Summary
In Gram-positive bacteria proteins are displayed on the cell surface using sortase enzymes. These cysteine transpeptidases join proteins bearing an appropriate sorting signal to strategically positioned amino groups on the cell surface. Working alone, or in concert with other enzymes, sortases either attach proteins to the cross-bridge peptide of the cell wall or they link proteins together to form pili. Because surface proteins play a fundamental role in microbial physiology and are frequently virulence factors, sortase enzymes have been intensely studied since their discovery a little more than a decade ago. Based on their primary sequences and functions sortases can be partitioned into distinct families called class A to F enzymes. Most bacteria elaborate their surfaces using more than one type of sortase that function non-redundantly by recognizing unique sorting signals within their protein substrates. Here we review what is known about the functions of these enzymes and the molecular basis of catalysis. Particular emphasis is placed on ‘pilin’ specific class C sortases that construct structurally complex pili. Exciting new data have revealed that these enzymes are amazingly promiscuous in the substrates that they can employ and that there is a startling degree of diversity in their mechanism of action. We also review recent data that suggest that sortases are targeted to specific sites on the cell surface where they work with other sortases and accessory factors to properly function.
Introduction
Sortase enzymes decorate the surfaces of Gram-positive bacteria with a diverse array of proteins that enable each microbe to effectively interact with its environment. They either ‘sort’ proteins to the cell surface by covalently joining them to the cell wall, or polymerize proteins to construct pili, multi-subunit hair-like fibres that extend from the cell surface to promote bacterial adhesion (Scott and Barnett, 2006; Scott and Zahner, 2006; Mandlik et al., 2008a; Proft and Baker, 2009; Clancy et al., 2010; Kline et al., 2010; Hendrickx et al., 2011). Sortases have attracted great interest as potential drug targets as many of the surface proteins they display are virulence factors (Suree et al., 2007; Maresso and Schneewind, 2008). They have also been developed into powerful molecular biology reagents to site-specifically attach proteins to a variety of biomolecules (Tsukiji and Nagamune, 2009; Proft, 2010; Popp et al., 2011). All sortase enzymes characterized to date function as cysteine transpeptidases that join proteins containing a cell wall sorting signal (CWSS) to an amino group located on the cell surface (Marraffini and Schneewind, 2006; Clancy et al., 2010). Although they are not essential for bacterial viability when cells are grown in rich media, sortases can be important virulence factors as they display surface proteins that mediate bacterial adhesion to host tissues, host cell entry, evasion and suppression of the immune response and acquisition of essential nutrients. The sorting reaction catalysed by the sortase A protein from Staphylococcus aureus (Sa-SrtA) is best understood and begins when a full-length precursor protein containing an amino terminal leader peptide is exported from the cytoplasm through the secretory pathway (Fig. 1A). The C-terminal CWSS is then processed by Sa-SrtA. The CWSS consists of a LPXTG motif (where X denotes any amino acid), followed by a segment of hydrophobic amino acids, and a tail composed primarily of positively charged residues. The C-terminal charged tail presumably retards export, positioning the protein for processing by the extracellular membrane associated Sa-SrtA enzyme. A highly conserved active site cysteine residue in Sa-SrtA then nucleophilically attacks the backbone carbonyl carbon of the threonine residue in the LPXTG motif, breaking the threonine and glycine peptide bond and creating a sortase-protein complex in which the components are linked via a thioacyl bond. The protein is then transferred by Sa-SrtA to the cell wall precursor lipid II, when the amino group in this molecule nucleophilically attacks the thioacyl linkage to create an isopeptide linked protein-lipid II product. Transglycosylation and transpeptidation reactions that synthesize the cell wall then incorporate this product into the peptidoglycan, where it is covalently linked to the cross-bridge peptide. Other sortases catalyse a similar transpeptidation reaction, but join remarkably different LPXTG motifs and amino groups.
Fig. 1.
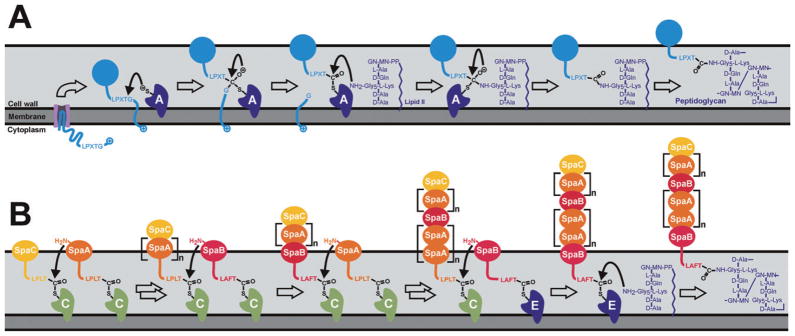
Mechanisms of sortase mediated attachment of surface proteins and pilus assembly at the bacterial cell wall.
A. The S. aureus housekeeping sortase A anchors surface proteins to the peptidoglycan. The precursor protein containing an amino terminal leader peptide is secreted across the membrane through the Sec pathway. The exported protein (light blue) is processed by the sortase enzyme (dark blue, labelled ‘A’), which recognizes the LPXTG sequence and cleaves the surface proteins between the threonine and glycine residues of the motif. The enzyme then recognizes the pentaglycine cross-bridge peptide of lipid II as the second substrate. Subsequent formation of a peptide bond between the carbonyl of the threonine and the free amino group of the cross-bridge peptide results in covalent attachment of the protein to lipid II. The surface protein is then fully incorporated into the cross-linked peptidoglycan via the transglycosylation and transpeptidation reactions during the bacterial cell wall synthesis. The sphere coloured light blue represents the folded form of the cell surface displayed protein.
B. Pilin-specific and housekeeping sortases assemble the SpaA pilus in C. diphtheria. The formation of complexes between the pilus-specific sortase C (light green) and the tip protein SpaC (light orange) initiates pilus assembly. The class C enzyme also recognizes the main pilin subunit SpaA (orange) forming SrtC–SpaA complexes. Nucleophilic attack by the free amino group originating from a lysine residue present in SpaA results in dissolution of the sortase–SpaC intermediate and the formation of a sortase–SpaA–SpaC complex. Repetition of this transpeptidation reaction results in pilus elongation. The class C sortase also incorporates the minor pilin SpaB (red) into the growing shaft by an analogous mechanism. Termination of pilus biogenesis is presumably initiated when the pilin polymer is transferred to the class E type housekeeping sortase (dark blue), which subsequently catalyses the nucleophilic attack by the amino group within lipid II. In the final assembly step the lipid II linked pilus is incorporated in the murein sacculus via normal cell wall biosynthesis.
