Abstract
CONTEXT
Brain-derived neurotrophic factor (BDNF) is suspected of being a causative factor in psychiatric disorders based on case reports or studies involving large structural anomalies.
OBJECTIVE
To determine the involvement of BDNF in human psychopathology
DESIGN
Case- Control study
SETTING
Microarray-based comparative genomic hybridization (aCGH) data from seven molecular diagnostic centers including 38, 550 affected subjects and 28, 705 unaffected subjects.
PATIENTS
Subjects referred to diagnostic screening centers for aCGH for physical or cognitive impairment.
MAIN OUTCOME MEASURE
Genomic copy number gains and losses
RESULTS
We report five individuals with psychopathology and genomic deletion of a critical region including BDNF. The defined critical region was never disrupted in control subjects or diagnostic cases without developmental abnormalities.
CONCLUSION
Hemizygosity of the BDNF region contributes to variable psychiatric phenotypes including anxiety, behavioral, and mood disorders.
Introduction
BDNF (brain-derived neurotrophic factor) is a nervous system growth factor that plays a critical role in synaptic modeling, neurodevelopment, and cell signaling1. It is a member of the nerve growth factor (NGF) family with structural similarity to NGF and neurotrophin 3/4, and structural differences distinct from the other nervous system growth factor families which include fibroblast growth factor, insulin-like growth factor, transforming growth factor-beta, and cytokine families2. While all nervous system growth factors support neurodevelopment, BDNF has been singularly implicated for its role in obesity, pain, and memory3–7. The protein is encoded by BDNF, located on the short arm of chromosome 14 at band p14, where a polymorphic variant at codon 66 specifies either Valine or Methionine and is thought to affect processing of proBDNF to BDNF. This locus has been considered as a risk factor for schizophrenia, major depression, ADHD, bipolar disorder and many other psychopathologies8, 9, primarily from association-based studies evaluating the non-synonymous Val66Met variant and studies comprising cases with deletions on 11p associated with deletions in WT1 and PAX610, 11.
BDNF sequencing studies in psychiatry and genomic copy loss studies support a link between BDNF with behavior and obesity. WAGR syndrome, a deletion syndrome of the short arm of chromosome 11 associated with Wilms tumor, aniridia, genitourinary anomalies and mental retardation in which deletions include PAX6 and WT1, sometimes includes larger deletions extending to BDNF. Two recent studies associated subjects with WAGR syndrome, with deletions extending to BDNF, with obesity, bipolar disorder, or ADHD10, 11. In support of a psychiatric phenotype due to copy loss at the BDNF locus, two independent case reports (three subjects in total) described obese patients who presented with complex neurobehavioral phenotypes12, 13. Further, a deep re-sequencing study of BDNF exons and flanking regions from subjects with major depression (MD) and controls, revealed several novel variants associated with MD, suggesting that genetic variation in BDNF may have an impact on mood14.
Molecular studies in rodents have supported a role for Bdnf in behavior, in particular through the finding that defective neuronal release of BDNF by in vivo knock-down leads to increased anxiety-like traits in mice15, 16, while heterozygous Bdnf knock-out mice do not display anxiety traits17, they are reported to be more aggressive and hyperphagic than wild-type mice18. BDNF has also been shown to have a key role in mediating social defeat stress in rodents19, in particular, it is required for the development of experience-dependent social aversion15. With respect to sensory systems, homozygous Bdnf knock-out mice show sensory deficits with decreased survival of sensory ganglia while sparing motor neuron development20, 21, in line with data from human WAGR patients with a BDNF deletion that suggest a deficit in nociception11. Together, data from rodents suggests that whole-organism deletion of BDNF leads to behavioral, sensory, and weight alterations, while deletion of BDNF specifically in brain areas associated with behavior lead to anxiety and aggression.
In view of the large number of association studies with suggestive evidence for BDNF polymorphisms in psychopathology, case reports describing large genomic alterations involving BDNF in subjects with psychiatric symptoms, and extensive phenotyping in animal models, we sought to better resolve the relationship between BDNF and psychopathology by identifying subjects with genomic copy number changes that include BDNF.
Methods
Table 1 summarizes all subjects used in this study. From Signature Genomics (SG), we analyzed a total of 26,144 probands studied using oligonucleotide-based whole-genome array-Comparative Genomic Hybridization (aCGH), using either a 105K-feature platform (SignatureChipOS version 1.0, custom-designed by SG, manufactured by Agilent Technologies, Santa Clara, CA) or a 135K-feature platform (SignatureChipOS version 2.0, custom-designed by SG, manufactured by RocheNimbleGen, Madison, WI), according to previously described methods22, 23. From this initial cohort, we divided subjects into those referred with an indication of a neurodevelopmental disorder (n = 14,616) and those referred with an indication for study that did not involve a known neurodevelopmental abnormality (n=11,528). Unlike the microarrays used to analyze controls, these specific SG platforms are incapable of detecting intragenic BNDF variations and are limited to whole-gene BDNF deletions, at a resolution of approximately 270 kb and 120 kb, respectively. The ethnic distribution in the samples from Signature Genomics Inc. was estimated from a sampling cross section previously described24. This sample (n=144 subjects, self reported) was composed of 75% Caucasian, 7% African-American, and 18% other. The gender distribution was 59% male, 41% female. The only alterations spanning BDNF observed in the SG group were WAGR syndrome patients (N=2), so there was no contribution to these analyses from this dataset, although they are included in all statistical analyses. The ethnicity of each patient described here with a copy gain or loss of BDNF was Caucasian.
Table 1.
