Abstract
We report an efficient approach for the chemical synthesis of Rhesus θ-defensin-1 (RTD-1) using Fmoc-based solid-phase peptide synthesis in combination with an intramolecular version of native chemical ligation. The corresponding linear thioester precursor was cyclized and folded in a one-pot reaction using reduced glutathione. The reaction was extremely efficiently yielding natively folded RTD-1 with minimal or no purification at all. This approach is fully compatible with the high throughput production of chemical libraries using this peptide scaffold.
Keywords: Defensin, Cyclic peptide, Antimicrobial peptide
Defensins are antimicrobial peptides that are important in the innate immune defense of mammals.1–3 Mammalian defensins have broad-spectrum antimicrobial activities including anti-viral, and have been shown to be involved in other defense mechanisms, such as immune modulation, neutralization of endotoxin, and anti-cancer activities, among others.3,4 Mammalian defensins are cationic cysteine-rich peptides with largely β-sheet structures stabilized by the presence of three disulfide bonds, and can be classified into three structurally distinct groups, α-, β- and θ-defensins. Despite differences in disulfide connectivities and the presence of an N-terminal α-helix segment in ²-defensins, α- and β-defensins adopt a similar overall fold. θ-Defensins, on the other hand, are backbone-cyclized octadecapeptides that are produced by the pair-wise, head-to-tail splicing of nonapeptides derived from α-defensin related precursors by an as yet unknown mechanism.1,5 θ-Defensins are only expressed in Old World monkeys and orangutans, 6–8 and to date, they remain the only known backbone-cyclized polypeptides expressed in animals.1 Interestingly, humans possess genes encoding θ-defensins, but they have lost the ability to produce the peptides due to a stop codon mutation within the signal sequence that prevents subsequent translation.9
Rhesus h-defensin-1 (RTD-1) was the first θ-defensin to be found in leukocyte extracts from Rhesus macaques.1 Other less abundant RTD variants, named RTD-2 through RTD-6, have also been found in Rhesus macaques more recenlty.10–12 θ-Defensins have broad antimicrobial activities in the presence of physiological concentrations of salt, divalent cations, and serum in vitro.1,13 Interestingly, they are also anti-fungal1 and have anti-viral activities against herpes simplex14 and HIV.9,15 In fact, chemically-synthesized θ-defensins derived from the corresponding human pseudogene sequences have been shown to protect human cells from infection by HIV-19 and have been evaluated as a topical anti-HIV agent for the prevention of HIV transmission.16–18 θ-Defensins also inactivate germinating anthrax spores and act as a competitive inhibitor of anthrax lethal factor protease.19
The structure of RTD-1 (Fig. 1A) is different from that of α- and β-defensins, exhibiting a backbone-cyclized double β-hairpin-like fold with two anti-parallel β-strands stabilized by three disulfide bonds in a ladder configuration (Fig. 1A).20 The circular backbone topology of θ-defensins is required for their biological activity as their linear counterparts show a decreased biological activity. The presence of three disulfides and a circular backbone topology makes θ-defensins a particularly stable peptide framework. Altogether, these features make these peptides an attractive molecular scaffold for the development of peptide-based therapeutics with optimized antimicrobial activity or for the introduction of novel biological activities.
Figure 1.
