Abstract
Objective:
To quantify the regional and global cerebral atrophy rates and assess acceleration rates in healthy controls, subjects with mild cognitive impairment (MCI), and subjects with mild Alzheimer disease (AD).
Methods:
Using 0-, 6-, 12-, 18-, 24-, and 36-month MRI scans of controls and subjects with MCI and AD from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database, we calculated volume change of whole brain, hippocampus, and ventricles between all pairs of scans using the boundary shift integral.
Results:
We found no evidence of acceleration in whole-brain atrophy rates in any group. There was evidence that hippocampal atrophy rates in MCI subjects accelerate by 0.22%/year2 on average (p = 0.037). There was evidence of acceleration in rates of ventricular enlargement in subjects with MCI (p = 0.001) and AD (p < 0.001), with rates estimated to increase by 0.27 mL/year2 (95% confidence interval 0.12, 0.43) and 0.88 mL/year2 (95% confidence interval 0.47, 1.29), respectively. A post hoc analysis suggested that the acceleration of hippocampal loss in MCI subjects was mainly driven by the MCI subjects that were observed to progress to clinical AD within 3 years of baseline, with this group showing hippocampal atrophy rate acceleration of 0.50%/year2 (p = 0.003).
Conclusions:
The small acceleration rates suggest a long period of transition to the pathologic losses seen in clinical AD. The acceleration in hippocampal atrophy rates in MCI subjects in the ADNI seems to be driven by those MCI subjects who concurrently progressed to a clinical diagnosis of AD.
Alzheimer disease (AD) is associated with higher rates of brain tissue loss than normal aging,1 with the rate in mild cognitive impairment (MCI) falling somewhere in between.2 This implies increases in the rate of tissue loss (i.e., acceleration) as the individual progresses from normal to MCI and then on to dementia due to AD.3,4 A sigmoidal model of tissue loss in AD5 has been proposed that suggests the rate of tissue loss first accelerates, then remains constant, and finally decelerates; however, detailed data about how the rates of regional and global atrophy change over time are lacking. Assessing how rates of atrophy change in the transition from normal aging through early cognitive impairment and on to dementia due to AD is important for understanding the presymptomatic period of AD, which is increasingly considered as a potential therapeutic window for disease-modification trials.
A number of studies have investigated acceleration in regional and global atrophy rates in normal aging, MCI, and clinically diagnosed AD, with most modeling tissue loss as a function of age or time from a particular clinical stage (e.g., at the time of AD dementia diagnosis or Mini-Mental State Examination [MMSE] score).3,4,6–8 Other studies have modeled the tissue loss as a function of time from study baseline.9–11 The Alzheimer's Disease Neuroimaging Initiative (ADNI) provides an opportunity to investigate the time course of atrophy because of its large size and consistent longitudinal imaging. Herein, we use data from the ADNI to quantify the regional and global cerebral atrophy rates and acceleration in rates in healthy controls and MCI subjects with up to 3 years of follow-up and subjects clinically diagnosed with mild AD with up to 2 years of follow-up.
METHODS
Subjects.
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.ucla.edu) (see appendix e-1 on the Neurology® Web site at www.neurology.org). For up-to-date information, see www.adni-info.org.
Our image data consisted of 0-, 6-, 12-, 18-, 24-, and 36-month T1-weighted magnetic resonance scans of healthy controls, subjects with MCI, and subjects with clinically diagnosed AD downloaded from the ADNI database (http://www.loni.ucla.edu/ADNI) over the course of the ADNI study, with the latest download in December 2011. The individual ADNI subject identifiers and time points used in this study can be found in appendix e-2.
Standard protocol approvals, registrations, and patient consents.
The ADNI was approved by the institutional review board at each site and was compliant with the Health Insurance Portability and Accountability Act. Written consent was obtained from all participants.
Image acquisition and preprocessing.
