Abstract
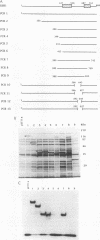
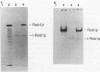
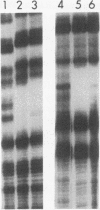
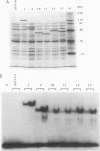
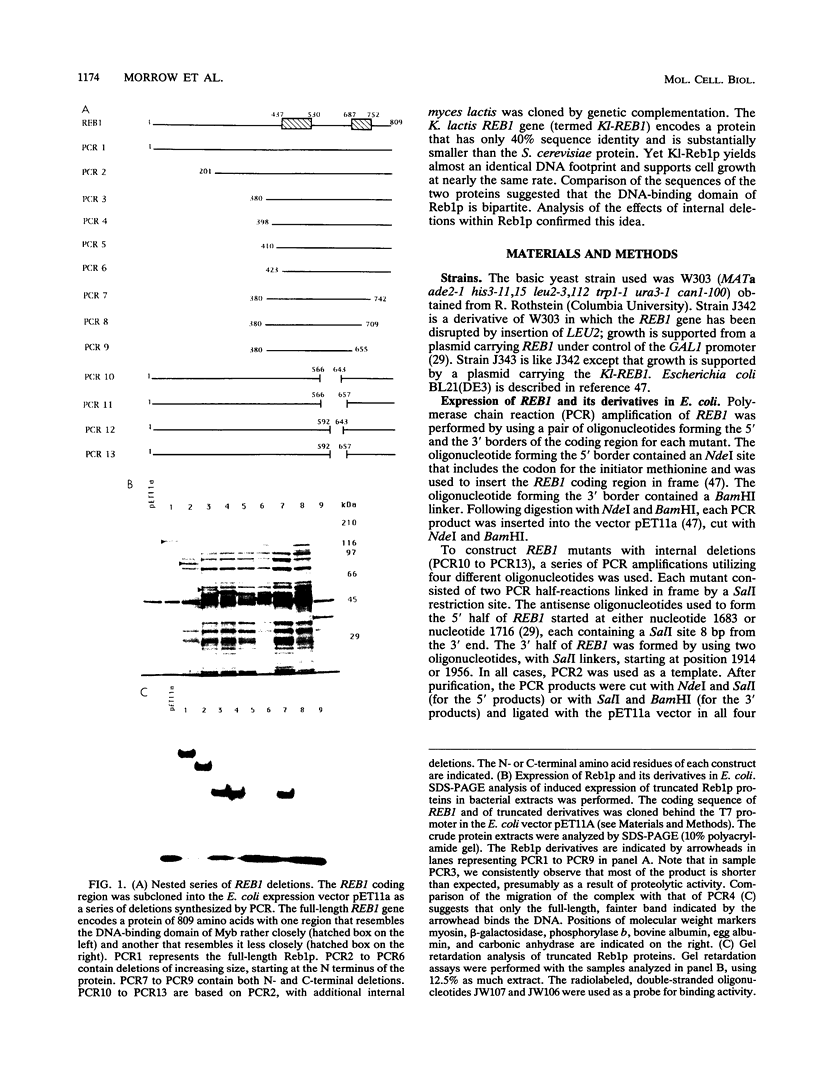
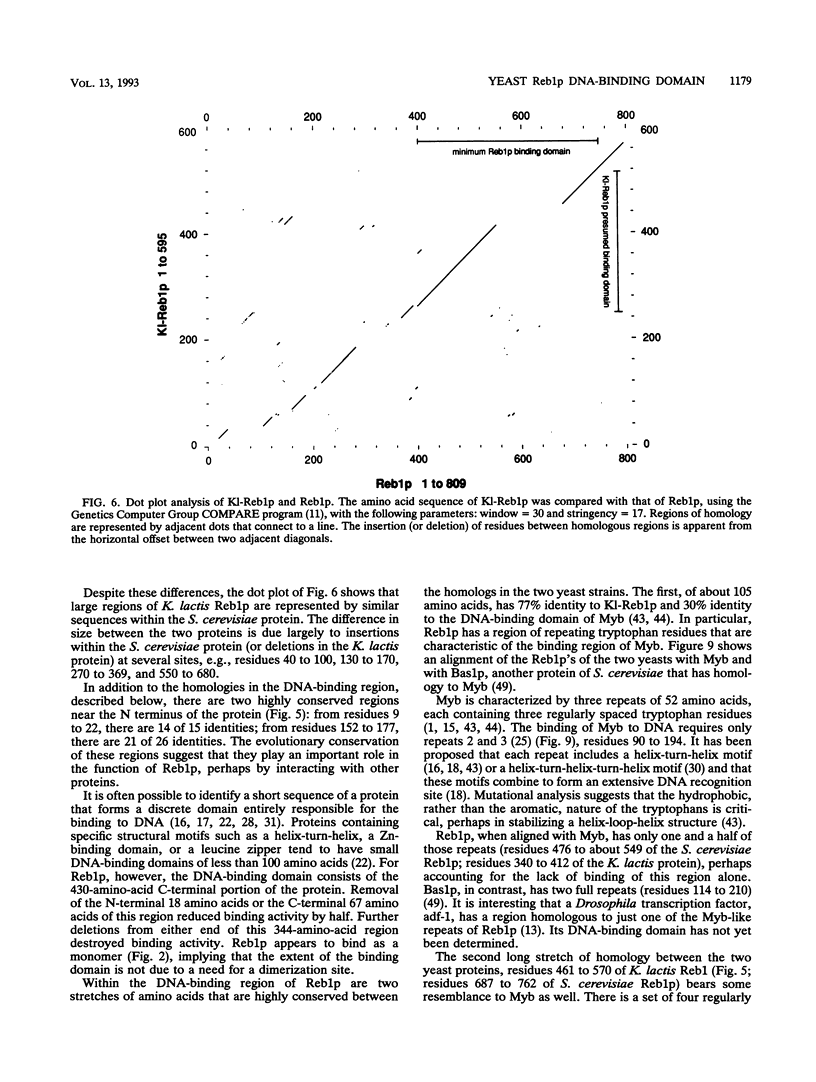
The REB1 gene encodes a DNA-binding protein (Reb1p) that is essential for growth of the yeast Saccharomyces cerevisiae. Reb1p binds to sites within transcriptional control regions of genes transcribed by either RNA polymerase I or RNA polymerase II. The sequence of REB1 predicts a protein of 809 amino acids. To define the DNA-binding domain of Reb1p, a series of 5' and 3' deletions within the coding region was constructed in a bacterial expression vector. Analysis of the truncated Reb1p proteins revealed that nearly 400 amino acids of the C-terminal portion of the protein are required for maximal DNA-binding activity. To further define the important structural features of Reb1p, the REB1 homolog from a related yeast, Kluyveromyces lactis, was cloned by genetic complementation. The K. lactis REB1 gene supports active growth of an S. cerevisiae strain whose REB1 gene has been deleted. The Reb1p proteins of the two organisms generate almost identical footprints on DNA, yet the K. lactis REB1 gene encodes a polypeptide of only 595 amino acids. Comparison of the two Reb1p sequences revealed that within the region necessary for the binding of Reb1p to DNA were two long regions of nearly perfect identity, separated in the S. cerevisiae Reb1p by nearly 150 amino acids but in the K. lactis Reb1p by only 40 amino acids. The first includes a 105-amino-acid region related to the DNA-binding domain of the myb oncoprotein; the second bears a faint resemblance to myb. The hypothesis that the DNA-binding domain of Reb1p is formed from these two conserved regions was confirmed by deletion of as many as 90 amino acids between them, with little effect on the DNA-binding ability of the resultant protein. We suggest that the DNA-binding domain of Reb1p is made up of two myb-like regions that, unlike myb itself, are separated by as many as 150 amino acids. Since Reb1p protects only 15 to 20 nucleotides in a chemical or enzymatic footprint assay, the protein must fold such that the two components of the binding site are adjacent.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anton I. A., Frampton J. Tryptophans in myb proteins. Nature. 1988 Dec 22;336(6201):719–719. doi: 10.1038/336719a0. [DOI] [PubMed] [Google Scholar]