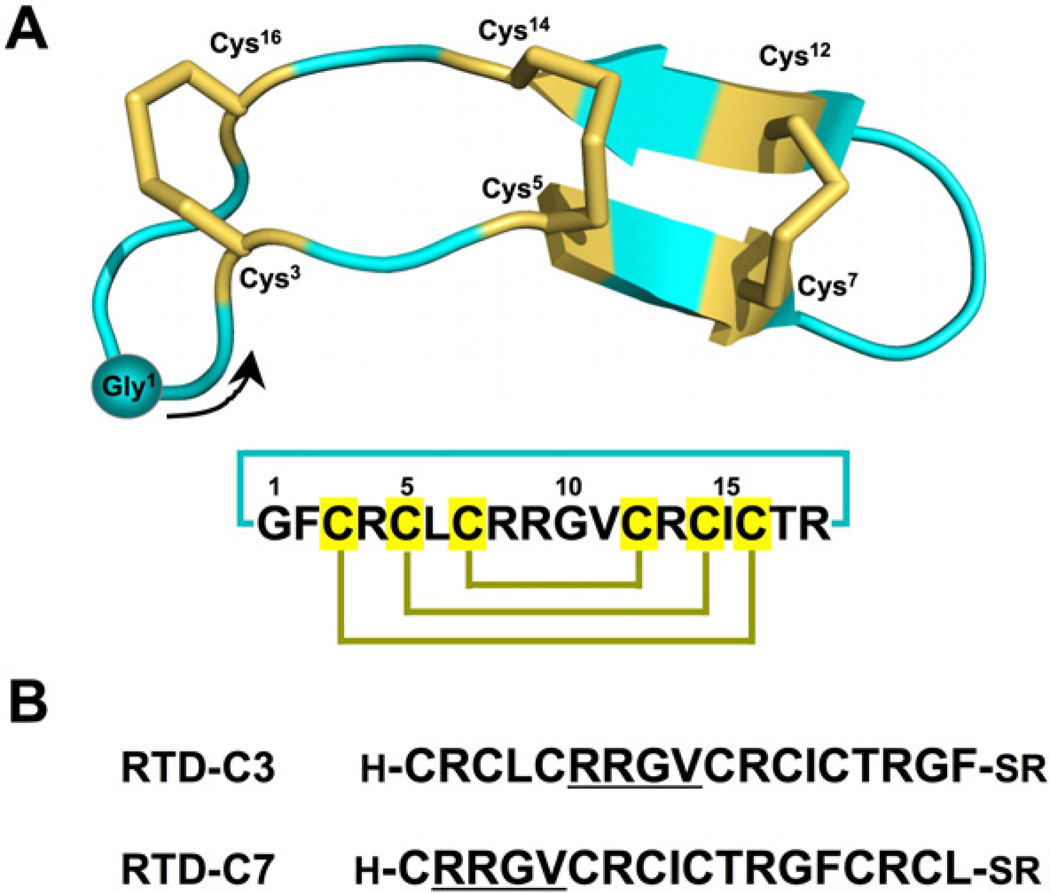
(A) Primary and tertiary structures of rhesus θ-defensin 1 (RTD-1) (PDB ID code: 1HVZ).20 The backbone cyclized peptide (connecting bond shown in blue) is stabilized by the three-disulfide bonds in a ladder formation (disulfide bonds shown in yellow). A solid circle is used to indicate the position of residue Gly1 with an arrow indicating the N-to-C direction of the polypeptide backbone. (B) Sequences of the linear peptide thioester precursors used in this work, RTD-C3 and RTD-C7. The sequence RRGV is underlined for reference.
The relatively small size of θ-defensins (18 residues) makes it possible to use chemical tools for the generation of large combinatorial libraries based on this scaffold for the screening and selection of optimized sequences for a particular biological activity. The chemical synthesis of θ-defensin RTD-1 has already been accomplished by either Fmoc-1 or Boc-based20 solid-phase peptide synthesis (SPPS). In both approaches, the fully reduced unprotected linear peptide was first folded under physiological conditions and then the N-terminal amino and C-terminal carboxylic groups were reacted using different coupling reagents.1,20 However, due to the use of non-chemoselective conditions during the backbone cyclization of the un-protected peptide, the ligation reaction required careful optimization and in both cases the final product had to be purified. Although this approach has been successful in the chemical production of different θ-defensins, the production of large libraries would require the optimization and purification of every potential member of the library making this approach impractical. In order to overcome this problem we decided to explore the use of intramolecular native chemical ligation (NCL)21,22 for the efficient production of cyclic h-defensins (Scheme 1). Intramolecular NCL requires the presence of an N-terminal Cys residue and a C-terminal α-thioester group in the same linear precursor molecule.23,24 The chemoselectivity of this intramolecular reaction is extremely exquisite at neutral pH producing the backbone cyclized peptide in almost quantitative yields.24–26
Scheme 1.
Diagram depicting the chemical synthesis of RTD-1 by Fmoc-based SPPS followed by GSH-induced cyclization/folding.
RTD-1 has six Cys residues that may be used for cyclization. In order to facilitate the cyclization reaction, we decided to use the Cys residues located in positions 3 and 7 (Fig. 1). These Cys residues do not posses a charged or β-branched residue N-terminally adjacent, which should facilitate the kinetics of the cyclization reaction. Accordingly, two different RTD-1 linear thioester precursors (RTDC3 and RTD-C7) were produced by Fmoc-based SPPS using a safety-catch sulfonamide linker (Scheme 1 and Fig. 1B).27,28 Activation of the sulfonamide peptide-resins with iodoacetonitrile followed by thiolytic cleavage with ethyl 3-mercaptopropionate provided the fully protected C-terminal thioester peptide precursors. Subsequent acidolysis with trifluoroacetic acid (TFA), and purification by precipitation with Et2O gave the corresponding unprotected linear thioester precursors with yields ≈10–15% based on the initial substitution of the resin. As shown in Figure 2A, HPLC analysis of the TFA crude for both linear precursors revealed in both cases the presence of a major peak that corresponded to the expected linear thioester precursor as determined by electro-spray mass spectrometry (ESMS) analysis (Fig. 2A). Other minor peaks present in both TFA crudes were identified by ES-MS as partially oxidized linear precursors.
