypes capable of causing toxicity in humans. Other assays for the detection of a single BoNT serotype are available with equivalent or better sensitivity compared with the mouse bioassay, including assays for the detection of BoNT/A (reviewed in Ref. [26]; see also Refs. [16,20–23,29,31,33,53]), BoNT/B [18,24,32,54], and BoNT/C [55]. Several assays sensitively measure two of the six serotypes, including a multiplexed bead-based immunoassay for BoNT/A and BoNT/B [17], a matrix-assisted laser desorption/ionization (MALDI)-based multiplex approach for BoNT/A and BoNT/B [56], and immunodetection of the substrate cleavage products of BoNT/A and BoNT/E [27]. There are a few immunoassays for the detection and differentiation of up to four of the six serotypes; however, not all of the included assays exhibit a low sensitivity [14,19,25]. Several highly sensitive assays that measure the endoprotease activity of multiple serotypes of botulinum toxin have been developed [28,30]. The BoTest assay uses fluorogenic substrate reporters that detect the proteolytic activity of BoNT serotypes A, B, D, E, F, and G. Two of the serotype assays are highly sensitive (A and E); however, the assays for serotypes B, D, F, and G are less sensitive [30], and not all serotype assays have been developed to be compatible with complex matrices [57]. The Endopep–MS assay is a mass spectrometric-based endopeptidase method for the detection and differentiation of BoNT serotypes with low sensitivities for BoNT serotypes A, B, E, and F and good compatibility with complex matrices [28].
To our knowledge, this is the first comprehensive BoNT detection assay to include BoNT serotypes C and D. The development of assays that distinguish BoNT serotypes C and D is especially difficult because of the overlapping sequencing similarities and the mosaic isoforms of these two serotypes. Type C botulism and type D botulism have been recorded in humans and are important diseases of wildlife and domestic animals [8]. Millions of waterfowl have reportedly died from botulism caused by BoNT/C [9], and both BoNT/C and BoNT/D are responsible for outbreaks of botulism in cattle and other domestic mammals and have had significant economic impacts [10,11]. Some traditional ELISAs have been developed for the detection of BoNT/C or BoNT/D, all with a sensitivity of approximately 250 pg/ml or higher [58–63]. A few ELISAs have been reported with better sensitivities but have not been tested in complex matrices [55,63,64] Therefore, the development of these highly sensitive BoNT/C and D assays fill a gap in the field of BoNT detection. The inclusion of BoNT/C and BoNT/D assays in the BoNT ELISA microarray allows for applications in veterinary medicine and may also be important for biodefense purposes.
Conclusion
ELISA-based protein antibody microarrays have been developed for the sensitive and simultaneous detection of BoNT serotypes A, B, C, D, E, and F. The assays were optimized with sensitivities better than for the standard mouse bioassay. Potential cross-reactivity was tested, and it was found that the assays could specifically detect and discriminate between individual BoNT serotypes. The multiplexed assay was evaluated in both clinical and food samples, and the sensitivity was consistent with that in buffer with femtomolar sensitivity and recoveries of more than 80%. These simultaneous and highly sensitive assays have the potential for high-throughput measurements of BoNT serotypes A, B, C, D, E, and F from a range of environmental, food, biodefense, and clinical samples. To test the potential of the BoNT protein microarray, we intend to analyze actual clinical and environmental samples.
Supplementary Material
Acknowledgments
This research was supported by the National Institute of Allergy and Infectious Diseases (NIAID) through awards U01 AI081895 (S.M.V.), U01 AI056493 (J.D.M.), U01 AI075443 (J.D.M.), and U54 AI065359 (J.D.M.). The Pacific Northwest National Laboratory (PNNL) is operated by Battelle for the U.S. Department of Energy (DOE) under contract (AC06-76RLO 1830). We thank David F. Lowry for helpful discussion concerning data analysis.
Footnotes
Abbreviations used: BoNT, botulinum neurotoxin; Lc, BoNT N-terminal 50-kDa light chain; Hc, BoNT C-terminal 100-kDa heavy chain; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; LOD, limit of detection; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; mAb, monoclonal antibody; scFv, single-chain variable fragment; HRP, horseradish peroxidase; Cy3, cyanine dye 3; PBS, phosphate-buffered saline.