Each individual was scanned with a number of sequences, but for this study we used the T1-weighted volumetric scans. Representative imaging parameters were as follows: repetition time = 2,300 milliseconds; inversion time = 1,000 milliseconds; echo time = 3.5 milliseconds; flip angle = 8°; field of view = 240 × 240 mm; and 160 sagittal 1.2-mm-thick slices and a 192 × 192 matrix yielding a voxel resolution of 1.25 × 1.25 × 1.2 mm, or 180 sagittal 1.2-mm-thick slices with a 256 × 256 matrix yielding a voxel resolution of 0.94 × 0.94 × 1.2 mm. The full details of the ADNI MRI protocol have been previously described,12 and are listed on the ADNI Web site (http://www.loni.ucla.edu/ADNI/Research/Cores/). Each examination underwent a quality-control evaluation at the Mayo Clinic (Rochester, MN). Quality control included inspection of each incoming image file for protocol compliance, clinically significant medical abnormalities, and image quality. The images also underwent internal quality control at the Dementia Research Centre, and those with significant motion artifacts causing severe blurring at the tissue boundaries were excluded from the current study. Therefore, only subjects with a usable baseline scan and at least one usable follow-up scan were included in the current study. The T1-weighted volumetric scans that passed the quality control were processed using the standard ADNI image processing pipeline, which included the corrections of gradient warping,13 B1 nonuniformity,14 intensity nonuniformity,15 and scanner-drift scaling.16
Volume-loss measurement.
Volume loss in the whole brain, hippocampus, and lateral ventricles between every pair of scans for each subject was measured using the robust boundary shift integral (referred to as KN-BSI),17,18 as described below. The whole brain and the hippocampi in each image were automatically delineated using multiple atlas propagation and segmentation,19,20 and the lateral ventricles were semiautomatically delineated using the Medical Image Display and Analysis Software.21 The images from all time points of each subject were affinely registered into a middle position,22 in order to provide consistent volume-loss estimation over multiple time points. Symmetric differential bias correction was performed on the transformed images.22,23 Volume loss between any 2 images was calculated by KN-BSI18 over the whole brain, the hippocampus, and the lateral ventricles.
Statistical analysis.
Baseline characteristics of subjects with usable scans were compared with those of subjects without usable scans, separately in each group, using t tests (for age), Wilcoxon rank sum tests (for MMSE), and Fisher exact tests (for gender and APOE ε4 status).
Separately for each group, annualized atrophy rates were calculated for 0–12, 12–24, and 24–36 months using the corresponding scan pairs, accounting for the interval in days between the scans. Whole-brain and hippocampal atrophy rates were expressed as percentage of initial volume, whereas the ventricular expansion rate was expressed as absolute loss in milliliters. We used the sum of the volume losses in the left and right hippocampi as the total loss in the hippocampus. Separately in each subject group, logistic regression models were used to investigate whether atrophy during 0–12 months was predictive of availability of scan pairs between 12–24 and 24–36 months.
To test for and estimate acceleration in atrophy rates, linear mixed models were fitted separately in each subject group to all of the available direct measures of loss between scans from 2 time points.24 The model contained fixed effects of time and time squared (to allow for acceleration), and random subject effects of time. The latter accommodates the repeated measures from subjects, and allows the (linear) atrophy rate to vary between subjects. No constant terms were included, consistent with an assumption that the loss between 2 scans acquired on the same day should on average be zero. We fitted models on the logarithmic scale for the whole brain and hippocampus, where zero acceleration corresponds to constant percentage atrophy per year and on the absolute scale for the ventricles, where zero acceleration implies a constant absolute loss per year. Models were fitted by maximum likelihood using the xtmixed command in Stata 12.1 (StataCorp, College Station, TX). Maximum likelihood estimates are consistent provided that atrophy measures are missing at random (i.e., the probability that a measurement is missing is independent of the measurement itself, given observed data).25
RESULTS
We downloaded MRI scans of 840 subjects, and excluded 74 subjects who had only baseline scans and 53 subjects who failed our internal quality control. Although there was evidence (p = 0.047) that APOE ε4 status of the excluded AD subjects (16 noncarriers, 18 heterozygotes, and 2 homozygotes) was different from the included AD subjects (49 noncarriers, 73 heterozygotes, and 34 homozygotes), there was no evidence that the 713 subjects included in this study were different from the 127 excluded subjects regarding age, MMSE score, gender, or APOE ε4 status in other groups (p > 0.10, all other tests). Table 1 shows the demographics of the 713 subjects and the number of scan pairs available (for consecutive scans) by subject group. As expected, the number of subjects with scans available decreased as the study progressed. Although not statistically significant, logistic regression analyses (run separately by subject group) suggested that those controls and AD subjects with higher whole-brain atrophy rates between 0 and 12 months were less likely to have a 12- to 24-month scan pair available (odds ratio for 1% increase in 0- to 12-month brain atrophy in controls 0.81 [95% confidence interval {CI} 0.42, 1.54], p = 0.52, and in AD subjects 0.70 [95% CI 0.42, 1.17], p = 0.18). There was also the suggestion that those MCI and AD subjects with higher rates of brain atrophy between 0 and 12 months were less likely to have a 24- to 36-month scan pair available (odds ratio in controls 0.71 [95% CI 0.41, 1.22], p = 0.21, and in MCI subjects 0.83 [95% CI 0.62, 1.13], p = 0.24). This suggests that those subjects who underwent scanning at 24 and 36 months may have systematically lower whole-brain atrophy rates than those who did not, and that estimates of atrophy rates in the latter part of the study based on those with scans available at the later time points may be biased as estimates of the population mean rate.