Figure 2.
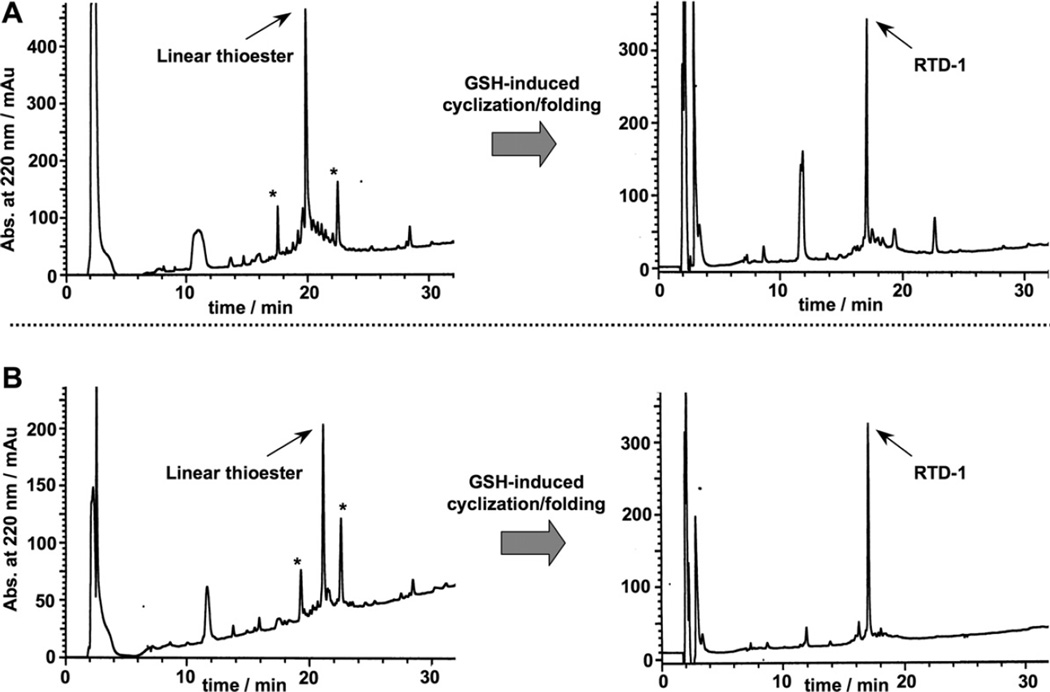
Reverse-phase C18-HPLC traces for the GSH-induced cyclization folding of purified precursors RTD-C3 (A) and RTD-C7 (B). The HPLC traces of the corresponding linear peptide thioester precursor TFA crudes are shown on the left. The thioester linear peptide is marked in each case with an arrow. Other minor peptide products are marked with an asterisk. These compounds were identified as partially oxidized linear peptides by ES-MS and disappear once the cyclization/folding is complete (right HPLC traces).
We next performed the backbone cyclization/folding in a one-pot reaction by diluting the corresponding unpurified peptide thioester precursors to a final peptide concentration of ≈25 µM in sodium phosphate buffer at pH 7.0 in the presence of 1 mM reduced glutathione (GSH). GSH has been shown to promote cyclization and concomitant folding when used in the production of several Cys-rich cyclic polypeptides.26,29–31 As shown in Figure 2A, the GSH-induced cyclization/folding reaction was very clean and efficient for both linear precursors, being complete in 24 h. In both cases the major peptide product was identified by ES-MS as the expected folded cyclized RTD-1 product (Fig. 2). It is worth noting, that the cyclization/folding of precursor RTD-C7 was extremely efficient producing a product with more than 90% of the peptide content being the desired cyclic folded product therefore requiring no purification (Fig. 2A). The yield for the one-pot cyclization and folding using precursor RTD-C3 was estimated by HPLC to be ≈80% (Fig. 2B).
The biological activity of pure synthetic RTD-1 was tested using an anthrax lethal factor (LF) protease inhibitory assay.32 Synthetic RTD-1 was able to inhibit LF with an IC50 of 419 ± 32 nM (Fig. 3A). This IC50 value corresponds to a Ki ≈ 0.4 µM under the conditions used in the inhibitory assay.33,34 This IC50 is similar to the values previously reported for native RTD-1,19,35 therefore confirming the biological activity of synthetic RTD-1.