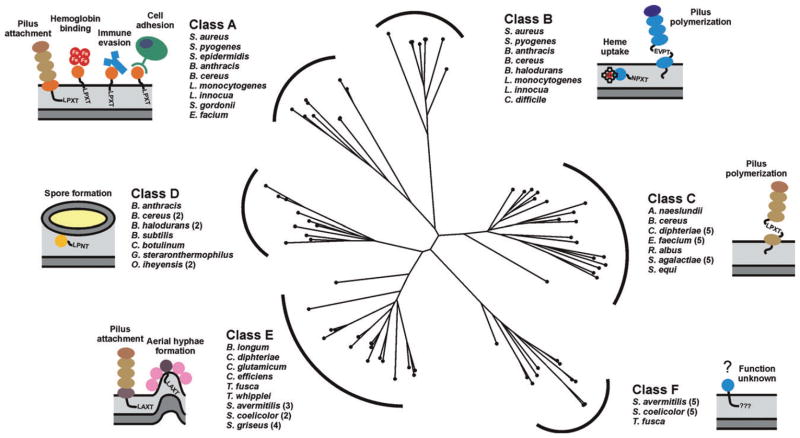
Since the discovery of Sa-SrtA a little more than decade ago by Schneewind and colleagues (Mazmanian et al., 1999), over 800 genes encoding related proteins have been identified in ~260 distinct bacterial species (Finn et al., 2010). The vast majority of sortases are found in Gram-positive bacteria that contain a conventional cell wall (they are absent in Mollicutes) (Pallen et al., 2001). Most bacterial species contain multiple sortase enzymes that have been named in an ad hoc manner (e.g. SrtA, SrtB, SrtC, etc.). To provide a framework in which to discuss their functions, we first grouped sortases from Gram-positive bacteria into families based upon their primary sequences (Fig. 2). Approximately 60% of all sortase proteins can be partitioned into six distinct families of enzymes that share related amino acid sequences. These include class A to D enzymes that have been described previously (Comfort and Clubb, 2004; Dramsi et al., 2006), and class E and F enzymes that are defined here. Experimental and bioinformatics analyses indicate members of each group recognize distinct CWSSs in which the LPXTG sequence is varied (hereafter called sorting signal motifs). Class A enzymes are present in Firmicutes and have been studied extensively. They appear to perform a housekeeping role in the cell as members of this group are capable of anchoring a large number of functionally distinct proteins to the cell wall. Class B enzymes are also present in Firmicutes and can have distinct functions. Some members of this group attach haem-receptors to the peptidoglycan, while others assemble pili. Class C enzymes are broadly distributed in Gram-positive bacteria and function as pilin polymerases that construct pili. Class D enzymes predominate in Bacilli and in Bacillus anthracis this type of enzyme anchors proteins to the cell wall that facilitate sporulation. Actinobacteria contain class E and F enzymes whose functions are largely unknown. In Corynebacterium diphtheriae a class E enzyme appears to perform a housekeeping function similar to class A enzymes (Ton-That and Schneewind, 2003), while class F enzymes have yet to be studied. Sortases are also present in a few Gram-negative and archaebacterial species, but the functions of these enzymes are unknown (Pallen et al., 2001; 2003; Comfort and Clubb, 2004). In this Microreview we discuss what is known about the functions and mechanism of transpeptidation of different types of sortase enzymes.
Fig. 2.
Phylogenic tree showing the relationships among the six classes of sortases from Gram-positive bacteria. A multiple sequence alignment based on pairwise constraints of a selected set of 73 sortase proteins was generated using the program COBALT and a phylogenetic tree constructed using the neighbour joining method (Papadopoulos and Agarwala, 2007). The analysed sortases can be partitioned into six distinct subfamilies based on their primary sequences. It should be noted that the class D and E enzymes described here are collectively referred to as a class D enzymes by Bierne and colleagues (Dramsi et al., 2005). Class D and E enzymes have also previously been referred to as subfamily-4 and -5 enzymes (Comfort and Clubb, 2004). The bacterial species associated with the enzyme classes A–F are listed and schematic representations of the main biological function of their corresponding sortase substrates are illustrated. The bacterial species and accession numbers of the amino acid sequences used for the alignment are as followed: Actionomyces naeslundii (AAC13546), Bacillus anthracis (NP_847260; NP_843215; NP_846988), Bacillus cereus (NP_834511; NP_830752; NP_830495; NP_834250; NP_832268), Bacillus halodurans (NP_244463; NP_244878; NP_244160), Bacillus subtilis (NP_388801), Bifidobacterium longum (NP_695779), Clostridium botulinum (YP_001254630), Clostridium difficile (YP_001089230), Corynebacterium diphtheriae (NP_940343; NP_940532; NP_940531; NP_938627;NP_940575; NP_938624), Corynebacterium efficiens (NP_739396), Corynebacterium glutamicum (NP_602126), Enterococcus faecium (ZP_05713110; ZP_05714716; ZP_05713995; ZP_05713369; ZP_00603528; ZP_05713288), Geobacillus sp. (YP_003672707), Listeria innocua (NP_470268; NP_471617), Listeria monocytogenes (NP_464454; NP_465705), Oceanobacillus iheyensis (NP_691253; NP_694114), Ruminococcus albus (ZP_06717336), Streptomyces avermitilis (NP_826393; NP_827770; NP_826840; NP_828474; NP_821266; NP_825513; NP_825514; NP_826383; NP_825510), Streptomyces coelicolor (NP_628037; NP_628038; NP_627928; NP_631498; NP_626722; NP_625233; NP_627070), Streptomyces griseus (YP_001825232; YP_001825235; YP_001826193; YP_001825236), Staphylococcus aureus (NP_375640; NP_374252), Staphylococcus epidermis (NP_765631), Streptococcus agalactiae (NP_687667; NP_687668; NP_687670; NP_688403; NP_688404), Streptococcus equi (YP_002746265), Streptococcus gordonii (YP_001450517), Streptococcus pyogenes (NP_802272; NP_801365), Thermobifida fusca (YP_290439; YP_288977), Tropheryma whipplei (NP_787692).
Sortases that attach proteins to the cell wall (class A, B, D and E enzymes)
Housekeeping class A enzymes
Nearly all bacteria in the Firmicutes subfamily contain a single class A sortase whose primary sequence is most closely related to the prototypical Sa-SrtA enzyme. Like Sa-SrtA, members of this group are believed to attach proteins to the cross-bridge peptide of the cell wall, presumably by first linking it to lipid II. Extensive experimental work on class A enzymes from different bacteria has revealed that each enzyme anchors a large number of functionally distinct proteins to the cell wall (Marraffini et al., 2006). This has led to the notion that they perform a housekeeping function in the cell, and is consistent with the finding that genes encoding class A enzymes are not genomically clustered with genes encoding potential protein substrates that contain a CWSS (Pallen et al., 2001; Comfort and Clubb, 2004). The busiest enzyme in this group appears to be the sortase A from Listeria monocytogenes, which based on a genome sequence analysis is predicted to display an astounding 43 distinct proteins (Boekhorst et al., 2005). Most surface proteins attached by class A enzymes contain a canonical LPXTG motif within their CWSS and have diverse functions that can promote bacterial adhesion, nutrient acquisition, host cell invasion, and immune evasion. Class A enzymes have attracted significant interest as potential drug targets because a number of clinically important pathogens use these sortases to display virulence factors and they are attenuated in their virulence if their srtA gene is eliminated (S. aureus, L. monocytogenes, Streptococcus pyogenes and Streptococcus pneumoniae among others) (Suree et al., 2007; Maresso and Schneewind, 2008).
Class B enzymes have diverse functions
Class B enzymes are widely distributed in Firmicutes and have primary sequences that are most closely related to the sortase B enzyme from S. aureus (Sa-SrtB) (Mazmanian et al., 2002). Although they share significant primary sequence homology, members of this group can have radically distinct functions. In some pathogenic microbes class B sortases attach haemoproteins to the cell wall (Mazmanian et al., 2002; 2003; Maresso and Schneewind, 2006; Maresso et al., 2006). In contrast, recent studies have shown that the class B enzymes in S. pyogenes assemble pili (Kang et al., 2011). This fundamental difference highlights the limits of assigning function strictly based on primary sequence homology and emphasizes the need for experimental work to decipher sortase function. Here we discuss class B enzymes that anchor haemoproteins, while pilin-specific class B sortases are discussed in the section reviewing functionally homologous class C enzymes.
In S. aureus and B. anthracis a single class B enzyme anchors the IsdC haemoprotein to the cell wall (Mazmanian et al., 2002; Maresso et al., 2006). In both microbes, IsdC is thought to facilitate haem capture from human haemoglobin by relaying haem from upstream haem-receptors to a membrane transporter complex that imports haem into the cell (Maresso and Schneewind, 2006; Maresso et al., 2006). In contrast to Sa-SrtA, Sa-SrtB and its substrate IsdC are only expressed in iron deplete conditions (Mazmanian et al., 2002). Class B enzymes recognize an unusual NP(Q/K)(T/S)(N/G/S)(D/A) sorting signal that differs markedly from the canonical LPXTG motif recognized by class A enzymes. Interestingly, the class A and B enzymes in S. aureus may attach their substrates to different sites within the cell wall. Sa-SrtA attaches its substrates to surface exposed sites that are heavily cross-linked. As demonstrated by protease sensitivity and immunoblot experiments, Sa-SrtB attaches IsdC to a buried site within the peptidoglycan that is not heavily cross-linked (Mazmanian et al., 2003; Marraffini and Schneewind, 2005). It has been hypothesized that the positional difference of the attached proteins results from Sa-SrtB’s ability to attach IsdC to preassembled peptidoglycan instead of lipid II, but this has not been demonstrated experimentally.