Table 1.
Demographics and MRI data of controls and subjects with MCI and AD for the main analysis, and MCI-S and MCI-P subjects for the post hoc analysisa
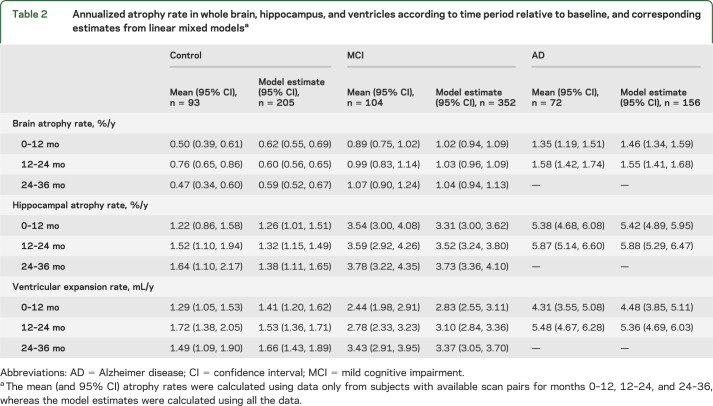
In table 2, we report mean (and 95% CI) atrophy rates using data only from subjects who had scan pairs available for all periods: 0–12, 12–24, and 24–36 months. Although these rates may be biased downward, they are more likely to give an unbiased representation of how rates change over time than if rates were presented for each period based on scan pairs available for that period. A further limitation of these estimates is that they are based on a much smaller subset of subjects, and consequently they are less precise. Table 2 also shows linear mixed model–based estimates of atrophy rate for the period, using information from all subjects. As expected, atrophy rates (and ventricular enlargement) are highest in AD subjects, lowest in controls, with MCI subjects in between. Without considering statistical significance, the means in table 2 suggest that atrophy rates (brain, hippocampal, and ventricular enlargement) are increasing in both the MCI and AD subjects over time. In controls, the only consistent increase is for hippocampal rates.
Table 2.
Annualized atrophy rate in whole brain, hippocampus, and ventricles according to time period relative to baseline, and corresponding estimates from linear mixed modelsa
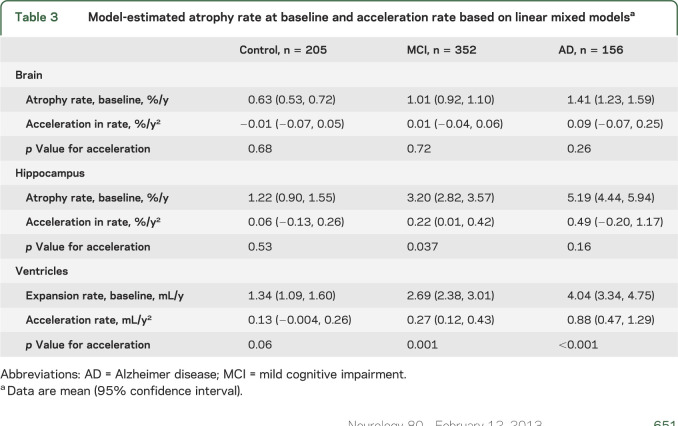
Table 3 shows the model-estimated atrophy rates and acceleration, and plots of the corresponding data on an individual basis are included in the supplemental material (see figures e-1 and e-2). For whole-brain atrophy, there was no evidence of acceleration (on average) in any group. Furthermore, the 95% CIs for the acceleration are narrow for controls and MCIs, suggesting we can be reasonably confident that if average whole-brain atrophy acceleration differs in truth from zero in these groups, it is very close to zero. There was statistically significant evidence of acceleration for hippocampal volume loss in MCI subjects (p = 0.037), with rates estimated to increase on average (across subjects) by 0.22%/year2 (95% CI 0.01, 0.42). There was evidence of acceleration in rates of ventricular enlargement in MCI (p = 0.001) and AD (p < 0.001) subjects, with rates estimated to increase by 0.27 mL/year2 (95% CI 0.12, 0.43) and 0.88 mL/year2 (95% CI 0.47, 1.29) in MCI and AD subjects, respectively.