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ab.2012.08.021.
References
- [1].Lamanna C. The most poisonous poison. Science. 1959;130:763–772. doi: 10.1126/science.130.3378.763. [DOI] [PubMed] [Google Scholar]
- [2].Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K. Botulinum toxin as a biological weapon: medical and public health management. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- [3].Gill DM. Bacterial toxins: a table of lethal amounts. Microbiol. Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sugiyama H. Clostridium botulinum neurotoxin. Microbiol. Rev. 1980;44:419–448. doi: 10.1128/mr.44.3.419-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].DasGupta BR, Sugiyama H. A common subunit structure in Clostridium botulinum type A, B, and E toxins. Biochem. Biophys. Res. Commun. 1972;48:108–112. doi: 10.1016/0006-291x(72)90350-6. [DOI] [PubMed] [Google Scholar]
- [6].Tonello F, Morante S, Rossetto O, Schiavo G, Montecucco C. Tetanus and botulism neurotoxins: a novel group of zinc–endopeptidases. Adv. Exp. Med. Biol. 1996;389:251–260. [PubMed] [Google Scholar]
- [7].Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- [8].Oguma K, Yokota K, Hayashi S, Takeshi K, Kumagai M, Itoh N, Tachi N, Chiba S. Infant botulism due to Clostridium botulinum type C toxin. Lancet. 1990;336:1449–1450. doi: 10.1016/0140-6736(90)93157-k. [DOI] [PubMed] [Google Scholar]
- [9].Sandler RJ, Rocke TE, Samuel MD, Yuill TM. Seasonal prevalence of Clostridium botulinum type C in sediments of a northern California wetland. J. Wildl. Dis. 1993;29:533–539. doi: 10.7589/0090-3558-29.4.533. [DOI] [PubMed] [Google Scholar]
- [10].Lindstrom M, Nevas M, Kurki J, Sauna-aho R, Latvala-Kiesila A, Polonen I, Korkeala H. Type C botulism due to toxic feed affecting 52,000 farmed foxes and minks in Finland. J. Clin. Microbiol. 2004;42:4718–4725. doi: 10.1128/JCM.42.10.4718-4725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nakamura K, Kohda T, Umeda K, Yamamoto H, Mukamoto M, Kozaki S. Characterization of the D/C mosaic neurotoxin produced by Clostridium botulinum associated with bovine botulism in Japan. Vet. Microbiol. 2010;140:147–154. doi: 10.1016/j.vetmic.2009.07.023. [DOI] [PubMed] [Google Scholar]
- [12].Eleopra R, Tugnoli V, Rossetto O, Montecucco C, De Grandis D. Botulinum neurotoxin serotype C: a novel effective botulinum toxin therapy in human. Neurosci. Lett. 1997;224:91–94. doi: 10.1016/s0304-3940(97)13448-6. [DOI] [PubMed] [Google Scholar]
- [13].Kautter DA, Solomon HM. Collaborative study of a method for the detection of Clostridium botulinum and its toxins in foods. J. Assoc. Off. Anal. Chem. 1977;60:541–545. [PubMed] [Google Scholar]
- [14].Ferreira JL. Comparison of amplified ELISA and mouse bioassay procedures for determination of botulinal toxins A, B, E, and F. J. AOAC Int. 2001;84:85–88. [PubMed] [Google Scholar]
- [15].Ferreira JL, Eliasberg SJ, Harrison MA, Edmonds P. Detection of preformed type A botulinal toxin in hash brown potatoes by using the mouse bioasssay and a modified ELISA test. J. AOAC Int. 2001;84:1460–1464. [PubMed] [Google Scholar]
- [16].Ozanich RM, Jr., Bruckner-Lea CJ, Warner MG, Miller K, Antolick KC, Marks JD, Lou J, Grate JW. Rapid multiplexed flow cytometric assay for botulinum neurotoxin detection using an automated fluidic microbead-trapping flow cell for enhanced sensitivity. Anal. Chem. 2009;81:5783–5793. doi: 10.1021/ac9006914. [DOI] [PubMed] [Google Scholar]