Gene knockout studies in S. aureus and L. monocytogenes indicate that their class B enzymes are needed to establish persistent infections, but that they are less important for virulence than their class A enzymes (Mazmanian et al., 2002; Jonsson et al., 2003; Newton et al., 2005). However, recent findings suggest that the Sa-SrtB enzyme could play a more prominent role in pathogenesis as S. aureus has been shown to have enhanced specificity for human haemoglobin such that murine models of infection may underestimate the pathogenic importance of the Sa-SrtB anchored IsdC protein and other iron-regulated surface determinant proteins that participate in the capture of haem-iron from human haemoglobin during infections (Pishchany et al., 2010). Sortases belonging to class B are also present in Clostridium perfringens, Clostridium difficile and Enterococcus faecalis. However, their functions remain unknown and the genes encoding these enzymes are not clustered with genes encoding putative sortase substrates.
Class D, E and F enzymes
Much less is known about members of the class D, E and F sortases. Class D enzymes are present in bacilli and have thus far only been characterized in B. anthracis (Marraffini and Schneewind, 2006; 2007). This microbe encodes a single class D enzyme (called Ba-SrtC) that attaches two proteins to the cell wall, BasH and BasI. Deletion of this enzyme reduces the efficiency of spore formation under oxygen limiting conditions, but is otherwise dispensable for pathogenesis in mice (Marraffini and Schneewind, 2006). Remarkably, the class D enzyme attaches each of its substrates to a distinct structure; it attaches BasI to the diaminopimelic acid moiety of the peptidoglycan of predivisional cells, while it attaches BasH exclusively to the forespore. The srtC and basI genes are coexpressed from the same operon before formation of the septum that divides the mother cell from the forespore enabling conventional anchoring of the BasI protein to the cell wall (Marraffini and Schneewind, 2006; 2007). In contrast, the basH gene is located elsewhere in the genome and is primarily expressed in the forespore after septum formation. BasH is then presumably attached to the fore-spore by class D enzymes that are transferred to the forespore from the mother cell’s membrane. Interestingly, B. anthracis contains class A and D sortase enzymes that recognize closely related sorting signals; both BasH and BasI contain a LPNTA sorting signal that differs only slightly from the canonical LP[A/N/K]TG sorting signal present in proteins anchored by B. anthracis SrtA. Nevertheless, the enzymes function non-redundantly suggesting that they may have evolved a high degree of specificity for their respective sorting signals (Marraffini and Schneewind, 2006). Class D enzymes in other bacilli may perform similar functions as genes encoding these enzymes are frequently clustered with genes encoding proteins with a LPNTA sorting signal motif, which in some instances share primary sequence homology with BasI and/or BasH. However, it remains unclear why certain species including B. cereus and Bacillus halodurans contain a second class D sortase gene that is not genomically clustered with potential protein substrate genes.
Instead of a class A enzyme, some high G + C bacterial species may use a class E enzyme as their housekeeping sortase (Ton-That and Schneewind, 2003). This idea is based on the finding that genes encoding class A and E enzymes are never found in the same organism, and similar to class A enzymes, the genes encoding class E enzymes are not positioned adjacent to genes encoding potential protein substrates (Comfort and Clubb, 2004). Comparative genome analyses suggest that class E enzymes recognize an LAXTG sorting signal, instead of the canonical LPXTG motif processed by class A enzymes (Comfort and Clubb, 2004). This unique specificity has been demonstrated for the class E enzyme in C. diphtheriae (named Cd-SrtF), which is the only class E enzyme that has been characterized experimentally (Chang et al., 2011). Interestingly, Cd-SrtF also attaches assembled pili to the cell wall, a function shared by some class A enzymes (discussed below) (Budzik et al., 2007; Swaminathan et al., 2007; Nobbs et al., 2008). In Streptomyces coelicolor, class E enzymes display chaplin surface proteins containing an LAXTG sorting signal, which presumably mediate aerial hyphae formation. (Claessen et al., 2003; Elliot et al., 2003; Capstick et al., 2007). Class F enzymes are present in S. coelicolor and other Actinobacteria, but their functions have yet to be explored (a total of 57 enzymes cluster into this family and are present in 20 bacterial species). It should be noted that in a previous study class D and E sortases were collectively grouped into a single family called class D sortases (Dramsi et al., 2005). However, based on our analyses class D and E enzymes share only limited primary sequence homology with one another and are predicted to process distinct sorting signals. Moreover, unlike class D enzymes, genes encoding class E enzymes are not genomically clustered near their substrates.
Sortases that assemble pili: class C enzymes
Gram-positive bacteria use class C enzymes to build pili that promote microbial adhesion and biofilm formation. The pili extend 0.2–3.0 μm from the cell surface and are assembled in a two-stage process. First, one or more class C enzymes form the long thin shaft of the pilus by linking together pilin subunits via isopeptide bonds. The base of the pilus is then anchored to the cell wall by a housekeeping sortase or, in some cases, the class C enzyme itself. Although the general features of this process appear to be conserved, there is great species-specific variation in pilus structure and the mechanism of biogenesis. For example, depending upon the organism both the number and type of sortase enzymes involved varies, and in some cases, accessory factors appear to be needed (Fig. 3). Pili in Gram-positive bacteria are constructed from either two or three types of pilin subunits. In two-component pili the shaft of the pilus is formed by multiple copies of a major pilin subunit, while the tip of the pilus contains a single copy of a minor ‘tip’ pilin subunit that typically functions as an adhesin. Three-component pili are similar, but they also contain a minor ‘basal’ pilin subunit that is covalently attached to the cell wall. Several transmission electron microscopy (EM) and immuno-gold labelling studies have led to the conclusion that the minor ‘basal’ pilin subunits are also interspersed throughout the shaft of the pilus, presumably because the sortase enzymes are promiscuous in the substrates they recognize. However, this issue is controversial as more recent high-resolution EM imaging of S. pneumoniae pili indicates that only the major pilin subunit is present in the shaft (discussed below). Interestingly, some pilin subunits within the pilus contain intra-protein isopeptide bonds that form spontaneously. These bonds presumably stabilize the structure of the pilus, but they have not been shown to be required for pilus biogenesis (Kang et al., 2007; Kang and Baker, 2009; Budzik et al., 2009a). Because of space restraints, we only summarize what is known about the functions of sortases in the best characterized pilus systems and refer the reader to several excellent reviews on this topic for more detailed information (Ton-That et al., 2004a; Scott and Zahner, 2006; Telford et al., 2006; Mandlik et al., 2008a; Proft and Baker, 2009; Kline et al., 2010; Hendrickx et al., 2011).
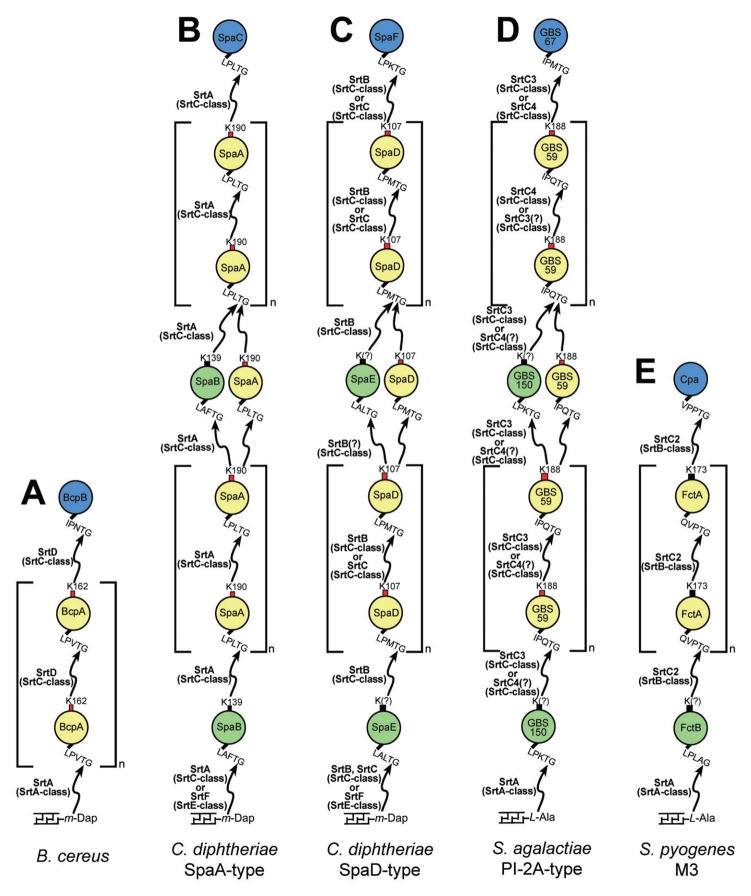
Fig. 3.