Table 3.
Model-estimated atrophy rate at baseline and acceleration rate based on linear mixed modelsa
Post hoc analysis.
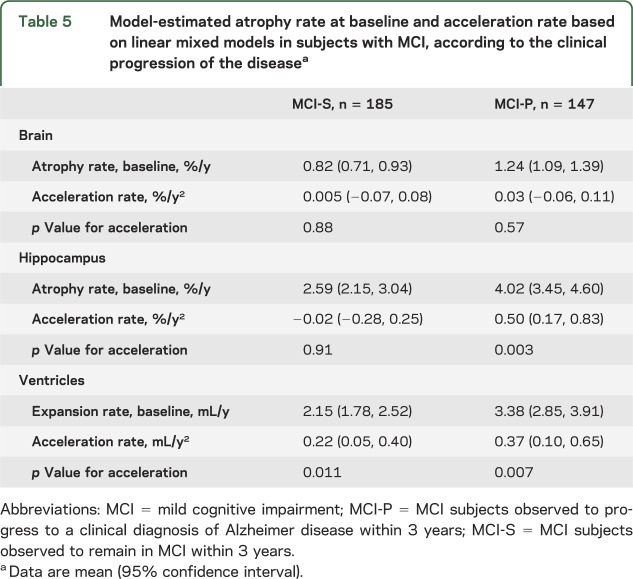
We hypothesized that the observed acceleration in hippocampal rates in MCI subjects was a manifestation of those MCI subjects who progressed to a clinical diagnosis of AD during the study. We therefore performed a post hoc analysis in 2 subgroups of MCI subjects: MCI-P (MCI subjects that progressed to a clinical diagnosis of AD within 3 years) and MCI-S (MCI subjects that remained in MCI within 3 years). Table 1 shows the demographics of the subjects according to the clinical progression of the disease, MMSE score at baseline, and the number of scan pairs available. The mean atrophy rates of the MCI-S subjects were more similar to controls, whereas the mean atrophy rates of MCI-P subjects were more similar to AD subjects (see table 4). There was statistically significant evidence of acceleration for hippocampal loss in MCI-P subjects (0.50%/year2 [95% CI 0.17, 0.83], p = 0.003), but not in MCI-S subjects (−0.02%/year2 [95% CI −0.28, 0.25], p = 0.91) (see table 5), and an interaction test formally comparing these showed evidence that they differed (p = 0.038). Although not statistically significantly different (p = 0.38), the estimated acceleration in ventricular expansion was also greater in the MCI-P subjects (0.37 mL/year2 [95% CI 0.10, 0.65], p = 0.007) than in the MCI-S subjects (0.22 mL/year2 [95% CI 0.05, 0.40], p = 0.011).
Table 4.
Annualized atrophy rate for subjects with MCI, according to the clinical progression of the diseasea
Table 5.
Model-estimated atrophy rate at baseline and acceleration rate based on linear mixed models in subjects with MCI, according to the clinical progression of the diseasea
DISCUSSION
Using data from the ADNI, we have investigated acceleration in rates of whole-brain and hippocampal atrophy and ventricular enlargement, using 3-year follow-up data on controls and subjects with MCI and 2-year follow-up data for subjects with AD. We have assessed acceleration using all available data by using linear mixed models. The clinically defined groups of subjects—controls, MCI, and AD—showed the expected group differences in mean rates of atrophy at baseline: brain atrophy rates were ∼2.4 times greater in the AD subjects than in the controls, ventricular enlargement was ∼3 times greater, and hippocampal atrophy was ∼4.3 times greater (table 3). The MCI group rates fell almost exactly halfway between the AD and control means for each of the measures.