Information on cases and controls1
| Site | Indications for Study |
N | Platform |
|---|---|---|---|
| Cases | |||
| SickKids | NDD, MCA only | 3,258 | Agilent 44K/180K |
| Children's Hospital |
All | 7,320 | Agilent 244K |
| Signature | NDD only | 14,616 | Signature ChipOS 105K/135K |
| Mayo | All | 13,135 | Agilent 180K |
| Harvard | Balanced chromosomal rearrangement with phenotype |
221 | Next Generation sequencing |
| Controls | |||
| ISC | Unaffected | 7,878 | Affymetrix 5.0/6.0 |
| Cooper et al.* | Unaffected | 6,113 | Affymetrix 6.0 |
| OHI | Unaffected | 1,234 | Affymetrix 6.0 |
| POPGEN | Unaffected | 1,123 | Affymetrix 6.0 |
| HapMap3 | Unaffected | 1,056 | Affymetrix 6.0 |
| SAGE | Unaffected | 1,287 | Illumina 1M |
| Shaikh et al. | Unaffected | 2,026 | Affymetrix 6.0 |
| DGV** | Unaffected | 7,988 | Multiple |
NDD = any neurodevelopmental disorder, behavioral, or neuropsychiatric disorder, including autism and autism spectrum disorder.
MCA = multiple congenital anomalies.
All = all indications for study included; precise phenotypes of all individuals were not available for further delineation of NDD.
Publicly available control data from Cooper et al. (2011) with WTCCC controls already analyzed in the ISC control set removed.
Controls from DGV filtered for overlap with other control studies presented.
See manuscript for references for each cohort
The clinical cytogenetics laboratory at The Hospital for Sick Children in Toronto, Canada screened patients using either Agilent 4×44K array25 or the 4×180K ISCA v2 microarray manufactured by Agilent and designed by Oxford Gene Technologies. Previously published CNV data from 11,509 controls genotyped with high resolution SNP microarrays were compiled from several subject groups including: 1,234 Affymetrix 6.0 controls from Ottawa26, 1,123 Affymetrix 6.0 controls from POPGEN26, 4,783 WTCCC Affymetrix 6.0 controls from WTCCC27, 1,056 HapMap3 Affymetrix 6.0 controls28, 1,287 Illumina 1M data from SAGE controls29, and 2,026 Hap550k controls30.
From the Developmental Genome Anatomy Project (DGAP) database (www.dgap.harvard.edu), we had access to information on 221 subjects, all of whom had a balanced chromosomal rearrangement. Identification of BDNF hemizygosity in one of these subjects (DGAP173) was assessed by an Agilent G3 1M array. Based on available karyotype information (t(2;11)(q11.2;p13), this deletion appears to be independent of the chromosomal rearrangement but we cannot rule out a more complex rearrangement involving both regions. Array processing for all other clinical diagnostic centers was done using commercially available Agilent 244K arrays, except for the Mayo Clinic, which used 180K Agilent arrays.
Control individuals were obtained from a variety of sources listed above as well as control data from the International Schizophrenia Consortium31, the Database of Genomic Variants32 (filtered for overlap with other studies), and those described and publicly available from Cooper and colleagues24, filtering WTCCC controls to avoid redundancy with the above control set. Table 1 describes these control subjects in more detail. All genomic coordinate positions are with reference to the human genome reference 18 (hg18). Statistical analyses were performed using Fisher’s exact test in the statistical package R.
Clinical diagnoses from all patients were performed by independent, qualified physicians who had seen the patient over a period of at least two years. We defined obesity as either BMI >30 kg/m2 or if it was specifically indicated by the primary caregiver. We defined overweight as a BMI >25 kg/m2. Psychiatric diagnoses were done using DSM-IV criteria by caregiver interviews with affected subjects. In all BDNF deletion cases, referring physicians were contacted and provided clinical information for all subjects, allowing for psychiatric phenotyping.
These studies were approved by the Institutional Review Boards of our institutions, and all caregivers for each subject gave informed consent.
Results
We screened microarray-based comparative genomic hybridization (aCGH) data for over 38,000 subjects from clinical diagnostic centers at Children’s Hospital Boston, The Hospital for Sick Children Toronto, the Mayo Clinic, Brigham and Women’s Hospital Boston, Manchester Academic Health Sciences Center, St. Justine Hospital Montreal, and Signature Genomics, Inc., for any subjects with copy number changes of the BDNF region (see Table 1 for complete description of all subject groups). We identified five subjects with deletions encompassing the entire BDNF gene and one subject with a duplication spanning BDNF (Figure 1 and Table 2). For all subjects, aCGH was used to initially identify BDNF copy changes and Figure 2 shows a visual example of aCGH data in subject 2 from this study. The deletion group displayed varied phenotypes that included neurodevelopmental, behavioral and mood disorders, in addition to being obese or overweight and insensitive to pain in some cases, as summarized in Table 2 and presented in greater detail below. The subject with a duplication also presented with developmental delay and dystonia, but no further information was available. Additional subjects identified with WAGR syndrome were excluded from this analysis (N=2 subjects from Signature Genomics) due to the very large number of genes in WAGR deletions, the severity of the associated neurodevelopmental phenotype33, and the inability to obtain any follow-up information on these subjects.