Figure 3.
Characterization of synthetic RTD-1. (A) Inhibition assay of RTD-1 against anthrax lethal factor (LF). LF activity was measured in the presence of increasing concentrations of peptide RTD-1 and it was determined as the rate LF cleaves a FRET-based substrate.32 (B) Summary of the1 H NMR assignments for the backbone protons: Δδ(1H) are the deviations in the chemical shifts of the main chain protons between the values obtained in this work for synthetic RTD-1 and those reported for RTD-1 at pH 6 (Table S1) and pH 4.5.20
Pure synthetic RTD-1 was also characterized by 2D-1H NMR to confirm its θ-defensin fold. The structure of RTD-1 has been previously elucidated by NMR in 10% MeCN aqueous buffer at pH 4.5.20 We have also recently characterized native folded recombinant 15N-labeled RTD-1 in phosphate buffer at pH 6.0.36 The chemical shifts of the assigned backbone amide and alpha protons (HNα and HCα) for synthetic RTD-1 were almost identical (≤.1 ppm) to those reported for RTD-1 at pH 6.0 (Fig. 3B and Table S1).36 We also compared the chemical shifts of synthetic RTD-1 with those reported by Craik for RTD-1 in 10% MeCN at pH 4.5.20 Again, no significant (≤03 ppm) differences were found in the backbone amide protons. We also saw a uniform shift rather than variable changes for the backbone alpha protons (≈0.2 ppm) at pH 6.0 (Fig. 3B and Table S1). Since backbone alpha protons usually reflect the secondary structure of the peptide backbone, this uniform offset in the resonances of the backbone HCα protons can be attributed to the different buffer conditions used in the two samples rather than changes in secondary structure. Altogether, these data confirm that synthetic RTD-1 adopts a native θ-defensin fold.
In summary, we have achieved the efficient synthesis of the θ-defensin RTD-1 using optimized Fmoc-based SPPS in combination with one-pot NCL-driven backbone cyclization with concomitant folding. We have shown that the GSH-induced cyclization/folding of linear precursor RTD-C7 is extremely clean not requiring the purification of the final cyclic folded θ-defensin. To our knowledge this represents the first chemical synthesis of RTD-1 using a chemical approach that is extremely efficient therefore requiring mini-mal or no purification at all and therefore fully compatible with the high throughput production of chemical libraries using this scaffold. These libraries could be used for the optimization of the already known properties of θ-defensins or for the introduction of novel biological activities into this backbone cyclized polypeptide scaffold.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Research Grant R01-GM090323 (J.A.C.) and NIH Grant 5R01GM085006-02 (A.S.); and by the Department of Defense Congressionally Directed Medical Research Program Grant PC09305 (J.A.C.).
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2012.02.080.
References and notes
- 1.Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. Science. 1999;286:498. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 2.Tanabe H, Yuan J, Zaragoza MM, Dandekar S, Henschen-Edman A, Selsted ME, Ouellette AJ. Infect. Immun. 2004;72:1470. doi: 10.1128/IAI.72.3.1470-1478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selsted ME, Ouellette AJ. Nat. Immunol. 2005;6:551. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 4.Lehrer RI. Nat. Rev. 2004;2:727. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 5.Selsted ME. Curr. Protein Pept. Sci. 2004;5:365. doi: 10.2174/1389203043379459. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen TX, Cole AM, Lehrer RI. Peptides. 2003;24:1647. doi: 10.1016/j.peptides.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 7.Garcia AE, Osapay G, Tran PA, Yuan J, Selsted ME. Infect. Immun. 2008;76:5883. doi: 10.1128/IAI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stegemann C, Tsvetkova EV, Aleshina GM, Lehrer RI, Kokryakov VN, Hoffmann R. Rapid Commun. Mass Spectrom. 2010;24:599. doi: 10.1002/rcm.4424. [DOI] [PubMed] [Google Scholar]