The role of sortase enzymes in pilus assembly in various Gram-positive bacteria. Gram-positive bacteria are diverse in their assembly of pili with respect to the number of pilin subunits, number of sortases, class of sortases, as well as the type of sorting signals, lysine residues and cell wall acceptors recognized. Red boxes indicate lysine residues within a WXXXVXVYPK pilin motif, while black boxes indicate the absence of an obvious pilin motif. Lack of biochemical data demonstrating which lysine residue is involved in pilus polymerization is indicated with (?).
The paradigm: the three-component spaA pilus from C. diphtheriae
The prototypical SpaA pilus in C. diphtheriae is assembled in a two-stage process (Figs 1B and 3B). First, a class C sortase (called Cd-SrtA) joins the pilin subunits (SpaA, SpaB and SpaC) to construct the pilus shaft (Ton-That and Schneewind, 2003). This structure is then attached to the cell wall by a housekeeping class E sortase (called Cd-SrtF) (Swaminathan et al., 2007). The shaft of the pilus is primarily formed by the main pilin subunit SpaA, and is decorated with two minor pilins that are present at lower abundance, SpaB and SpaC. The SpaC pilin is located at the tip of the pilus, while SpaB is primarily located at its base, but also infrequently interspersed throughout the shaft (Ton-That and Schneewind, 2003; Mandlik et al., 2008b). In the working model of the assembly process put forth by Ton-That and colleagues, assembly begins with the export of the tip protein SpaC, which contains an LPLTG sorting signal that is cleaved between the threonine and glycine residues by the class C enzyme (Guttilla et al., 2009). This forms a sortase–SpaC complex wherein a thioacyl bond connects the sortase active site cysteine residue to the threonine carbonyl carbon in SpaC. Similarly, another copy of this class C enzyme recognizes a LPLTG sorting signal in the main pilin subunit, SpaA, forming a thioacyl sortase–SpaA intermediate. These acyl intermediates are then thought to react with one another when the free amino group originating from a lysine residue present in SpaA (Lys190) attacks the acyl bond in the sortase–SpaC intermediate. Lys190 is located in a ‘pilin’ motif (residues WXXXVX-VYPK) that is frequently conserved in other main pilin subunits (Ton-That and Schneewind, 2003). Its nucleophilic attack on the acyl bond results in dissolution of the sortase–SpaC intermediate and the creation of a sortase–SpaA–SpaC complex in which the SpaA and SpaC components are covalently linked to each other by an isopeptide bond (the carbonyl carbon in the threonine residue in the sorting signal motif of SpaC is joined to the ε-amino group of Lys190 in SpaA) (Ton-That and Schneewind, 2003). The nascent pilus is elongated through repetition of this transpeptidation reaction, with additional SpaA pilin subunits presumably entering the assembly reaction pre-loaded onto a sortase in the form of an acyl intermediate. Evidence that pilin subunits enter the reaction as sortase attached acyl intermediates is indirect. It is based upon the observation that only small amounts of polymerized pili are released into the solvent in wild-type cells, presumably because during the assembly process the nascent pilus is always covalently linked to a membrane associated sortase via a thioacyl bond, Furthermore, biochemical data demonstrate that C. diphtheriae class C enzymes form stable complexes with pilin subunits (Guttilla et al., 2009). However, it is also conceivable that SpaA proteins that have yet to be processed by sortase might serve as substrates in the reaction instead of SpaA proteins that are acyl linked to sortase. In this alternative model, the side-chain of Lys190 within a membrane associated SpaA protein containing an intact CWSS would nucleophilically attack the acyl bond that joins the pilin polymer to the sortase. As a result, the growing pilus would become temporally associated with the membrane via the C-terminus of the recently added SpaA subunit. The polymer would then be transferred to a class C sortase when the enzyme cleaved the threonine residue in the LPLTG motif of the terminal membrane associated SpaA protein. As in the aforementioned ‘preloaded’ model that assumes that SpaA enters the assembly reaction as a sortase–SpaA acyl intermediate, a repetition of this process would grow the pilus from its base.
The class C enzyme also attaches the minor pilin SpaB to the SpaA pilus. SpaB does not contain a pilin motif, but alanine substitution of Lys139 within this protein abolishes its cross-linking to the SpaA pilus, suggesting this lysine is linked to the threonine residue within the sorting signal motif of SpaA (Mandlik et al., 2008b). The minor pilin SpaC lacks a lysine residue that can be used by the class C enzyme as a nucleophile, explaining why it is only found at the tip of the pilus. Fascinatingly, incorporation of SpaB can act as a molecular switch to promote pilus anchoring to the cell wall (Mandlik et al., 2008b). In accordance with the model put forth by Ton-That and colleagues, this occurs when the class E sortase (called Cd-SrtF) cleaves the LAFTG sorting signal to form a Cd-SrtF-SpaB acyl intermediate (Guttilla et al., 2009). In a reaction that is presumably catalysed by the class C enzyme, Lys139 within the SpaB component of the Cd–SrtF–SpaB acyl intermediate attacks the acyl bond that joins the terminal SpaA subunit of the pilus polymer to the catalytic cysteine residue of the class C enzyme. This transfers the pilus fibre to Cd-SrtF and terminates the polymerization reaction because Cd-SrtF is incapable of using lysine residues located within the pilin subunits as a nucleophile, but instead transfers the pilus to the free amino group located within lipid II (Swaminathan et al., 2007). Cd-SrtF can then recognize the LAFTG sorting signal within SpaB and attach the completed pilus to the free amino group within lipid II. In the final assembly step, cell wall biogenesis reactions then incorporate the lipid II linked pilus into the murein sacculus.
Pilus assembly appears to be a stochastic process, as both the number and length of pili is determined by the relative abundance of the major and minor pilin subunits (Swierczynski and Ton-That, 2006; Mandlik et al., 2008b; Quigley et al., 2009). For example, while it has not been shown in the SpaA pilus, in the related C. diphtheriae SpaH pilus, an increase in the amount of SpaH, the major pilin component, results in longer pilus fibres. As the SpaH pilus also contains a minor basal pilin that can be incorporated by its class C sortase that can terminate the assembly reaction (described below), increased amounts of the major pilin subunit presumably enable it to out-compete the minor basal pilin for access to the sortase thereby causing longer pili to be produced. Furthermore, the prevalence of the minor tip pilin used to construct the S. pyogenes T3 pilus (further discussed below) has been shown to influence the number of pili that are displayed suggesting that it competes with the major pilin subunit in this system to gain access to the sortase (Swierczynski and Ton-That, 2006; Quigley et al., 2009). Although they play a key structural role, the minor pilin subunits in many pili are not required for pilus biogenesis. For example, in the SpaA pilus, neither SpaB nor SpaC are required for SpaA polymerization, albeit anchoring of the pilus to the cell wall is inefficient when SpaB is absent (Ton-That and Schneewind, 2003; Mandlik et al., 2008b). SpaA, along with major pilin subunits from many other pilin systems, contains a highly conserved E-box motif, which when mutated prevents SpaA pilus formation (Ton-That et al., 2004b). This motif does not appear to be directly involved in the sortase catalysed transpeptidation reaction, but a glutamic acid residue within it catalyses the formation of an intra-protein isopeptide bond that stabilizes the pilin subunit (Kang et al., 2009).