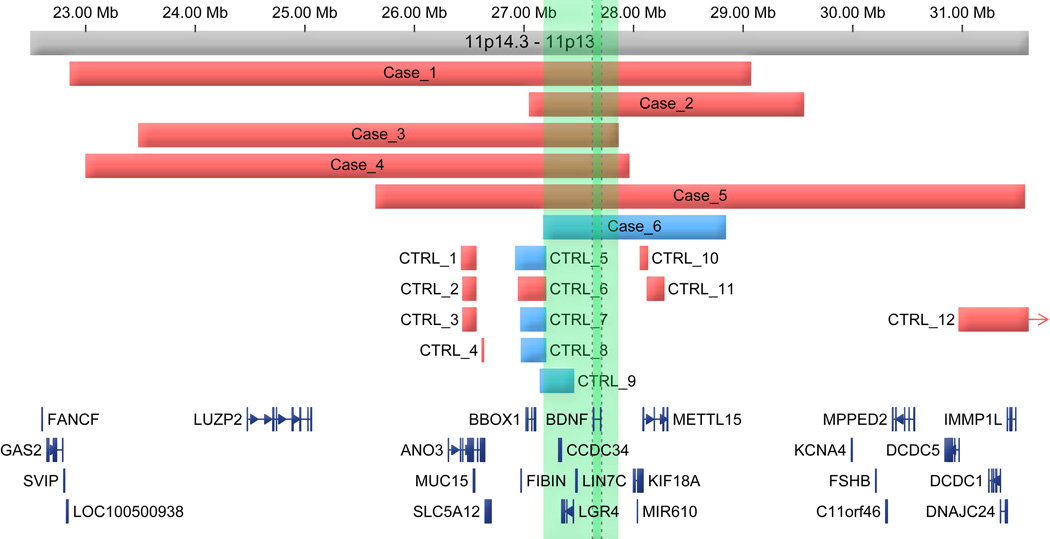
Figure 1. All cases and controls (CTRL) with copy gains (blue) or losses (red) near the BDNF locus.
Megabase marks (Mb) represent hg18 build coordinates. Green shading represents identified critical region while darker green shading corresponds specifically to the genomic location of BDNF.
Table 2.
Characteristics of current subjects as well as those previously reported to have alterations in BDNF.
| Subject | Age/sex | Genotype | Nociception | Psychopathology | Overweight/obese | Mental Dx |
|---|---|---|---|---|---|---|
| 1 | 10/M | Deletion: 22,858,513–29,066,320 | Pain insensitivity | ADHD, anxiety disorder, aggressive behaviors | BMI=22.6 | PDD-NOS |
| 2 | 21/F | Deletion: 27, 050, 622–29, 550, 113 | self—injurious behaviors | Major depression, generalized anxiety disorder | BMI=51.7 | Mild MR |
| 3 | 2.75/M | Deletion: 23,484,198–27,857,928 | N/A | Impaired behavior | mother BMI= 39.5 proband BMI=27.4 | GDD in proband; ID in mother |
| 4 | 16/M | Deletion: 23,002,186–27,956,720 | N/A | Adjustment disorder, Major depression, generalized anxiety disorder | BMI= 50.5 | PDD |
| 5 | 7/M | Deletion: 25,649,116–31,566,599 | Pain insensitivity | Anxiety, ADHD, temper tantrums, intolerance to frustration | BMI=28.3 | Moderate MR |
| 6 | 3/F | Duplication: 27,179,904–28,837,666 | N/A | Not reported | No | Moderate MR and dystonia |
| 42Ref | 13/M | Deletion | N/A | Not reported | Yes | MR |
| 12Ref | 25/F | Deletion | N/A | Mood disturbances, obsessive–compulsive behavior, temper tantrums, intolerance to frustration requiring antipsychotic medications | Yes | Mild/moderate MR |
| 12Ref | 14/F | Deletion | N/A | Chronic anxiety, poor acceptance of change, logorrhea, echolaly, poor social interactions, labile mood, and bouts of aggressiveness and motor agitation that required treatment with risperidone | Yes | PDD: Moderate/severe MR |
| 13Ref | 9/F | Position effect due to an inversion | Pain insensitivity | Complex neurobehavioral phenotype, repetitive behaviors, extreme hyperactivity, no concept of danger | Yes | Low IQ |
Abbreviations: ADHD: Attention Deficit Hyperactivity Disorder; PDD-NOS: Pervasive Developmental Delay, not objectively specified; MR: Mental retardation; GDD: Global Developmental Delay; ID: intellectual disability IQ: Intelligence quotient. All genomic coordinates according to human reference hg18.
Figure 2. A ~2.5 Mb deletion in Subject 2, including BDNF.
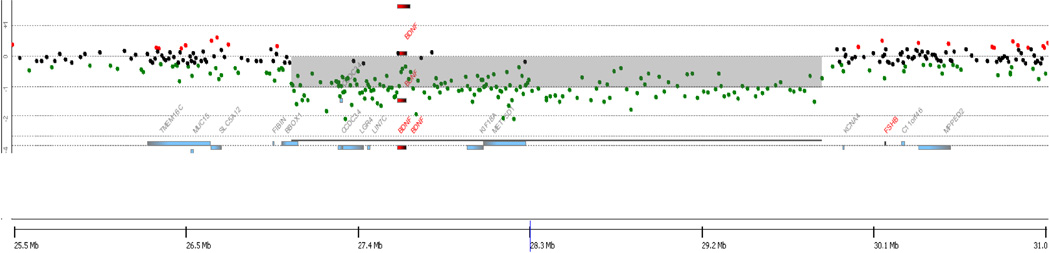
aCGH results demonstrating a deletion on chromosome 11. Gray shading represents the predicted size of the deletion, while individual probes from the array are represented by dots colored black, green, or red. Probes colored green represent decreased probe intensity from DNA from Subject 2, reflecting copy loss. hg18 build coordinates are shown on the X-axis.