- [17].Pauly D, Kirchner S, Stoermann B, Schreiber T, Kaulfuss S, Schade R, Zbinden R, Avondet MA, Dorner MB, Dorner BG. Simultaneous quantification of five bacterial and plant toxins from complex matrices using a multiplexed fluorescent magnetic suspension assay. Analyst. 2009;134:2028–2039. doi: 10.1039/b911525k. [DOI] [PubMed] [Google Scholar]
- [18].Scotcher MC, Cheng LW, Stanker LH. Detection of botulinum neurotoxin serotype B at sub mouse LD50 levels by a sandwich immunoassay and its application to toxin detection in milk. PLoS One. 2010;5:e11047. doi: 10.1371/journal.pone.0011047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sharma SK, Ferreira JL, Eblen BS, Whiting RC. Detection of type A, B, E, and F Clostridium botulinum neurotoxins in foods by using an amplified enzyme-linked immunosorbent assay with digoxigenin-labeled antibodies. Appl. Environ. Microbiol. 2006;72:1231–1238. doi: 10.1128/AEM.72.2.1231-1238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stanker LH, Merrill P, Scotcher MC, Cheng LW. Development and partial characterization of high-affinity monoclonal antibodies for botulinum toxin type A and their use in analysis of milk by sandwich ELISA. J. Immunol. Methods. 2008;336:1–8. doi: 10.1016/j.jim.2008.03.003. [DOI] [PubMed] [Google Scholar]
- [21].Varnum SM, Warner MG, Dockendorff B, Anheier NC, Jr., Lou J, Marks JD, Smith LA, Feldhaus MJ, Grate JW, Bruckner-Lea CJ. Enzyme-amplified protein microarray and a fluidic renewable surface fluorescence immunoassay for botulinum neurotoxin detection using high-affinity recombinant antibodies. Anal. Chim. Acta. 2006;570:137–143. doi: 10.1016/j.aca.2006.04.047. [DOI] [PubMed] [Google Scholar]
- [22].Volland H, Lamourette P, Nevers MC, Mazuet C, Ezan E, Neuburger LM, Popoff M, Creminon C. A sensitive sandwich enzyme immunoassay for free or complexed Clostridium botulinum neurotoxin type A. J. Immunol. Methods. 2008;330:120–129. doi: 10.1016/j.jim.2007.11.006. [DOI] [PubMed] [Google Scholar]
- [23].Ahn-Yoon S, DeCory TR, Durst RA. Ganglioside–liposome immunoassay for the detection of botulinum toxin. Anal. Bioanal. Chem. 2004;378:68–75. doi: 10.1007/s00216-003-2365-4. [DOI] [PubMed] [Google Scholar]
- [24].Chiao DJ, Shyu RH, Hu CS, Chiang HY, Tang SS. Colloidal gold-based immunochromatographic assay for detection of botulinum neurotoxin type B. J. Chromatogr. B. 2004;809:37–41. doi: 10.1016/j.jchromb.2004.05.033. [DOI] [PubMed] [Google Scholar]
- [25].Rivera VR, Gamez FJ, Keener WK, White JA, Poli MA. Rapid detection of Clostridium botulinum toxins A, B, E, and F in clinical samples, selected food matrices, and buffer using paramagnetic bead-based electrochemiluminescence detection. Anal. Biochem. 2006;353:248–256. doi: 10.1016/j.ab.2006.02.030. [DOI] [PubMed] [Google Scholar]
- [26].Grate JW, Ozanich RM, Warner MG, Marks JD, Bruckner-Lea CJ. Advances in assays and analytical approaches for botulinum–toxin detection. Trends Anal. Chem. 2010;29:1137–1156. [Google Scholar]
- [27].Jones RG, Ochiai M, Liu Y, Ekong T, Sesardic D. Development of improved SNAP25 endopeptidase immuno-assays for botulinum type A and E toxins. J. Immunol. Methods. 2008;329:92–101. doi: 10.1016/j.jim.2007.09.014. [DOI] [PubMed] [Google Scholar]