The class C enzyme that assembles the SpaA pilus is amazingly promiscuous, as it can recognize two distinct sorting signals (LPLTG in SpaA and SpaC, and LAFTG in SpaB) and it can employ lysine residues that originate from different proteins (either Lys190 within the pilin motif of SpaA or Lys139 in SpaB) (Fig. 3B). In addition, in the absence of the housekeeping sortase Cd-SrtF, the class C enzyme attaches the pilus to the cell wall, albeit at a reduced rate (Swaminathan et al., 2007). This indicates that it is able to use diaminopimelic acid as a nucleophile when it is located in the cross-bridge peptide of lipid II and/or in preassembled peptidoglycan. This ability may function as a fail-safe mechanism enabling pilus attachment if the housekeeping sortase is not available. Interestingly, when the SpaB sorting signal motif (LAFTG) is altered to the SpaA sorting signal moiety (LPLTG), the class C sortase was more successful in attaching the pilus to the cell wall in the absence of the class E sortase (Chang et al., 2011). This suggests that the LAFTG sorting signal motif of SpaB is preferentially recognized by the class E enzyme, while the LPLTG motif in SpaA and SpaC is a better substrate for the class C enzyme. Very recently, the class C and E enzymes have been shown to work conjunctively to attach SpaB-SpaC heterodimers to the cell wall (Chang et al., 2011). Mechanistically, this process is related to the pilus assembly reaction, with the class C enzyme linking SpaB via its K139 residue to the CWSS of SpaC. The class E enzyme then specifically recognizes the LAFTG sorting signal in SpaB to attach the heterodimer to the cell wall. Assembly of a heterodimer consisting of two disparate pilin adhesins may be a novel strategy for corynebacteria to form a lasting intimate zone of adhesion with their host cells.
Two-component pili
Pili in B. cereus, Actinomyces spp., and the FCT-1 S. pyogenes pilus systems are less complex and contain only two types of pilin subunits (Mora et al., 2005; Budzik et al., 2007; Mishra et al., 2007). The pilus in B. cereus is prototypical and consists of a major pilin protein BcpA that forms the shaft, and a minor tip pilin, BcpB (Fig. 3A). These proteins are linked together by a single class C enzyme (called Bc-SrtD) (Budzik et al., 2009b). It cleaves related sorting signals within BcpA (LPVTG) and BcpB (IPNTG), and catalyses a transpeptidation that joins the threonine residues in each signal to the side-chain of Lys162 in BcpA (located within a pilin motif) (Budzik et al., 2008a). In a striking contrast to the three-component pili, two-component pili do not contain a minor basal pilin subunit. Assembly is terminated by a class A enzyme (called Bc-SrtA), which transfers the pilus fibre to the cell wall by joining the sorting signal within the basal BcpA subunit to the diaminopimelic acid moiety within lipid II and/or the cell wall (Budzik et al., 2008b). Sortase specificity for the sorting signal appears to regulate the assembly process, as the class A enzyme is unable to process the IPNTG signal within BcpB, facilitating usage of this tip pilin by the class C enzyme. Interestingly, in the absence of the housekeeping enzyme pili are not covalently attached to the cell wall, indicating that, unlike the SpaA pilus in C. diphtheria, in B. cereus the class C enzyme is unable to attach the pilus to the cell wall and is reliant on the housekeeping sortase to perform this task (Budzik et al., 2007).
Assembling pili using two class C enzymes
Gram-positive bacteria frequently use two class C sortase enzymes to assemble pili from three distinct pilin subunits. C. diphtheriae expresses SpaD- and SpaH-type pili in addition to the SpaA-type pili (Gaspar and Ton-That, 2006; Swierczynski and Ton-That, 2006). These separate pilus types each use their own specific sortases, and no cross-reactivity has been observed in spite of the similarities between their respective sorting signals. The SpaD pilus is assembled by two class C enzymes (called Cd-SrtB and Cd-SrtC), and consists of a main pilin, SpaD, a minor tip pilin, SpaF, and a basal pilin, SpaE, which is also interspersed throughout the shaft (Fig. 3C). The class C sortases appear to have partially redundant functions as each can polymerize SpaD and attach the SpaF tip pilin. However, only Cd-SrtB can incorporate SpaE presumably by joining the lysine within SpaE to the threonine residue within the CWSS of SpaD (Gaspar and Ton-That, 2006). This suggests that both enzymes are able to use the lysine residue within the SpaD pilin motif as a nucleophile and that each can recognize the LPMTG and LPKTG sorting signals within SpaD and SpaF respectively. The assembly process is completed when the terminal SpaE subunit in the fibre is transferred to the housekeeping class E enzyme, which then incorporates the fibre into the cell wall (Swaminathan et al., 2007). This mechanism again highlights the substrate promiscuity of class C enzymes, as Cd-SrtB presumably uses distinct lysine residues located within the SpaD and SpaE proteins as nucleophiles, and it also recognizes distinct sorting signals within all three pilin subunits to form acyl intermediates. The SpaH pilus appears to be assembled using a similar mechanism, as it is also constructed from three pilin subunits (SpaG, SpaH and SpaI) by two class C enzymes (Cd-SrtD and Cd-SrtE) (Swierczynski and Ton-That, 2006).
Pilus assembly in Streptococcus agalactiae follows a similar pattern as the SpaD pilus in C. diphtheria, but is slightly more complicated (Fig. 3D). Its P1-2a pilus is formed from a major pilin, GBS59, which contains a pilin motif, and two minor pilins, GBS67 and GBS150 (Dramsi et al., 2006; Rosini et al., 2006). Two class C enzymes, Sag-SrtC3 and Sag-SrtC4, function redundantly to polymerize GBS59. Interestingly, recent work suggests that each class C enzyme predominantly incorporates a specific minor pilin into the pilus, with significantly reduced ability to incorporate the other minor pilin. Sag-SrtC4 incorporates the minor pilin GBS67 found at the pilus tip, and Sag-SrtC3 specifically incorporates GBS150, which is found within the shaft and at its base (Rosini et al., 2006). Low-resolution EM images also reveal that GBS67 is incorporated throughout the pilus shaft; however, this could be artefactual (discussed below). There is conflicting data regarding the role of the class C enzymes in this system, as Dramsi et al. have proposed that they function redundantly to incorporate both minor pilins (Dramsi et al., 2006). The class C enzymes in this system are unable to attach the pilus to the cell wall, which instead requires the class A housekeeping sortase that recognizes the LPKTG sorting signal present within GBS150 (Nobbs et al., 2008). S. agalactiae also expresses an additional three-component pilus system, PI-1, whose subunits share primary sequence homology with the components of the aforementioned PI-2a pilus. This pilus is also assembled by two class C enzymes (Sag-SrtC1 and Sag-SrtC2) that operate redundantly to polymerize the main pilin, suggesting that it is constructed in a similar manner as the P1-2a pilus (Dramsi et al., 2006; Rosini et al., 2006).
Streptococcus pneumoniae pili are even more complicated, as the pilus encoded from the rlrA pathogenicity islet is formed by three pilin proteins (RrgA, RrgB and RrgC) and three sortases (Spn-SrtC-1, Spn-SrtC-2 and Spn-SrtC-3) (Hava et al., 2003; LeMieux et al., 2006). Transmission EM images have shown that the shaft of the pilus is primarily composed of RrgB, and that the minor pilins RrgA and RrgC are located at the tip and base of the pilus respectively (Hilleringmann et al., 2009). RrgA may also be integrated throughout the pilus (LeMieux et al., 2006; Falker et al., 2008) and can form heterodimers with RrgC similar to what is seen in C. diphtheriae (LeMieux et al., 2008). Spn-SrtC-1 recognizes each of the individual subunits and polymerizes mature pilus structures in vivo, and can readily polymerize RrgB subunits in vitro (Falker et al., 2008; Manzano et al., 2008). The role of Spn-SrtC-2 is less clear, with some research groups reporting an ability to polymerize the major pilin subunit, RrgB, along with the ancillary RrgA (Falker et al., 2008), while others report no RrgB polymerization function by Spn-SrtC-2 alone (Manzano et al., 2008). Spn-SrtC-3 has been implicated in pilus localization on the cellular surface, suggesting a direct role in anchoring the constructed pilus to the cell wall (Falker et al., 2008). As with Spn-SrtC-2, the ability of Spn-SrtC-3 to polymerize specific pilus subunits has not yet been fully elucidated. Unlike pili in C. diphtheriae, B. cereus and S. agalactiae, the housekeeping sortase is not required for anchoring the S. pneumoniae pili to the cellular surface (LeMieux et al., 2008). This may be due to the inability of the Spn-SrtA enzyme to recognize the sorting signals on each of the S. pneumoniae pilin subunits, and because all three SrtC enzymes have demonstrated an ability to attach pilin subunits to the peptidoglycan (Neiers et al., 2009).