Hippocampal atrophy in subjects with MCI was estimated to accelerate by 0.22%/year2 (p = 0.037) on average. Furthermore, in the subjects with MCI who subsequently were given a diagnosis of AD, the acceleration was twice this at 0.50%/year2 (p = 0.003) and was very similar to that seen in the AD group (0.49%/year2, p = 0.16). This suggests that the observed acceleration of hippocampal loss in MCI subjects is mainly driven by the MCI subjects that were observed to progress to clinical AD within 3 years. Although the estimated hippocampal acceleration in AD subjects was moderately large, there was no statistically significant evidence that it differed from zero. However, this may be because of reduced power due to the shorter period of follow-up (only 2 years as opposed to 3 for the MCI group) and/or greater heterogeneity in the AD group, e.g., a proportion of AD subjects may no longer be accelerating in hippocampal atrophy as suggested in the sigmoid model.5 As for ventricular expansion, the rates in MCI and AD subjects were estimated to accelerate at 0.27 and 0.88 mL/year2, respectively.
Previous studies investigating the time course of hippocampal volume loss as a function of time from baseline have reported conflicting results.9,10,26 Using 0-, 6-, and 12-month magnetic resonance scans from the ADNI, hippocampal atrophy rates were estimated to accelerate by 26.5 mm3/year2 (or 1.6%) in subjects with AD and 12.1 mm3/year2 (or 0.6%) in subjects with MCI, but remained constant in controls.9 Another study found that hippocampal atrophy rate in controls (aged 49–85 years) accelerated from 30 mm3/year to 124 mm3/year in approximately 15 months,10 although the annual percentage change from the first to second follow-up was approximately 4%, which was similar to the rates in AD subjects reported by an ADNI study.9 Such acceleration may be driven by the older control subjects. In addition, accelerating hippocampal atrophy was found in the control, MCI, and AD subjects with AD-like CSF molecular profiles using 0-, 6-, and 12-month magnetic resonance scans from ADNI.11 However, using 0-, 6-, and 12-month magnetic resonance scans from the controls and MCI subjects in ADNI, the intercept of the regression line assuming no acceleration fitted to the volume loss of the hippocampus at 6 and 12 months was close to zero.26 This implies that the atrophy rates in controls and MCI subjects stay nearly the same over 1 year. Differences in results between studies regarding acceleration may be attributable to differences in the number of subjects, the number of time points, and the hippocampal segmentation protocol used. To address the latter, the “harmonization of hippocampal segmentation” project was recently set up to develop a harmonized protocol that aims to reduce the present heterogeneity in hippocampal segmentation protocols.27
Many studies have investigated the time course of brain tissue loss as a function of age or other characteristics.3,4,6–8,28,29 Using data from young adult familial AD patients, hippocampal atrophy was estimated to accelerate by 0.40%/year2 (95% CI 0.17, 0.63), which is similar to our estimate of 0.49%/year2.4 In the same study, the authors found evidence that whole-brain atrophy rates accelerated by 0.26%/year2 (95% CI 0.16, 0.37), an estimate that falls just outside the 95% CI we obtained in this study. This might be attributable to a more aggressive onset to the familial disease—perhaps consistent with an age at onset that is some 30 or 40 years earlier than the age at onset in late-onset sporadic AD in ADNI—or to the fact that the clinically defined AD (dementia thought to be due to AD) group in ADNI inevitably contains a proportion of subjects who do not have AD pathologically.
Recently it was hypothesized that the dynamics of cerebral tissue loss within individuals follows a nonlinear and sigmoidal trajectory.5 Several studies have attempted to fit sigmoidal models for the trajectory of cerebral tissue loss (e.g., hippocampus and cortical thickness) using cross-sectional data from ADNI.11,30,31 The sigmoidal model implies that atrophy rates within a subject initially increase from zero, remain constant for a period, and then eventually decrease to zero. In the current study, we modeled the trajectory of cerebral tissue loss in each disease group using longitudinal data from ADNI over a 2- or 3-year period. Our linear mixed model allowed for atrophy rates to vary between subjects, but assumed that any acceleration was the same for all subjects. Further analysis with longer follow-up will permit models to be fitted that allow acceleration (or deceleration) in rates to vary both within and between subjects, which may provide evidence regarding the validity of different models of progression.