- [28].Kalb SR, Moura H, Boyer AE, McWilliams LG, Pirkle JL, Barr JR. The use of Endopep–MS for the detection of botulinum toxins A, B, E, and F in serum and stool samples. Anal. Biochem. 2006;351:84–92. doi: 10.1016/j.ab.2006.01.027. [DOI] [PubMed] [Google Scholar]
- [29].Pellett S, Tepp WH, Toth SI, Johnson EA. Comparison of the primary rat spinal cord cell (RSC) assay and the mouse bioassay for botulinum neurotoxin type A potency determination. J. Pharmacol. Toxicol. Methods. 2010;61:304–310. doi: 10.1016/j.vascn.2010.01.003. [DOI] [PubMed] [Google Scholar]
- [30].Ruge DR, Dunning FM, Piazza TM, Molles BE, Adler M, Zeytin FN, Tucker WC. Detection of six serotypes of botulinum neurotoxin using fluorogenic reporters. Anal. Biochem. 2011;411:200–209. doi: 10.1016/j.ab.2011.01.002. [DOI] [PubMed] [Google Scholar]
- [31].Bagramyan K, Barash JR, Arnon SS, Kalkum M. Attomolar detection of botulinum toxin type A in complex biological matrices. PLoS One. 2008;3:e2041. doi: 10.1371/journal.pone.0002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ferracci G, Marconi S, Mazuet C, Jover E, Blanchard MP, Seagar M, Popoff M, Leveque C. A label-free biosensor assay for botulinum neurotoxin B in food and human serum. Anal. Biochem. 2011;410:281–288. doi: 10.1016/j.ab.2010.11.045. [DOI] [PubMed] [Google Scholar]
- [33].Frisk ML, Tepp WH, Johnson EA, Beebe DJ. Self-assembled peptide monolayers as a toxin sensing mechanism within arrayed microchannels. Anal. Chem. 2009;81:2760–2767. doi: 10.1021/ac802707u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zangar RC, Varnum SM, Covington CY, Smith RD. A rational approach for discovering and validating cancer markers in very small samples using mass spectrometry and ELISA microarrays. Dis. Markers. 2004;20:135–148. doi: 10.1155/2004/754640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zangar RC, Daly DS, White AM. ELISA microarray technology as a high-throughput system for cancer biomarker validation. Expert Rev. Proteomics. 2006;3:37–44. doi: 10.1586/14789450.3.1.37. [DOI] [PubMed] [Google Scholar]
- [36].Nowakowski A, Wang C, Powers DB, Amersdorfer P, Smith TJ, Montgomery VA, Sheridan R, Blake R, Smith LA, Marks JD. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc. Natl. Acad. Sci. USA. 2002;99:11346–11350. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Amersdorfer P, Wong C, Chen S, Smith T, Deshpande S, Sheridan R, Finnern R, Marks JD. Molecular characterization of murine humoral immune response to botulinum neurotoxin type A binding domain as assessed by using phage antibody libraries. Infect. Immun. 1997;65:3743–3752. doi: 10.1128/iai.65.9.3743-3752.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Razai A, Garcia-Rodriguez C, Lou J, Geren IN, Forsyth CM, Robles Y, Tsai R, Smith TJ, Smith LA, Siegel RW, Feldhaus M, Marks JD. Molecular evolution of antibody affinity for sensitive detection of botulinum neurotoxin type A. J. Mol. Biol. 2005;351:158–169. doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- [39].Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat. Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- [40].Levy R, Forsyth CM, LaPorte SL, Geren IN, Smith LA, Marks JD. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J. Mol. Biol. 2007;365:196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fischer A, Garcia-Rodriguez C, Geren I, Lou J, Marks JD, Nakagawa T, Montal M. Molecular architecture of botulinum neurotoxin E revealed by single particle electron microscopy. J. Biol. Chem. 2008;283:3997–4003. doi: 10.1074/jbc.M707917200. [DOI] [PubMed] [Google Scholar]