Accessory factors and class B enzymes are involved in pilus biogenesis
In nearly all pilus systems studied to date, the minor pilins are dispensable for polymerization of the main pilin subunit that forms the shaft of the pilus (C. diphtheriae, B. cereus, S. pneumoniae, S. pyogenes and S. agalactiae). Interestingly, a two-component pilus in Streptococcus suis appears to be the exception to this rule. Similar to the B. cereus paradigm, it contains a minor pilin that is located at the tip of the pilus (Sgp2) and a major pilin that forms the shaft (Sgp1). When Sgp2 is deleted, there is a significant decrease in the amount of polymerized Sgp1, a phenotype that is similar to knocking out the class C enzyme (Ss-SrtG) (Okura et al., 2011). This suggests that the minor pilin Sgp2 functions as a chaperone for Sgp1, or that its presence, through an unknown mechanism, is required to initiate the polymerization of Sgp1. In a surprising example of convergent evolution, a class B enzyme, not a class C enzyme, assembles pili in S. pyogenes (Kang et al., 2011). The FCT-3 pilus in S. pyogenes serotype M3 is constructed by a single class B sortase, Sp-SrtC2, which polymerizes the major pilin subunit Tee3/FctA and attaches the minor tip pilin Cpa (Fig. 3E) (Quigley et al., 2009). This system also contains a minor pilin FctB that contains a LPLAG sorting signal. Its location and sortase dependent incorporation into the pilus has not been extensively explored. However, based on amino acid sequence homology, it is likely located at the base of the pilus (Smith et al., 2010). Interestingly, assembly of the FCT-3 pilus also requires the SipA2/LepA protein that shares sequence homology with signal peptidases, but is presumably inactive as it is missing key active sites residues (Zahner and Scott, 2008). It has been proposed that SipA2/LepA functions as a chaperone that associates with Tee3/FctA, reminiscent of pilus assembly in Gram-negative bacteria. Sp-SrtC2 highlights the diversity of sortases, as the side-chain amino group of K173 within the main pilin Tee3/FctA that it joins to other subunits is not located within a pilin motif. In addition, Cpa and Tee3/FctA contain very distinct signals, VPPTG and QVPTG, respectively, which are nevertheless recognized by Sp-SrtC2. The use of peptidase-like proteins as chaperones may be common to many pilus systems as a SipA homologue is also required for the assembly of the PI-2 pilus in S. pneumoniae (Bagnoli et al., 2008) and fimbrial gene clusters in Actinomyces naeslundii also contain genes encoding peptidase-like proteins (Mishra et al., 2007).
Class C sortase substrate recognition
Across the multiple species and pilin systems studied to date, the pilin polymerizing sortases demonstrate a unique variance in their ability to recognize a variety of sorting signals and amino groups. For example, cell wall attachment of pili in S. agalactiae relies on the housekeeping sortase as the class C sortase is unable to do this, while in S. pneumoniae the housekeeping sortase is unnecessary for cell wall attachment (LeMieux et al., 2008; Nobbs et al., 2008). The pilin polymerizing sortases also have little ability to function outside of their specific system. For example, Cd-SrtA is able to polymerize SpaA, but not SpaH, even though both major pilin subunits have identical LPLTG motifs (Swierczynski and Ton-That, 2006). There are obviously other factors beyond the CWSS that control how sortases function and recognize specific substrates. This is likely the case for other types of sortases as domain switching experiments performed by Barnett and co-workers have shown that both the class A and pilin polymerase class B enzymes in S. pyogenes recognize features in their protein substrates in addition to their respective sorting signals (Barnett et al., 2004). The recent crystal structure of FctB, the anchoring pilin subunit from S. pyogenes, has provided insight into how minor pilins are recognized and attached to the cell wall. FctB contains an Ig-like domain with striking resemblance to the N-terminus of the previously solved S. pyogenes major pilin, with nearly identical placement of the intermolecular isopeptide bond-forming lysine (Kang et al., 2007; Linke et al., 2010). This helps explain how a single sortase can recognize both lysines on the major and anchoring pilin subunits. Unique to FctB, however, is a long proline-rich C-terminal tail forming a polyproline-II (PPII) helix before the start of the sorting signal. In the major pilin, the sorting signal begins immediately after the globular domain, with little separation (Linke et al., 2010). Closer examination of other pilin sequences revealed that most anchoring pilin subunits have a proline-rich tail separating the globular domain from the sorting signal (Linke et al., 2010). This difference may allow the housekeeping sortase to preferentially utilize the anchoring pilin subunits, while the pilin polymerizing sortase prefers the sorting signal more proximal to the globular domain (see discussion on SrtC structure below).
Some class C sortases also promiscuously integrate pilin subunits. In many species, the basal pilin is also intermittently integrated into the pilus shaft rather than terminating polymerization and resulting in cell wall anchoring (Fig. 3). For example, in C. diphtheriae, SpaB is generally necessary for cell wall anchoring, but it is not sufficient, as in some cases it can be incorporated into the pilus by being attached to a SpaA molecule (Fig. 3B). This process occurs less frequently than SpaA incorporation, as immuno-gold microscopy experiments indicate that only small amounts of SpaB are interspersed throughout the pilus shaft (Ton-That and Schneewind, 2003). This presumably occurs because Cd-SrtA occasionally recognizes the LAFTG sorting signal of SpaB and attaches it to the pilin motif lysine within SpaA. It is unclear how frequently the basal pilin is incorporated in the pilus shaft; however, it has been reported in other pilin systems, including SpaD from C. diphtheriae (Gaspar and Ton-That, 2006), S. agalactiae (Rosini et al., 2006) and S. pneumoniae (LeMieux et al., 2006; Falker et al., 2008). In some cases, the minor tip pilin has also been seen interspersed throughout the pilus shaft. However, this issue is controversial, as more recent higher resolution EM studies of a pilus from S. pneumoniae have shown that its minor tip and basal pilin subunits are only present at the termini (LeMieux et al., 2008; Hilleringmann et al., 2009). For the case of the minor tip pilin, its presence within the shaft of the pilus in low-resolution EM images could be artefactual and caused by the bundling of pili of differing lengths. The presence of basal minor pilin in these EM images presumably is not artificial, as bundling would not explain its positioning distal to the cell wall. Instead, the presence of the basal minor pilin subunits within the shaft presumably occurs because the sortase enzymes that assemble them are promiscuous in the substrates that they recognize.
Molecular basis of catalysis – why so slow?
Given their key roles in displaying virulence factors and assembling pili, considerable effort has been put forth to understand the catalytic mechanism of transpeptidation. This process is best understood for Sa-SrtA, as the transpeptidation reaction it catalyses can be replicated in vitro using peptide substrates (Marraffini et al., 2006; Clancy et al., 2010). Structural and biochemical experiments have shown that Sa-SrtA adopts an eight-stranded β-barrel structure that houses three essential active site residues: His120, Cys184 and Arg197 (Fig. 4). Transpeptidation proceeds through a ping-pong mechanism that is initiated when the LPXTG sorting signal binds to a large groove adjacent to the active site (Huang et al., 2003; Frankel et al., 2005). An NMR structure of Sa-SrtA covalently bound to an analogue of the sorting signal revealed that binding occurs through an induced-fit mechanism (Suree et al., 2009). A large active loop, called the β6/β7 loop, undergoes a disordered–ordered transition so as to partially encapsulate the leucine residue within the sorting signal. The active site thiol of Cys184 then nucleophilically attacks the carbonyl carbon of the substrate threonine residue, forming a transient tetrahedral oxyanion intermediate that may be stabilized by Arg197 (Huang et al., 2003; Frankel et al., 2005). This intermediate rearranges into a more stable thioacyl enzyme-substrate complex after breakage of the threonine-glycine peptide bond. The terminal amine group within the pentaglycine branch of lipid II then nucleophilically attacks the carbonyl carbon of the threonine, initially generating a second tetrahedral intermediate that subsequently rearranges into the final product. How lipid II is recognized remains a mystery, but based on NMR data, binding of the sorting signal may induce a structural change that unmasks a second groove on the enzyme that interacts with this substrate (Suree et al., 2009). The side-chain of His120 is positioned within this groove, and may function as a general acid to protonate the substrate leaving group of the first tetrahedral intermediate, and/or as a general base that deprotonates the amine group of the lipid II nucleophile (Frankel et al., 2005; 2007). Other sortases likely use a similar mechanism to catalyse transpeptidation as the active site arginine, cysteine and histidine residues are conserved.