Subject 1 was identified with a BDNF deletion at chr11:22,858,513–29,066,320 and a small deletion at chr19: 61453936–61530271 intersecting the testes-specific gene ZSCAN5A. The 10-year-old male has been diagnosed with pervasive developmental disorder not otherwise specified, attention deficit hyperactivity disorder (ADHD), anxiety, behavioral issues (e.g., constantly hitting head against the wall), and mood dysregulation. At four years of age, his condition regressed markedly and to date he has been treated with escitalopram (Lexapro), aripiprazole (Abilify), citalopram (Celexa), guanfacine (Tenex), methylphenidate (Ritalin), atomoxetine (Strattera), and clonidine. His height and weight at age nine were 138.7cm and 43.5kg (95th–97th percentile), respectively, with a BMI of 22.6 (see online-only Figure S1 for a weight chart for this subject taken at different time points showing a progression towards obesity). He is extremely aggressive and parental report notes that the subject does not complain of pain when accidents occur. Array results were confirmed using clinically available Multiplex Ligation-dependent Probe Amplification probes targeting BDNF (MRC-Holland; SALSA MLPA P219).
Subject 2 (DGAP173) is a 21-year-old female with a karyotype of 46,XX,t(2;11)(q11.2;p13) who also has a 2.5Mb deletion (Chr11: 27, 050, 622–29, 550, 113) on chromosome 11 that includes BDNF (Figure 2). Array CGH results were confirmed using clinically available Multiplex Ligation-dependent Probe Amplification probes targeting BDNF. She has mild developmental delay (combined language and motor delay), major depression, generalized anxiety, sleep disturbance (sleep apnea), self-injurious behaviors, agitation, and tantrums. In 2009 at age 19, she weighed 167.6 kg and had a height of 180.1cm, with a body mass index of 51.7. Her head circumference was 61cm, which is outside of the normal adult range of 55–58cm. She has male pattern hirsutism (thought to be associated with a tentative diagnosis of polycystic ovarian syndrome, maternally inherited) and has had only a single period with no further menstruation even with trials of oral contraceptive pills. Impaired glucose tolerance without evidence of type 2 diabetes, poor lipid profile with elevated triglycerides and total cholesterol and low HDL, elevated testosterone, some deepening of the voice, and history of one non-febrile seizure at two years of age, were also noted. Her skin was remarkable for eczema, moles, and skin tags. She has dysmorphic features including bilateral epicanthal folds giving a saddle appearance to the nasal bridge, a small nose, complex malocclusion with upper teeth more narrow and frontal than lower. Morphologically, she has somewhat short hands, slightly hyperkeratotic and sweaty palms, fifth finger brachydactyly and clinodactyly, minor extension limitation of the right elbow, hypoplastic toenails, short feet, and copper-colored verrucous lesions in intertriginous regions (acanthosis nigricans versus epidermal nevi) present on the back, chest and neck.
Subject 3 was identified with a maternally inherited deletion at chr11:23,484,198–27,857,928. He was referred for investigation at 2 years 9 months of age for severe receptive and expressive speech delay. He has impaired social, play, and behavioral skills as well as global developmental delay and a duplex left kidney. He is a large child with weight of 29.3 kg, height of 103.5 cm and a BMI of 27.4, all of which are greater than the 97th percentile. His head circumference is 52 cm, which is considered within the normal range at age 2.75 years, but is at the 94th percentile. Family history is of note in that his mother has intellectual difficulties. Her height was 171 cm and weight of 114.3 kg and BMI of 39.5. No further information is available for her. FISH analysis confirmed she has the same deletion. Maternal grandparents are of normal intellect and growth, and FISH analyses were normal.
Subject 4 is a 16 year-old male whose 36-week gestation was notable for the umbilical cord being wrapped around his neck. 180K Agilent microarray screen revealed a chr11:23,002,186–27,956,720 (HG18) de novo deletion. He has hypercholesterolemia, a fatty liver, hypertension, and is prediabetic. At an assessment done at age 16, he was 151 kg, 1.73 meters, and had a BMI of 50.1. He has speech delay and pervasive developmental disorder, an IQ/DQ of 58. With respect to psychopathology, he has been diagnosed with an adjustment disorder (mixed disturbance of emotion and conduct) depressive disorder, and anxiety disorder. FISH confirmed the array results using RP11–1150I2.
Subject 5 is male with a disruption in BDNF (chr11:25,649,116–31,566,599). No other genetic anomalies were detected in this subject, initially ascertained through learning difficulties, severe speech and language delay, and obesity (BMI 28.3 at 5 years 10 months; >97th percentile). He has a statement of special educational need and at 4.5 years his overall general conceptual ability is limited (score on the British Abilities Scale BAS II was 47 (<0.4%ile) in keeping with a severe learning disability); he was reported to be able to write his name at six years of age. He has poor fine motor skills and poor problem solving skills. With respect to sensory systems he has hyperacusis and a high pain threshold. He is described as having inappropriate toddler-like tantrums triggered by not getting his own way or not being able to eat when he wishes. He has sleeping difficulties and is currently taking melatonin. A strengths and difficulties questionnaire completed by his teacher at age five years noted very high scores for overall stress, hyperactivity and attentional difficulties and high scores for difficulties getting along with other children.
Subject 6 has a BDNF duplication and was indicated for screening because of developmental delay and dystonia (chr11:27,179,904–28,837,666). No further information was available on this subject.