Our analysis has a number of strengths and limitations. Statistical power to detect acceleration in atrophy rates is strongly affected by the length of time over which subjects are followed, which was relatively short, particularly for the AD subjects. Despite this, our estimates of acceleration have relatively narrow CIs. A proportion of subjects in each group had missing scans at one or more follow-up time points, causing a loss of information and potentially biasing our results. However, we have used a statistical modeling technique that accommodates such missingness under the missing at random assumption. We have used a consistent atrophy measurement technique that treats each time point equally, which avoids bias toward any particular time point. Our analysis has used data from a large number of subjects with standardized and consistent protocols used for imaging and diagnosis.
Overall, we have found evidence of acceleration in hippocampal atrophy rates in subjects with MCI in the ADNI. This acceleration seems to be driven by those subjects with MCI who subsequently received a clinical diagnosis of AD dementia within 3 years. The rate of acceleration of hippocampal atrophy is relatively slow compared with the difference in atrophy rates in controls and clinical AD. This suggests a long period of transition to the pathologic losses seen in clinical AD, provided acceleration in rates is constant. Rates of ventricular enlargement were found to increase over time in both MCI and AD subjects. Our findings give further motivation for future longitudinal studies that will enable investigation of how atrophy rates evolve throughout the course of AD.
Supplementary Material
ACKNOWLEDGMENT
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (NIH grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the NIH (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514. The research was supported by the National Institute for Health Research Biomedical Research Unit in Dementia based at UCLH/UCL, the Medical Research Council, Alzheimer's Research UK, and ADNI. The Dementia Research Centre is an Alzheimer's Research UK Co-ordinating Centre and has also received equipment funded by the Alzheimer's Research UK. The authors thank the image analysts and the research associates in the Dementia Research Centre for their help in the study, and the ADNI study subjects and investigators for their participation.
Glossary
- AD
Alzheimer disease
- ADNI
Alzheimer's Disease Neuroimaging Initiative
- CI
confidence interval
- MCI
mild cognitive impairment
- MCI-P
mild cognitive impairment–progressed
- MCI-S
mild cognitive impairment–static
- MMSE
Mini-Mental State Examination
- KN-BSI
robust boundary shift integral
Footnotes
Supplemental data at www.neurology.org
AUTHOR CONTRIBUTIONS
Dr. Leung performed the image analysis and wrote the manuscript. Dr. Bartlett performed the statistical analysis, gave advice, and edited the manuscript. Dr. Barnes, Ms. Manning, and Dr. Ourselin gave advice and edited the manuscript. Dr. Fox conceived the study, gave advice, and edited the manuscript.
STUDY FUNDING
No targeted funding reported.
DISCLOSURE
K. Leung, J. Bartlett, J. Barnes, E. Manning, and S. Ourselin report no disclosure. N. Fox reports that his research has received payment for consultancy or for image analysis services from Avid/Lilly, BMS, Elan/Janssen, GE, Lundbeck, and Pfizer/Wyeth. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Fox NC, Schott JM. Imaging cerebral atrophy: normal ageing to Alzheimer's disease. Lancet 2004;363:392–394. [DOI] [PubMed] [Google Scholar]
- 2.Jack CR, Jr, Shiung MM, Gunter JL, et al. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology 2004;62:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jack CR, Jr, Weigand SD, Shiung MM, et al. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology 2008;70(19 pt 2):1740–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridha BH, Barnes J, Bartlett JW, et al. Tracking atrophy progression in familial Alzheimer's disease: a serial MRI study. Lancet Neurol 2006;5:828–834. [DOI] [PubMed] [Google Scholar]
- 5.Jack CR, Jr, Knopman DS, Jagust WJ, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol 2010;9:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Driscoll I, Davatzikos C, An Y, et al. Longitudinal pattern of regional brain volume change differentiates normal aging from MCI. Neurology 2009;72:1906–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald CR, McEvoy LK, Gharapetian L, et al. Regional rates of neocortical atrophy from normal aging to early Alzheimer disease. Neurology 2009;73:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson NE, Moore MM, Dame A, et al. Trajectories of brain loss in aging and the development of cognitive impairment. Neurology 2008;70:828–833. [DOI] [PubMed] [Google Scholar]
- 9.Schuff N, Woerner N, Boreta L, et al. MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain 2009;132(pt 4):1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raz N, Ghisletta P, Rodrigue KM, Kennedy KM, Lindenberger U. Trajectories of brain aging in middle-aged and older adults: regional and individual differences. Neuroimage 2010;51:501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabuncu MR, Desikan RS, Sepulcre J, et al. The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Arch Neurol 2011;68:1040–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jack CR, Jr, Bernstein MA, Fox NC, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008;27:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jovicich J, Czanner S, Greve D, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage 2006;30:436–443. [DOI] [PubMed] [Google Scholar]
- 14.Narayana PA, Brey WW, Kulkarni MV, Sievenpiper CL. Compensation for surface coil sensitivity variation in magnetic resonance imaging. Magn Reson Imaging 1988;6:271–274. [DOI] [PubMed] [Google Scholar]
- 15.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998;17:87–97. [DOI] [PubMed] [Google Scholar]
- 16.Gunter JL, Bernstein MA, Borowski BJ, et al. Validation testing of the MRI calibration phantom for the Alzheimer's Disease Neuroimaging Initiative Study. ISMRM 14th Scientific Meeting and Exhibition; 2006.