- [42].Garcia-Rodriguez C, Geren IN, Lou J, Conrad F, Forsyth C, Wen W, Chakraborti S, Zao H, Manzanarez G, Smith TJ, Brown J, Tepp WH, Liu N, Wijesuriya S, Tomic MT, Johnson EA, Smith LA, Marks JD. Neutralizing human monoclonal antibodies binding multiple serotypes of botulinum neurotoxin. Protein Eng. Des. Sel. 2011;24:321–331. doi: 10.1093/protein/gzq111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lou J, Geren I, Garcia-Rodriguez C, Forsyth CM, Wen W, Knopp K, Brown J, Smith T, Smith LA, Marks JD. Affinity maturation of human botulinum neurotoxin antibodies by light chain shuffling via yeast mating. Protein Eng. Des. Sel. 2010;23:311–319. doi: 10.1093/protein/gzq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gonzalez RM, Seurynck-Servoss SL, Crowley SA, Brown M, Omenn GS, Hayes DF, Zangar RC. Development and validation of sandwich ELISA microarrays with minimal assay interference. J. Proteome Res. 2008;7:2406–2414. doi: 10.1021/pr700822t. [DOI] [PubMed] [Google Scholar]
- [45].Woodbury RL, Varnum SM, Zangar RC. Elevated HGF levels in sera from breast cancer patients detected using a protein microarray ELISA. J. Proteome Res. 2002;1:233–237. doi: 10.1021/pr025506q. [DOI] [PubMed] [Google Scholar]
- [46].Varnum SM, Woodbury RL, Zangar RC. A protein microarray ELISA for screening biological fluids. Methods Mol. Biol. 2004;264:161–172. doi: 10.1385/1-59259-759-9:161. [DOI] [PubMed] [Google Scholar]
- [47].Daly DS, White AM, Varnum SM, Anderson KK, Zangar RC. Evaluating concentration estimation errors in ELISA microarray experiments. BMC Bioinformatics. 2005;6:17. doi: 10.1186/1471-2105-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kalb SR, Lou J, Garcia-Rodriguez C, Geren IN, Smith TJ, Moura H, Marks JD, Smith LA, Pirkle JL, Barr JR. Extraction and inhibition of enzymatic activity of botulinum neurotoxins/A1, /A2, and /A3 by a panel of monoclonal anti–BoNT/A antibodies. PLoS One. 2009;4:e5355. doi: 10.1371/journal.pone.0005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kalb SR, Garcia-Rodriguez C, Lou J, Baudys J, Smith TJ, Marks JD, Smith LA, Pirkle JL, Barr JR. Extraction of BoNT/A, /B, /E, and /F with a single, high affinity monoclonal antibody for detection of botulinum neurotoxin by Endopep–MS. PLoS One. 2010;5:e12237. doi: 10.1371/journal.pone.0012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Grate JW, Warner MG, Ozanich RM, Jr., Miller KD, Colburn HA, Dockendorff B, Antolick KC, Anheier NC, Jr., Lind MA, Lou J, Marks JD, Bruckner-Lea CJ. Renewable surface fluorescence sandwich immunoassay biosensor for rapid sensitive botulinum toxin detection in an automated fluidic format. Analyst. 2009;134:987–996. doi: 10.1039/b900794f. [DOI] [PubMed] [Google Scholar]
- [51].Warner MG, Grate JW, Tyler A, Ozanich RM, Miller KD, Lou J, Marks JD, Bruckner-Lea CJ. Quantum dot immunoassays in renewable surface column and 96-well plate formats for the fluorescence detection of botulinum neurotoxin using high-affinity antibodies. Biosens. Bioelectron. 2009;25:179–184. doi: 10.1016/j.bios.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Webb RP, Smith TJ, Wright PM, Montgomery VA, Meagher MM, Smith LA. Protection with recombinant Clostridium botulinum C1 and D binding domain subunit (Hc) vaccines against C and D neurotoxins. Vaccine. 2007;25:4273–4282. doi: 10.1016/j.vaccine.2007.02.081. [DOI] [PubMed] [Google Scholar]