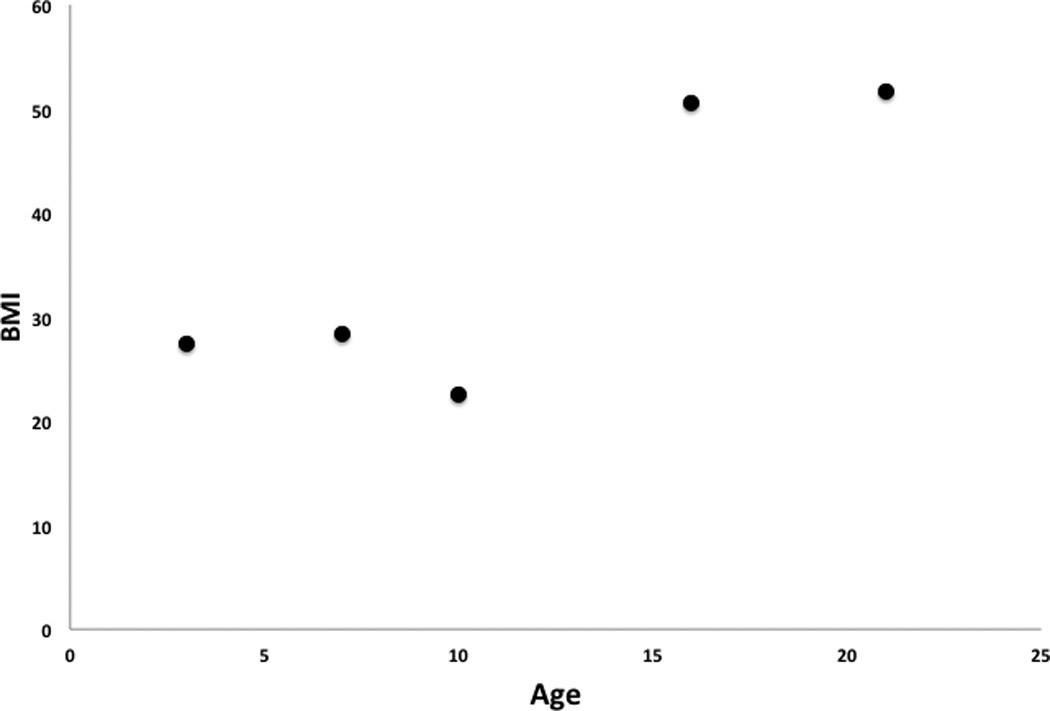
There was a notable relationship between age and BMI in subjects with a BDNF deletion, strongly supporting a role for a deletion in this region and obesity. Specifically, while all subjects were overweight at a young age, older subjects had even higher BMIs, suggesting a progression towards increasing obesity (BMI vs age, Pearson = 0.86, p=0.06), with a particular increase after the later teen years (Figure 3). We were able to further support the hypothesis that people with a BDNF deletion have increased BMIs over time by acquiring data from a single subject (subject 1) who received multiple assessments over time. Supporting on-line figure S1 shows the increase in BMI over time compared to age standards.
Figure 3. Subjects with deletions in BDNF have increasing BMI’s over time.
While each of the BDNF-containing deletions reported here disrupted multiple genes, the critical region of overlap included only BBOX1, CCDC34, LGR4, BDNF and LIN7C (Figure 1). We therefore attempted to narrow the critical region responsible for the mood and behavior phenotypes by examining structural variations in datasets from individuals without a comparable phenotype. We found no structural variations affecting BDNF in CNV data from 28, 705 control individuals with high resolution chromosomal microarrays (Figure 1 and Table 1), despite the superior resolution of these platforms relative to those used to analyze most of the cases. There was also no disruption of the BDNF locus from clinical diagnostic cases not reported to have a neurological abnormality (n=11,528) assayed through SG's Genoglyphix Chromosome Aberration Database. Collectively, though disruption of this locus was rare, we find a nominally significant burden of dosage alterations spanning BDNF in cases compared to all controls (Fisher’s exact test p = 0.042) as well as the combination of controls and clinical diagnostic cases without a neurodevelopmental abnormality (n = 40, 233; p = 0.014). Similar results were obtained if we restricted analyses to only those cases with deletion of the locus (p = 0.076 and 0.028, respectively). There was evidence for deletion of BBOX1, as well as for duplication of BBOX1, CCDC34 and LGR4, though there were no disruptions of LIN7C. CCDC34 has previously been reported as disrupted in a case of translocation34 without an associated neurodevelopmental phenotype. Taken together, these findings indicate that deletions encompassing BDNF are rare, but when they occur they are highly penetrant in producing a distinct phenotypic spectrum which includes behavioral/psychiatric traits due to alterations in BDNF, LIN7C, LGR4 or some combination of these genes.
Comment
This study represents the largest and highest genomic resolution study to date investigating the role of BDNF in psychopathology. Previous reports identified single cases with large deletions encompassing BDNF or cases with BDNF deletions and WAGR syndrome, where one study identified four different WAGR syndrome subjects with behavioral disturbances10. The current study included over 38,000 probands collected internationally and found five subjects with BDNF deletions with heterogeneous, but always psychiatric, phenotypes. Despite being the most extensive study to date of the role of BDNF in psychopathology, this study should be considered supportive of the role of BDNF in psychopathology and not unequivocal, as the critical region included two other potentially causative genes affected in all BDNF deletion cases. Nonetheless, animal data and analysis of the function of these two genes in the critical region strongly suggest that BDNF hemizygosity leads to psychopatholgy.
Mouse studies of LGR4 and LIN7C orthologues suggest a less central role for these genes in behavior. Lgr4 knock-out mice show embryonic lethality, thought to be due to its fundamental role in organogenesis, particularly of the kidney and the sex organs35, 36. Subject 3 in our study had a duplex left kidney and subject 2 had polycystic ovary syndrome. Notably, expression of Lgr4 is largely absent from brain except in the olfactory bulb and periventricular area; expression is highest in kidney, gall bladder, heart, bone and spinal cord37. Thus, LGR4 hemizygosity is unlikely to contribute to psychopathology in humans, but could account for other observed abnormalities. Lin7c (aka MALS-3) has a role in maintaining cell polarity during development in the mouse38 though two paralogues, Lin7a and Lin7b, are suspected of being able to compensate for Lin7c deficiency39. Distribution of Lin7c expression in mouse brain is low compared to Lin7a and Lin7b and is restricted to the dentate gyrus, cerebellum, and superior colliculus. In contrast, Lin7a and Lin7b are abundantly expressed in other brain regions, especially cortex and dentate gyrus39. While this expression pattern does not suggest a primary role for LIN7C hemizygosity in psychopathology, such a contribution, alone or in interaction with BDNF, cannot be excluded.