- Aurora R., Herr W. Segments of the POU domain influence one another's DNA-binding specificity. Mol Cell Biol. 1992 Feb;12(2):455–467. doi: 10.1128/mcb.12.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns S. M., Lane D. J., Sogin M. L., Bibeau C., Weisburg W. G. Evolutionary relationships among pathogenic Candida species and relatives. J Bacteriol. 1991 Apr;173(7):2250–2255. doi: 10.1128/jb.173.7.2250-2255.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender T. P., Kuehl W. M. Murine myb protooncogene mRNA: cDNA sequence and evidence for 5' heterogeneity. Proc Natl Acad Sci U S A. 1986 May;83(10):3204–3208. doi: 10.1073/pnas.83.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedenkapp H., Borgmeyer U., Sippel A. E., Klempnauer K. H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988 Oct 27;335(6193):835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- Brandl C. J., Struhl K. A nucleosome-positioning sequence is required for GCN4 to activate transcription in the absence of a TATA element. Mol Cell Biol. 1990 Aug;10(8):4256–4265. doi: 10.1128/mcb.10.8.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I., Lue N. F., Buchman A. R., LaPointe J. W., Lorch Y., Kornberg R. D. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev. 1990 Apr;4(4):503–514. doi: 10.1101/gad.4.4.503. [DOI] [PubMed] [Google Scholar]
- Choe S. Y., Schultz M. C., Reeder R. H. In vitro definition of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1992 Jan 25;20(2):279–285. doi: 10.1093/nar/20.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques. 1989 May;7(5):514–520. [PubMed] [Google Scholar]
- Deshler J. O., Larson G. P., Rossi J. J. Kluyveromyces lactis maintains Saccharomyces cerevisiae intron-encoded splicing signals. Mol Cell Biol. 1989 May;9(5):2208–2213. doi: 10.1128/mcb.9.5.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989 Nov 24;246(4933):1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- England B. P., Admon A., Tjian R. Cloning of Drosophila transcription factor Adf-1 reveals homology to Myb oncoproteins. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):683–687. doi: 10.1073/pnas.89.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Frampton J., Gibson T. J., Ness S. A., Döderlein G., Graf T. Proposed structure for the DNA-binding domain of the Myb oncoprotein based on model building and mutational analysis. Protein Eng. 1991 Dec;4(8):891–901. doi: 10.1093/protein/4.8.891. [DOI] [PubMed] [Google Scholar]
- Frampton J., Leutz A., Gibson T., Graf T. DNA-binding domain ancestry. Nature. 1989 Nov 9;342(6246):134–134. doi: 10.1038/342134a0. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Kim P. S. Modular structure of transcription factors: implications for gene regulation. Cell. 1991 May 31;65(5):717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- Gabrielsen O. S., Sentenac A., Fromageot P. Specific DNA binding by c-Myb: evidence for a double helix-turn-helix-related motif. Science. 1991 Sep 6;253(5024):1140–1143. doi: 10.1126/science.1887237. [DOI] [PubMed] [Google Scholar]