- [53].Liu YY, Rigsby P, Sesardic D, Marks JD, Jones RG. A functional dual-coated (FDC) microtiter plate method to replace the botulinum toxin LD50 test. Anal. Biochem. 2012;425:28–35. doi: 10.1016/j.ab.2012.02.038. [DOI] [PubMed] [Google Scholar]
- [54].Wictome M, Newton K, Jameson K, Hallis B, Dunnigan P, Mackay E, Clarke S, Taylor R, Gaze J, Foster K, Shone C. Development of an in vitro bioassay for Clostridium botulinum type B neurotoxin in foods that is more sensitive than the mouse bioassay. Appl. Environ. Microbiol. 1999;65:3787–3792. doi: 10.1128/aem.65.9.3787-3792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Jones RG, Liu Y, Sesardic D. New highly specific botulinum type C1 endopeptidase immunoassays utilising SNAP25 or Syntaxin substrates. J. Immunol. Methods. 2009;343:21–27. doi: 10.1016/j.jim.2009.01.001. [DOI] [PubMed] [Google Scholar]
- [56].Kull S, Pauly D, Stormann B, Kirchner S, Stammler M, Dorner MB, Lasch P, Naumann D, Dorner BG. Multiplex detection of microbial and plant toxins by immunoaffinity enrichment and matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 2010;82:2916–2924. doi: 10.1021/ac902909r. [DOI] [PubMed] [Google Scholar]
- [57].Piazza TM, Blehert DS, Dunning FM, Berlowski-Zier BM, Zeytin FN, Samuel MD, Tucker WC. In vitro detection and quantification of botulinum neurotoxin type E activity in avian blood. Appl. Environ. Microbiol. 2011;77:7815–7822. doi: 10.1128/AEM.06165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Montgomery VA, Smith LA. Diagnostic and possible therapeutic application of a monoclonal antibody (14G8) directed against botulinum type C neurotoxin. Hybridoma (Larchmt.) 2011;30:209–216. doi: 10.1089/hyb.2010.0109. [DOI] [PubMed] [Google Scholar]
- [59].Thomas RJ. Detection of Clostridium botulinum types C and D toxin by ELISA. Aust. Vet. J. 1991;68:111–113. doi: 10.1111/j.1751-0813.1991.tb00769.x. [DOI] [PubMed] [Google Scholar]
- [60].Hunter LC, Miller JK, Poxton IR. The association of Clostridium botulinum type C with equine grass sickness: a toxicoinfection? Equine Vet J. 1999;31:492–499. doi: 10.1111/j.2042-3306.1999.tb03857.x. [DOI] [PubMed] [Google Scholar]
- [61].Zechmeister TC, Kirschner AK, Fuchsberger M, Gruber SG, Suess B, Rosengarten R, Pittner F, Mach RL, Herzig A, Farnleitner AH. Prevalence of botulinum neurotoxin C1 and its corresponding gene in environmental samples from low and high risk avian botulism areas. ALTEX. 2005;22:185–195. [PubMed] [Google Scholar]
- [62].Rocke TE, Smith SR, Nashold SW. Preliminary evaluation of a simple in vitro test for the diagnosis of type C botulism in wild birds. J. Wildl. Dis. 1998;34:744–751. doi: 10.7589/0090-3558-34.4.744. [DOI] [PubMed] [Google Scholar]
- [63].Gessler F, Hampe K, Schmidt M, Bohnel H. Immunomagnetic beads assay for the detection of botulinum neurotoxin types C and D. Diagn. Microbiol. Infect. Dis. 2006;56:225–232. doi: 10.1016/j.diagmicrobio.2006.04.014. [DOI] [PubMed] [Google Scholar]
- [64].Gessler F, Hampe K, Bohnel H. Sensitive detection of botulinum neurotoxin types C and D with an immunoaffinity chromatographic column test. Appl. Environ. Microbiol. 2005;71:7897–7903. doi: 10.1128/AEM.71.12.7897-7903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.