The presence of psychiatric manifestations in subjects with BDNF-associated deletion is consistent with previously reported cases, as delineated in Table 2, along with their associated neurodevelopmental and behavioral phenotypes. Taken together, this collection of subjects support the conclusion that gross disruption of BDNF in humans is associated with psychopathology, being obese or overweight, and, at least sometimes, pain insensitivity - phenotypes consistent with data from manipulation of Bdnf in rodents. No information was available for three deletion subjects with respect to pain insensitivity (one of the subjects with a BDNF deletion is reported to engage in self-injurious behavior, a phenotype frequently associated with pain insensitivity in individuals with intellectual disability40), so we cannot draw a conclusion concerning the universality of pain insensitivity, but follow-up studies are warranted. Both the overweight/obese and nociceptive phenotypes in humans are also supported by a study of WAGR patients; while those with deletions that extended to BDNF were more likely to be obese and insensitive to pain11 than those without BDNF deletion.
The consensus phenotype for individuals with a deletion in BDNF suggests that young children are hyperactive, anxious, and have an intolerance to change. As subjects age, they likely develop more pronounced anxiety and mood disorders, exemplified by the 16 year-old and 21 year-old subjects with major depressive disorder and generalized anxiety disorder, and by a 25 year-old female with mood disturbances from a previous report12. Identification of a single locus that may be linked to major depression or anxiety highlights the heterogeneity of these psychiatric diseases - most subjects with major depression do not have deletions in BDNF for example - and the need to possibly re-assess how clinical categorization proceeds41.
Chromosomal aberrations at genomic loci that associate with mental retardation are common, but hemizygosity of a locus that can affect a spectrum of phenotypes including mood is less common, and the mechanisms that could contribute to such phenotypic diversity remain to be elucidated. Deeper investigation of the regulation of BDNF and of the molecular actions of the transcribed product will be required to better understand how hemizygosity at this locus contributes to psychopathology.
Supplementary Material
Acknowledgements
This work was funded by a grant from Canadian Institute of Health Research to CE, grants from the NIH GM061354 (CCM, JFG), HD065286 (JFG), MH087123 (MET) and the Simons Foundation Autism Research Initiative (JFG), and the National Basic Research Program of China (973 Program) (2010CB529601) (BLW), the Science and Technology Council of Shanghai (09JC1402400 and 09ZR1404500) (BLW). We are deeply thankful to all patients, families, and referring physicians who participated in this study. We are also grateful to the members of the ISC, WTCCC, Evan Eichler, and Bradley Coe for their invaluable contribution of control resources.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Klein R, Smeyne RJ, Wurst W, Long LK, Auerbach BA, Joyner AL, Barbacid M. Targeted disruption of the trkB neurotrophin receptor gene results in nervous system lesions and neonatal death. Cell. 1993 Oct 8;75(1):113–122. [PubMed] [Google Scholar]
- 2.Nagahara AH, Tuszynski MH. Potential therapeutic uses of BDNF in neurological and psychiatric disorders. Nature reviews. Drug discovery. 2011 Mar;10(3):209–219. doi: 10.1038/nrd3366. [DOI] [PubMed] [Google Scholar]
- 3.Anand P. Neurotrophic factors and their receptors in human sensory neuropathies. Progress in brain research. 2004;146:477–492. doi: 10.1016/S0079-6123(03)46030-5. [DOI] [PubMed] [Google Scholar]
- 4.Lu B, Gottschalk W. Modulation of hippocampal synaptic transmission and plasticity by neurotrophins. Progress in brain research. 2000;128:231–241. doi: 10.1016/S0079-6123(00)28020-5. [DOI] [PubMed] [Google Scholar]
- 5.Krishnan V, Nestler EJ. Linking molecules to mood: new insight into the biology of depression. The American journal of psychiatry. 2010 Nov;167(11):1305–1320. doi: 10.1176/appi.ajp.2009.10030434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merighi A, Salio C, Ghirri A, Lossi L, Ferrini F, Betelli C, Bardoni R. BDNF as a pain modulator. Progress in neurobiology. 2008 Jul;85(3):297–317. doi: 10.1016/j.pneurobio.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Rios M, Fan G, Fekete C, Kelly J, Bates B, Kuehn R, Lechan RM, Jaenisch R. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Molecular endocrinology. 2001 Oct;15(10):1748–1757. doi: 10.1210/mend.15.10.0706. [DOI] [PubMed] [Google Scholar]
- 8.Neves-Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy JL. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. American journal of human genetics. 2002 Sep;71(3):651–655. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZY, Bath K, McEwen B, Hempstead B, Lee F. Impact of genetic variant BDNF (Val66Met) on brain structure and function. Novartis Foundation symposium. 2008;289:180–188. doi: 10.1002/9780470751251.ch14. discussion 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinawi M, Sahoo T, Maranda B, Skinner SA, Skinner C, Chinault C, Zascavage R, Peters SU, Patel A, Stevenson RE, Beaudet AL. 11p14.1 microdeletions associated with ADHD, autism, developmental delay, and obesity. American journal of medical genetics. Part A. 2011 Jun;155A(6):1272–1280. doi: 10.1002/ajmg.a.33878. [DOI] [PubMed] [Google Scholar]