Fig. 4.
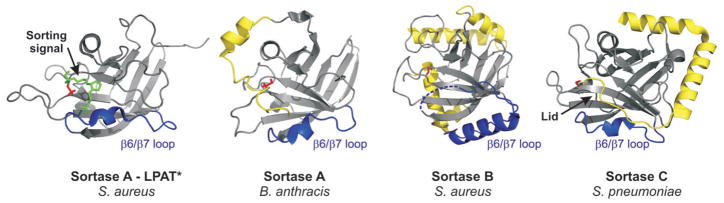
Sortase structure overview. Representative structures of sortase enzymes from different classes are shown. Class A: SrtA from S. aureus in complex with the sorting signal analogue LPAT* (green) (PDB code 2KID) and SrtA from B. anthracis (PDB code 2KW8). Class B: SrtB from S. aureus (PDB code 1NG5). Class C: SrtC1 from S. pneumoniae (PDB code 3G66). The β6/β7 loops are coloured in blue. Unique structural features are highlighted in yellow: the N-terminal extension in B. anthracis SrtA, the additional helices in SrtB from S. aureus and the flexible lid in S. pneumoniae SrtC1. The catalytic cysteine residues are highlighted in red and shown in a stick representation.
Atomic structures of representative class A, B and C enzymes have revealed interesting structural variations that could contribute to function (Fig. 4) (Ilangovan et al., 2001; Zhang et al., 2004; Zong et al., 2004a,b; Manzano et al., 2008; 2009; Neiers et al., 2009; Race et al., 2009; Suree et al., 2009; Weiner et al., 2010; Cozzi et al., 2011; Kang et al., 2011; Lu et al., 2011; Persson, 2011). Perhaps not surprisingly, class C enzymes that assemble pili exhibit some of the largest structural differences with Sa-SrtA. Crystal structures have revealed that these enzymes contain a novel polypeptide segment called a ‘lid’ that contacts the active site and masks the presumed binding site for the sorting signal (Manzano et al., 2008; Neiers et al., 2009; Cozzi et al., 2011; Lu et al., 2011; Persson, 2011). S. agalactiae SrtC1 proteins containing mutations in the lid display similar in vitro activity compared with the wild-type enzyme (Cozzi et al., 2011), and recent studies of S. pneumoniae SrtC-1 suggest that its lid is unstable in the apo-enzyme (Manzano et al., 2009). This has led to the hypothesis that the lid has a regulatory function, restricting sorting signal access to the active site until other factors or substrates involved in pilus biogenesis dislodge it (Cozzi et al., 2011). Interestingly, not all class C enzymes contain a lid, suggesting that other regulatory mechanisms may be at work for these sortases. This issue is only beginning to be explored, but studies thus far have shown that enzymes lacking a lid can require additional factors for proper pilus assembly. For example, the S. suis SrtC enzyme does not contain a lid and the assembly reaction it mediates requires the minor pilin subunit Spg2 to initiate pilus formation (Lu et al., 2011; Okura et al., 2011). The pilin polymerase class B enzyme from S. pyogenes also doesn’t contain a lid and requires the SipA2/LepA protein to construct the pilus (Zahner and Scott, 2008; Kang et al., 2011).
Class B sortases also exhibit unique structural variations that may enable them to recognize unusual sorting signals (Zhang et al., 2004; Zong et al., 2004b; Kang et al., 2011). In the structures of these enzymes the short β6/β7 loop present in class A and C sortases is replaced with a large extended α-helical segment. In Sa-SrtB, this segment contains residues important for recognizing its novel NPQTN sorting signal, as domain swapping experiments that replaced the β6/β7 loop in Sa-SrtA with the corresponding extended loop in Sa-SrtB changed the specificity profile towards that of Sa-SrtB (Bentley et al., 2007). Interestingly, the recently determined structure of the pilin polymerase class B enzyme from S. pyogenes also contains an extended α-helical segment, which is presumably involved in recognizing an unusual EVPTG sorting signal (Kang et al., 2011). Structural studies of several class A enzymes have also revealed interesting variations that may be important for function. In addition to Sa-SrtA, the structures of the class A enzymes from B. anthracis and S. pyogenes have been determined (Race et al., 2009; Weiner et al., 2010). Surprisingly, both enzymes appear to contain a preformed binding pocket for the sorting signal, as the β6/β7 loop in these enzymes is structurally ordered in the absence of their substrate. Thus, unlike Sa-SrtA, these enzymes may recognize the LPXTG sorting signal through a lock-and-key mechanism. Interestingly, the B. anthracis class A enzyme also contains an N-terminal α-helical appendage that contacts the imidazole ring of the active site histidine residue (Weiner et al., 2010). It may participate in the recognition of lipid II, or could have a regulatory function similar to the lid structures found in class C enzymes. Clearly, much work needs to be done to understand how these structural differences impact sortase function.
A common feature shared by sortase enzymes studied to date is that they have low enzymatic activity in vitro. For the case of Sa-SrtA, McCafferty and colleagues have proposed that key active site residues in this enzyme are improperly ionized (Frankel et al., 2005). To be active, the Cys184 and His120 side-chains presumably need to be in their ionized thiolate and immidazolium forms respectively. However, based on pKa measurements, only ~ 0.1% of the Sa-SrtA exists in this state; in the remainder of the enzymes the cysteine and histidine residues are uncharged (Connolly et al., 2003; Frankel et al., 2005). The available structural data suggest that other sortases may have a similar problem that reduces their activity, because in all structures determined to date the active site histidine and cysteine residues are positioned too far apart from one another to form a thiolate-immidazolium ion pair, and are thus presumably uncharged. This has recently been substantiated for the B. anthracis SrtA enzyme, as pKa measurements indicate that its active site histidine is predominantly uncharged at physiological pH (Weiner et al., 2010). On the cell surface, sortase enzymes may need to associate with other factors to be fully functional. These factors may facilitate catalysis by properly guiding the substrates into the enzyme active site and/or they could alter the electrostatic environment such that the active site cysteine was converted to its nucleophilic thiolate form. This notion is compatible with biochemical data that have shown that some sortases require the Spa2/LepA protein to be fully functional (Zahner and Scott, 2008; Okura et al., 2011).