- Gonda T. J., Gough N. M., Dunn A. R., de Blaquiere J. Nucleotide sequence of cDNA clones of the murine myb proto-oncogene. EMBO J. 1985 Aug;4(8):2003–2008. doi: 10.1002/j.1460-2075.1985.tb03884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves P. M., Maurer K., Mager W. H., Planta R. J. Kluyveromyces contains a functional ABF1-homologue. Nucleic Acids Res. 1992 May 11;20(9):2211–2215. doi: 10.1093/nar/20.9.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter H., Kavety B., Vandekerckhove J., Kiefer F., Gallwitz D. Sequence, expression and mutational analysis of BAF1, a transcriptional activator and ARS1-binding protein of the yeast Saccharomyces cerevisiae. EMBO J. 1989 Dec 20;8(13):4265–4272. doi: 10.1002/j.1460-2075.1989.tb08612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Henry Y. A., Chambers A., Tsang J. S., Kingsman A. J., Kingsman S. M. Characterisation of the DNA binding domain of the yeast RAP1 protein. Nucleic Acids Res. 1990 May 11;18(9):2617–2623. doi: 10.1093/nar/18.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K. M., Reakes C. F., Watson R. J. Characterization of the sequence-specific interaction of mouse c-myb protein with DNA. EMBO J. 1990 Jan;9(1):161–169. doi: 10.1002/j.1460-2075.1990.tb08092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Ju Q. D., Morrow B. E., Warner J. R. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol. 1990 Oct;10(10):5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei-Ishii C., Sarai A., Sawazaki T., Nakagoshi H., He D. N., Ogata K., Nishimura Y., Ishii S. The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J Biol Chem. 1990 Nov 15;265(32):19990–19995. [PubMed] [Google Scholar]
- Katzen A. L., Kornberg T. B., Bishop J. M. Isolation of the proto-oncogene c-myb from D. melanogaster. Cell. 1985 Jun;41(2):449–456. doi: 10.1016/s0092-8674(85)80018-0. [DOI] [PubMed] [Google Scholar]
- Kulkens T., Riggs D. L., Heck J. D., Planta R. J., Nomura M. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 1991 Oct 11;19(19):5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkens T., van Heerikhuizen H., Klootwijk J., Oliemans J., Planta R. J. A yeast ribosomal DNA-binding protein that binds to the rDNA enhancer and also close to the site of Pol I transcription initiation is not important for enhancer functioning. Curr Genet. 1989 Dec;16(5-6):351–359. doi: 10.1007/BF00340714. [DOI] [PubMed] [Google Scholar]
- Kulkens T., van der Sande C. A., Dekker A. F., van Heerikhuizen H., Planta R. J. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992 Dec;11(12):4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. J., Sweet D. J., Pelham H. R. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell. 1990 Jun 29;61(7):1359–1363. doi: 10.1016/0092-8674(90)90699-f. [DOI] [PubMed] [Google Scholar]