- 11.Han JC, Liu QR, Jones M, Levinn RL, Menzie CM, Jefferson-George KS, Adler-Wailes DC, Sanford EL, Lacbawan FL, Uhl GR, Rennert OM, Yanovski JA. Brain-derived neurotrophic factor and obesity in the WAGR syndrome. The New England journal of medicine. 2008 Aug 28;359(9):918–927. doi: 10.1056/NEJMoa0801119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bremond-Gignac D, Crolla JA, Copin H, Guichet A, Bonneau D, Taine L, Lacombe D, Baumann C, Benzacken B, Verloes A. Combination of WAGR and Potocki-Shaffer contiguous deletion syndromes in a patient with an 11p11.2-p14 deletion. European journal of human genetics : EJHG. 2005 Apr;13(4):409–413. doi: 10.1038/sj.ejhg.5201358. [DOI] [PubMed] [Google Scholar]
- 13.Gray J, Yeo GS, Cox JJ, Morton J, Adlam AL, Keogh JM, Yanovski JA, El Gharbawy A, Han JC, Tung YC, Hodges JR, Raymond FL, O'Rahilly S, Farooqi IS. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes. 2006 Dec;55(12):3366–3371. doi: 10.2337/db06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licinio J, Dong C, Wong ML. Novel sequence variations in the brain-derived neurotrophic factor gene and association with major depression and antidepressant treatment response. Archives of general psychiatry. 2009 May;66(5):488–497. doi: 10.1001/archgenpsychiatry.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, Graham D, Tsankova NM, Bolanos CA, Rios M, Monteggia LM, Self DW, Nestler EJ. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006 Feb 10;311(5762):864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 16.Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006 Oct 6;314(5796):140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacQueen GM, Ramakrishnan K, Croll SD, Siuciak JA, Yu G, Young LT, Fahnestock M. Performance of heterozygous brain-derived neurotrophic factor knockout mice on behavioral analogues of anxiety, nociception, and depression. Behavioral neuroscience. 2001 Oct;115(5):1145–1153. doi: 10.1037//0735-7044.115.5.1145. [DOI] [PubMed] [Google Scholar]
- 18.Lyons WE, Mamounas LA, Ricaurte GA, Coppola V, Reid SW, Bora SH, Wihler C, Koliatsos VE, Tessarollo L. Brain-derived neurotrophic factor-deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proceedings of the National Academy of Sciences of the United States of America. 1999 Dec 21;96(26):15239–15244. doi: 10.1073/pnas.96.26.15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnan V, Han MH, Graham DL, Berton O, Renthal W, Russo SJ, Laplant Q, Graham A, Lutter M, Lagace DC, Ghose S, Reister R, Tannous P, Green TA, Neve RL, Chakravarty S, Kumar A, Eisch AJ, Self DW, Lee FS, Tamminga CA, Cooper DC, Gershenfeld HK, Nestler EJ. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007 Oct 19;131(2):391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Ernfors P, Lee KF, Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994 Mar 10;368(6467):147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- 21.Jones KR, Farinas I, Backus C, Reichardt LF. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994 Mar 25;76(6):989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballif BC, Theisen A, McDonald-McGinn DM, Zackai EH, Hersh JH, Bejjani BA, Shaffer LG. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin Genet. 2008 Nov;74(5):469–475. doi: 10.1111/j.1399-0004.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- 23.Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, Rosenfeld JA, Lamb AN, Sahoo T. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010 Nov;18(11):1196–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nature genetics. 2011 Sep;43(9):838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baldwin EL, Lee JY, Blake DM, Bunke BP, Alexander CR, Kogan AL, Ledbetter DH, Martin CL. Enhanced detection of clinically relevant genomic imbalances using a targeted plus whole genome oligonucleotide microarray. Genetics in medicine : official journal of the American College of Medical Genetics. 2008 Jun;10(6):415–429. doi: 10.1097/GIM.0b013e318177015c. [DOI] [PubMed] [Google Scholar]
- 26.Lionel AC, Crosbie J, Barbosa N, Goodale T, Thiruvahindrapuram B, Rickaby J, Gazzellone M, Carson AR, Howe JL, Wang Z, Wei J, Stewart AF, Roberts R, McPherson R, Fiebig A, Franke A, Schreiber S, Zwaigenbaum L, Fernandez BA, Roberts W, Arnold PD, Szatmari P, Marshall CR, Schachar R, Scherer SW. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Science translational medicine. 2011 Aug 10;3(95):95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- 27.Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, Holmes C, Marchini JL, Stirrups K, Tobin MD, Wain LV, Yau C, Aerts J, Ahmad T, Andrews TD, Arbury H, Attwood A, Auton A, Ball SG, Balmforth AJ, Barrett JC, Barroso I, Barton A, Bennett AJ, Bhaskar S, Blaszczyk K, Bowes J, Brand OJ, Braund PS, Bredin F, Breen G, Brown MJ, Bruce IN, Bull J, Burren OS, Burton J, Byrnes J, Caesar S, Clee CM, Coffey AJ, Connell JM, Cooper JD, Dominiczak AF, Downes K, Drummond HE, Dudakia D, Dunham A, Ebbs B, Eccles D, Edkins S, Edwards C, Elliot A, Emery P, Evans DM, Evans G, Eyre S, Farmer A, Ferrier IN, Feuk L, Fitzgerald T, Flynn E, Forbes A, Forty L, Franklyn JA, Freathy RM, Gibbs P, Gilbert P, Gokumen O, Gordon-Smith K, Gray E, Green E, Groves CJ, Grozeva D, Gwilliam R, Hall A, Hammond N, Hardy M, Harrison P, Hassanali N, Hebaishi H, Hines S, Hinks A, Hitman GA, Hocking L, Howard E, Howard P, Howson JM, Hughes D, Hunt S, Isaacs