- 9.Cole AM, Hong T, Boo LM, Nguyen T, Zhao C, Bristol G, Zack JA, Waring AJ, Yang OO, Lehrer RI. Proc. Natl. Acad. Sci. U.S.A. 2002;1813:99. doi: 10.1073/pnas.052706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonova L, Kokryakov VN, Aleshina G, Hong T, Nguyen T, Zhao C, Waring AJ, Lehrer RI. J. Leukoc. Biol. 2001;70:461. [PubMed] [Google Scholar]
- 11.Tran D, Tran PA, Tang YQ, Yuan J, Cole T, Selsted ME. J. Biol. Chem. 2002;277:3079. doi: 10.1074/jbc.M109117200. [DOI] [PubMed] [Google Scholar]
- 12.Tongaonkar P, Tran P, Roberts K, Schaal J, Osapay G, Tran D, Ouellette AJ, Selsted ME. J. Leukoc. Biol. 2011;89:283. doi: 10.1189/jlb.0910535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran D, Tran P, Roberts K, Osapay G, Schaal J, Ouellette A, Selsted ME. Antimicrob. Agents Chemother. 2008;52:944. doi: 10.1128/AAC.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasin B, Wang W, Pang M, Cheshenko N, Hong T, Waring AJ, Herold BC, Wagar EA, Lehrer RI. J. Virol. 2004;78:5147. doi: 10.1128/JVI.78.10.5147-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penberthy WT, Chari S, Cole AL, Cole AM. Cell. Mol. Life Sci. 2011;68:2231. doi: 10.1007/s00018-011-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munk C, Wei G, Yang OO, Waring AJ, Wang W, Hong T, Lehrer RI, Landau NR, Cole AM. AIDS Res. Hum. Retroviruses. 2003;19:875. doi: 10.1089/088922203322493049. [DOI] [PubMed] [Google Scholar]
- 17.Owen SM, Rudolph D, Wang W, Cole AM, Sherman MA, Waring AJ, Lehrer RI, Lal RB. J. Pept. Res. 2004;63:469. doi: 10.1111/j.1399-3011.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 18.Gallo SA, Wang W, Rawat SS, Jung G, Waring AJ, Cole AM, Lu H, Yan X, Daly NL, Craik DJ, Jiang S, Lehrer RI, Blumenthal R. J. Biol. Chem. 2006;281:18787. doi: 10.1074/jbc.M602422200. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Mulakala C, Ward SC, Jung G, Luong H, Pham D, Waring AJ, Kaznessis Y, Lu W, Bradley KA, Lehrer RI. J. Biol. Chem. 2006;281:32755. doi: 10.1074/jbc.M603614200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trabi M, Schirra HJ, Craik DJ. Biochemistry. 2001;40:4211. doi: 10.1021/bi002028t. [DOI] [PubMed] [Google Scholar]
- 21.Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Science. 1994;266:776. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 22.Dawson PE, Kent SB. Annu. Rev. Biochem. 2000;69:923. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 23.Camarero JA, Muir TW. Chem. Commun. 1997;1997:1369. [Google Scholar]
- 24.Camarero JA, Muir TW. J. Am. Chem. Soc. 1999;121:5597. [Google Scholar]
- 25.Camarero JA, Pavel J, Muir TW. Angew. Chem. Int. Ed. 1998;37:347. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Kimura RH, Tran AT, Camarero JA. Angew. Chem. Int. Ed. 2006;45:973. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- 27.Ingenito R, Bianchi E, Fattori D, Pessi A. J. Am. Chem. Soc. 1999;121:11369. [Google Scholar]
- 28.Shin Y, Winans KA, Backes BJ, Kent SBH, Ellman JA, Bertozzi CR. J. Am. Chem. Soc. 1999;121:11684. [Google Scholar]
- 29.Austin J, Wang W, Puttamadappa S, Shekhtman A, Camarero JA. Chembiochem. 2009;10:2663. doi: 10.1002/cbic.200900534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Austin J, Kimura RH, Woo YH, Camarero JA. Amino Acids. 2010;38:1313. doi: 10.1007/s00726-009-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Contreras J, Elnagar AYO, Hamm-Alvarez S, Camarero JA. J. Controlled Release. 2011;155:134. doi: 10.1016/j.jconrel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura RH, Steenblock ER, Camarero JA. Anal. Biochem. 2007;369:60. doi: 10.1016/j.ab.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Y, Prusoff WH. Biochem. Pharmacol. 1973;22:3099. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 34.Tonello F, Ascenzi P, Montecucco C. J. Biol. Chem. 2003;278:40075. doi: 10.1074/jbc.M306466200. [DOI] [PubMed] [Google Scholar]
- 35.Kawakami T, Ohta A, Ohuchi M, Ashigai H, Murakami H, Suga H. Nat. Chem. Biol. 2009;5:888. doi: 10.1038/nchembio.259. [DOI] [PubMed] [Google Scholar]
- 36.Gould A, Li Y, Majumder S, Garcia AE, Carlsson P, Shekhtman A, Camarero JA. Mol. Biosyst. 2012;8:1359. doi: 10.1039/c2mb05451e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.