- Majello B., Kenyon L. C., Dalla-Favera R. Human c-myb protooncogene: nucleotide sequence of cDNA and organization of the genomic locus. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9636–9640. doi: 10.1073/pnas.83.24.9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow B. E., Johnson S. P., Warner J. R. Proteins that bind to the yeast rDNA enhancer. J Biol Chem. 1989 May 25;264(15):9061–9068. [PubMed] [Google Scholar]
- Morrow B. E., Ju Q., Warner J. R. Purification and characterization of the yeast rDNA binding protein REB1. J Biol Chem. 1990 Dec 5;265(34):20778–20783. [PubMed] [Google Scholar]
- Musters W., Knol J., Maas P., Dekker A. F., van Heerikhuizen H., Planta R. J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989 Dec 11;17(23):9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Wienand U., Peterson P. A., Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987 Dec 1;6(12):3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle J. E., Holmberg S. A REB1-binding site is required for GCN4-independent ILV1 basal level transcription and can be functionally replaced by an ABF1-binding site. Mol Cell Biol. 1992 Dec;12(12):5516–5526. doi: 10.1128/mcb.12.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode P. R., Sweder K. S., Oegema K. F., Campbell J. L. The gene encoding ARS-binding factor I is essential for the viability of yeast. Genes Dev. 1989 Dec;3(12A):1926–1939. doi: 10.1101/gad.3.12a.1926. [DOI] [PubMed] [Google Scholar]
- Rosson D., Reddy E. P. Nucleotide sequence of chicken c-myb complementary DNA and implications for myb oncogene activation. Nature. 1986 Feb 13;319(6054):604–606. doi: 10.1038/319604a0. [DOI] [PubMed] [Google Scholar]
- Saikumar P., Murali R., Reddy E. P. Role of tryptophan repeats and flanking amino acids in Myb-DNA interactions. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8452–8456. doi: 10.1073/pnas.87.21.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H., Kanei-Ishii C., Nagase T., Nakagoshi H., Gonda T. J., Ishii S. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5758–5762. doi: 10.1073/pnas.86.15.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sturm R. A., Herr W. The POU domain is a bipartite DNA-binding structure. Nature. 1988 Dec 8;336(6199):601–604. doi: 10.1038/336601a0. [DOI] [PubMed] [Google Scholar]
- Tice-Baldwin K., Fink G. R., Arndt K. T. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science. 1989 Nov 17;246(4932):931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- Wang H., Nicholson P. R., Stillman D. J. Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol Cell Biol. 1990 Apr;10(4):1743–1753. doi: 10.1128/mcb.10.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]