JD, Jain M, Jewell DP, Johnson T, Jolley JD, Jones IR, Jones LA, Kirov G, Langford CF, Lango-Allen H, Lathrop GM, Lee J, Lee KL, Lees C, Lewis K, Lindgren CM, Maisuria-Armer M, Maller J, Mansfield J, Martin P, Massey DC, McArdle WL, McGuffin P, McLay KE, Mentzer A, Mimmack ML, Morgan AE, Morris AP, Mowat C, Myers S, Newman W, Nimmo ER, O'Donovan MC, Onipinla A, Onyiah I, Ovington NR, Owen MJ, Palin K, Parnell K, Pernet D, Perry JR, Phillips A, Pinto D, Prescott NJ, Prokopenko I, Quail MA, Rafelt S, Rayner NW, Redon R, Reid DM, Renwick, Ring SM, Robertson N, Russell E, St Clair D, Sambrook JG, Sanderson JD, Schuilenburg H, Scott CE, Scott R, Seal S, Shaw-Hawkins S, Shields BM, Simmonds MJ, Smyth DJ, Somaskantharajah E, Spanova K, Steer S, Stephens J, Stevens HE, Stone MA, Su Z, Symmons DP, Thompson JR, Thomson W, Travers ME, Turnbull C, Valsesia A, Walker M, Walker NM, Wallace C, Warren-Perry M, Watkins NA, Webster J, Weedon MN, Wilson AG, Woodburn M, Wordsworth BP, Young AH, Zeggini E, Carter NP, Frayling TM, Lee C, McVean G, Munroe PB, Palotie A, Sawcer SJ, Scherer SW, Strachan DP, Tyler-Smith C, Brown MA, Burton PR, Caulfield MJ, Compston A, Farrall M, Gough SC, Hall AS, Hattersley AT, Hill AV, Mathew CG, Pembrey M, Satsangi J, Stratton MR, Worthington J, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand W, Parkes M, Rahman N, Todd JA, Samani NJ, Donnelly P. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010 Apr 1;464(7289):713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010 Sep 2;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, Piven J, Ponting CP, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Sequeira AF, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stein O, Sykes N, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Webber C, Weksberg R, Wing K, Wittemeyer K, Wood S, Wu J, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Devlin B, Ennis S, Gallagher L, Geschwind DH, Gill M, Haines JL, Hallmayer J, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Scherer SW, Sutcliffe JS, Betancur C. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010 Jul 15;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O'Hara R, Casalunovo T, Conlin LK, D'Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome research. 2009 Sep;19(9):1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008 Sep 11;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nature genetics. 2004 Sep;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 33.Miller RW, Fraumeni JF, Jr, Manning MD. Association of Wilms's Tumor with Aniridia, Hemihypertrophy and Other Congenital Malformations. The New England journal of medicine. 1964 Apr 30;270:922–927. doi: 10.1056/NEJM196404302701802. [DOI] [PubMed] [Google Scholar]
- 34.Kutsche K, Glauner E, Knauf S, Pomarino A, Schmidt M, Schroder B, Nothwang H, Schuler H, Goecke T, Kersten A, Althaus C, Gal A. Cloning and characterization of the breakpoint regions of a chromosome 11;18 translocation in a patient with hamartoma of the retinal pigment epithelium. Cytogenetics and cell genetics. 2000;91(1–4):141–147. doi: 10.1159/000056835. [DOI] [PubMed] [Google Scholar]
- 35.Mohri Y, Oyama K, Akamatsu A, Kato S, Nishimori K. Lgr4-deficient mice showed premature differentiation of ureteric bud with reduced expression of Wnt effector Lef1 and Gata3. Developmental dynamics : an official publication of the American Association of Anatomists. 2011 Jun;240(6):1626–1634. doi: 10.1002/dvdy.22651. [DOI] [PubMed] [Google Scholar]
- 36.Mendive F, Laurent P, Van Schoore G, Skarnes W, Pochet R, Vassart G. Defective postnatal development of the male reproductive tract in LGR4 knockout mice. Developmental biology. 2006 Feb 15;290(2):421–434. doi: 10.1016/j.ydbio.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 37.Mazerbourg S, Bouley DM, Sudo S, Klein CA, Zhang JV, Kawamura K, Goodrich LV, Rayburn H, Tessier-Lavigne M, Hsueh AJ. Leucine-rich repeat-containing, G protein-coupled receptor 4 null mice exhibit intrauterine growth retardation associated with embryonic and perinatal lethality. Molecular endocrinology. 2004 Sep;18(9):2241–2254. doi: 10.1210/me.2004-0133. [DOI] [PubMed] [Google Scholar]
- 38.Srinivasan K, Roosa J, Olsen O, Lee SH, Bredt DS, McConnell SK. MALS-3 regulates polarity and early neurogenesis in the developing cerebral cortex. Development. 2008 May;135(10):1781–1790. doi: 10.1242/dev.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misawa H, Kawasaki Y, Mellor J, Sweeney N, Jo K, Nicoll RA, Bredt DS. Contrasting localizations of MALS/LIN-7 PDZ proteins in brain and molecular compensation in knockout mice. The Journal of biological chemistry. 2001 Mar 23;276(12):9264–9272. doi: 10.1074/jbc.M009334200. [DOI] [PubMed] [Google Scholar]
- 40.Barron JL, Sandman CA. Self-injurious behavior and stereotypy in an institutionalized mentally retarded population. Applied research in mental retardation. 1984;5(4):499–511. doi: 10.1016/s0270-3092(84)80041-7. [DOI] [PubMed] [Google Scholar]
- 41.Holtzheimer PE, Mayberg HS. Stuck in a rut: rethinking depression and its treatment. Trends in neurosciences. 2011 Jan;34(1):1–9. doi: 10.1016/j.tins.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gul D, Ogur G, Tunca Y, Ozcan O. Third case of WAGR syndrome with severe obesity and constitutional deletion of chromosome (11)(p12p14) American journal of medical genetics. 2002 Jan 1;107(1):70–71. doi: 10.1002/ajmg.10013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.