- 17.Freeborough PA, Fox NC. The boundary shift integral: an accurate and robust measure of cerebral volume changes from registered repeat MRI. IEEE Trans Med Imaging 1997;16:623–629. [DOI] [PubMed] [Google Scholar]
- 18.Leung KK, Clarkson MJ, Bartlett JW, et al. Robust atrophy rate measurement in Alzheimer's disease using multi-site serial MRI: tissue-specific intensity normalization and parameter selection. Neuroimage 2010;50:516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung KK, Barnes J, Modat M, et al. Brain MAPS: an automated, accurate and robust brain extraction technique using a template library. Neuroimage 2011;55:1091–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung KK, Barnes J, Ridgway GR, et al. Automated cross-sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and Alzheimer's disease. Neuroimage 2010;51:1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeborough PA, Fox NC, Kitney RI. Interactive algorithms for the segmentation and quantitation of 3-D MRI brain scans. Comput Methods Programs Biomed 1997;53:15–25. [DOI] [PubMed] [Google Scholar]
- 22.Leung KK, Ridgway GR, Ourselin S, Fox NC. Consistent multi-time-point brain atrophy estimation from the boundary shift integral. Neuroimage 2012;59:3995–4005. [DOI] [PubMed] [Google Scholar]
- 23.Lewis EB, Fox NC. Correction of differential intensity inhomogeneity in longitudinal MR images. Neuroimage 2004;23:75–83. [DOI] [PubMed] [Google Scholar]
- 24.Frost C, Kenward MG, Fox NC. The analysis of repeated ‘direct' measures of change illustrated with an application in longitudinal imaging. Stat Med 2004;23:3275–3286. [DOI] [PubMed] [Google Scholar]
- 25.Verbeke G, Molenberghs G. Linear Mixed Models for Longitudinal Data. New York: Springer; 2000. [Google Scholar]
- 26.Yushkevich PA, Avants BB, Das SR, Pluta J, Altinay M, Craige C. Bias in estimation of hippocampal atrophy using deformation-based morphometry arises from asymmetric global normalization: an illustration in ADNI 3 T MRI data. Neuroimage 2010;50:434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: a mandatory step for wide clinical use. Alzheimers Dement 2011;7:171–174. [DOI] [PubMed] [Google Scholar]
- 28.Chan D, Janssen JC, Whitwell JL, et al. Change in rates of cerebral atrophy over time in early-onset Alzheimer's disease: longitudinal MRI study. Lancet 2003;362:1121–1122. [DOI] [PubMed] [Google Scholar]
- 29.Mori E, Lee K, Yasuda M, et al. Accelerated hippocampal atrophy in Alzheimer's disease with apolipoprotein E epsilon4 allele. Ann Neurol 2002;51:209–214. [DOI] [PubMed] [Google Scholar]
- 30.Schuff N, Tosun D, Insel PS, et al. Nonlinear time course of brain volume loss in cognitively normal and impaired elders. Neurobiol Aging 2010;33:845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caroli A, Frisoni GB. The dynamics of Alzheimer's disease biomarkers in the Alzheimer's Disease Neuroimaging Initiative cohort. Neurobiol Aging 2010;31:1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.