Colocalizing protein secretion, anchoring and cell wall synthesis machinery may increase the efficiency of protein display
Several recent findings suggest that efficiency and fidelity of protein anchoring is increased by coordinating this process with protein secretion and cell wall synthesis. The secretion translocon in Gram-positive bacteria is localized to discrete sites. In rod-shaped Bacillus subtilis, protein secretion appears to be restricted to spiral-like clusters along the longitudinal axis of the cell (Campo et al., 2004), while in streptococcal cells single or multiple distinct secretion foci have been detected adjacent to the newly forming septum (Rosch and Caparon, 2004; Carlsson et al., 2006). Immuno-gold electron micrographs indicate that sortases colocalize with the SecA component of the translocation machinery at these foci, suggesting that protein secretion and anchoring occur at the same site in streptococci and enterococci (Hu et al., 2008; Kline et al., 2009). The efficiency of the sorting reaction may also be increased by localizing it to sites where the cell wall is being synthesized and where the lipid II substrate is presumably most abundant. This has been observed in S. pyogenes as its class A enzyme predominately localizes to the septum, with its presence peaking during septation and then decreasing after cell division (Raz and Fischetti, 2008). Similarly, in dividing S. aureus cells proteins attached to the cell wall by Sa-SrtA are mainly deposited at two to four foci close to a layer of newly synthesized cell wall. Cell growth and expansion then generate a more diffuse distribution of the proteins in a ring-like pattern (DeDent et al., 2007; 2008). Interestingly, in S. aureus some proteins may also be anchored to a second site that is on the cell pole distal to the septum. The final destination of Sa-SrtA attached proteins seems to be determined by their signal peptides, with proteins harbouring a conventional signal sequence at their N-termini being directed to peripheral sites, while proteins containing a signal peptide with an YSIRK/GS motif are retained at sites close to the septum where their attachment is presumably coupled to cell wall synthesis (DeDent et al., 2008). Sequence elements within sortases may also function to properly direct them to specific sites on the cell surface, as deletion of the positively charged residues at the C-terminus of the class C enzyme from E. faecalis resulted in decreased focal localization of the enzyme and disrupted pilus biogenesis (Kline et al., 2009). Interestingly, deletion of srtA or srtC genes involved in pilin biogenesis also caused their respective substrates to accumulate at a single focus, suggesting that the CWSS or another sequence element may act to retain the substrates at the secretion locus before them being processed by a sortase. In C. diphtheria, the major and minor pilins that form the SpaA pilus colocalize with their cognate sortase, and led to the proposal by Ton-That and colleagues that pilus biogenesis in Gram-positive bacteria occurs within highly organized ‘pilusosome’ assembly centres (Guttilla et al., 2009). Formation of sortase oligomers could further function to increase the efficiency of the sorting reaction. Sa-SrtA dimers detected by gel electrophoresis under native conditions and size exclusion chromatography were reported to display increased activity compared with monomeric proteins (Lu et al., 2007). Sortase dimerization could be disrupted by amino acid substitutions in the putative interaction surface (Zhu et al., 2008). However, it remains to be seen whether sortases also interact on the cell surface and whether formation of such oligomers facilitates catalysis in vivo.
Conclusion
Members of the sortase enzyme superfamily decorate the surfaces of Gram-positive bacteria with proteins that play key roles in microbial physiology and pathogenesis. Genome sequencing efforts have identified nearly a thousand sortase homologues whose functions are only beginning to be discovered. All sortases characterized to date are cysteine transpeptidases that either covalently join proteins to the cell wall peptidoglycan, or link proteins together to construct pili. Based on their primary sequences around ~60% of sortase homologues in Gram-positive bacteria can be clustered into six families, called class A to F enzymes. Class A and C sortases have been studied extensively. Most class A enzymes appear to have a housekeeping role in the cell as members of this group attach a large number of functionally distinct proteins to the cell wall, while all class C enzymes studied to date assemble pili. The functions of other types of sortases are less clear as only a few representative members have been investigated, and in some instances related enzymes within a given class have been shown to have distinct functions. In the future, it will therefore be important to move beyond classification schemes that are based solely on overall primary sequence homology by identifying specific enzyme features that confer function. In particular, structure-function studies will need to be performed to identify enzyme determinants that control substrate specificity. This will bring to light how sortases selectively route different proteins to the cell surface by recognizing their unique sorting signals, and reveal how they can function to either attach proteins to the cell wall or assemble pili. It will also be critical to learn the molecular basis through which sortases work with other enzymes and accessory factors on the cell surface, which will require obtaining a better understanding of the targeting and retention mechanisms of these proteins as well as the generation of robust in vitro systems to study pilus assembly. Many surprises likely await as hundreds of sortase homologues can’t be classified into the aforementioned subfamilies suggesting that they will be found to perform novel functions on the cell surface.
Acknowledgments
Because of space limitations we were unable to fully discuss all sortase-related research and apologize to those investigators whose work has not been mentioned here. We would like to thank Dr. Marie Elliot and the three anonymous reviewers of this manuscript for very helpful comments. This work was supported by Swiss National Science Foundation Fellowship PBEZP3-124281 (T.S.), Ruth L. Kirschstein National Research Service Award GM07185 (E.W.) and National Institutes of Health Grant AI52217 (R.T.C).
References
- Bagnoli F, Moschioni M, Donati C, Dimitrovska V, Ferlenghi I, Facciotti C, et al. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J Bacteriol. 2008;190:5480–5492. doi: 10.1128/JB.00384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett TC, Patel AR, Scott JR. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J Bacteriol. 2004;186:5865–5875. doi: 10.1128/JB.186.17.5865-5875.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley ML, Gaweska H, Kielec JM, McCafferty DG. Engineering the substrate specificity of Staphylococcus aureus Sortase A. The beta6/beta7 loop from SrtB confers NPQTN recognition to SrtA. J Biol Chem. 2007;282:6571–6581. doi: 10.1074/jbc.M610519200. [DOI] [PubMed] [Google Scholar]
- Boekhorst J, de Been MW, Kleerebezem M, Siezen RJ. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J Bacteriol. 2005;187:4928–4934. doi: 10.1128/JB.187.14.4928-4934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Marraffini LA, Schneewind O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol. 2007;66:495–510. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- Budzik JM, Marraffini LA, Souda P, Whitelegge JP, Faull KF, Schneewind O. Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci USA. 2008a;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Oh SY, Schneewind O. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J Biol Chem. 2008b;283:36676–36686. doi: 10.1074/jbc.M806796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Poor CB, Faull KF, Whitelegge JP, He C, Schneewind O. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc Natl Acad Sci USA. 2009a;106:19992–19997. doi: 10.1073/pnas.0910887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Oh SY, Schneewind O. Sortase D forms the covalent bond that links BcpB to the tip of Bacillus cereus pili. J Biol Chem. 2009b;284:12989–12997. doi: 10.1074/jbc.M900927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, et al. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- Capstick DS, Willey JM, Buttner MJ, Elliot MA. SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol Microbiol. 2007;64:602–613. doi: 10.1111/j.1365-2958.2007.05674.x. [DOI] [PubMed] [Google Scholar]
- Carlsson F, Stalhammar-Carlemalm M, Flardh K, Sandin C, Carlemalm E, Lindahl G. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442:943–946. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- Chang C, Mandlik A, Das A, Ton-That H. Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol Microbiol. 2011;79:1236–1247. doi: 10.1111/j.1365-2958.2010.07515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FG, et al. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy KW, Melvin JA, McCafferty DG. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers. 2010;94:385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfort D, Clubb RT. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun. 2004;72:2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly KM, Smith BT, Pilpa R, Ilangovan U, Jung ME, Clubb RT. Sortase from Staphylococcus aureus does not contain a thiolate-imidazolium ion pair in its active site. J Biol Chem. 2003;278:34061–34065. doi: 10.1074/jbc.M305245200. [DOI] [PubMed] [Google Scholar]
- Cozzi R, Malito E, Nuccitelli A, D’Onofrio M, Martinelli M, Ferlenghi I, et al. Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus. FASEB J. 2011;25:1874–1886. doi: 10.1096/fj.10-174797. [DOI] [PubMed] [Google Scholar]
- DeDent A, Bae T, Missiakas DM, Schneewind O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 2008;27:2656–2668. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDent AC, McAdow M, Schneewind O. Distribution of protein A on the surface of Staphylococcus aureus. J Bacteriol. 2007;189:4473–4484. doi: 10.1128/JB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, et al. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 2003;17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falker S, Nelson AL, Morfeldt E, Jonas K, Hultenby K, Ries J, et al. Sortase-mediated assembly and surface topology of adhesive pneumococcal pili. Mol Microbiol. 2008;70:595–607. doi: 10.1111/j.1365-2958.2008.06396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel BA, Kruger RG, Robinson DE, Kelleher NL, McCafferty DG. Staphylococcus aureus sortase transpeptidase SrtA: insight into the kinetic mechanism and evidence for a reverse protonation catalytic mechanism. Biochemistry. 2005;44:11188–11200. doi: 10.1021/bi050141j. [DOI] [PubMed] [Google Scholar]
- Frankel BA, Tong Y, Bentley ML, Fitzgerald MC, McCafferty DG. Mutational analysis of active site residues in the Staphylococcus aureus transpeptidase SrtA. Biochemistry. 2007;46:7269–7278. doi: 10.1021/bi700448e. [DOI] [PubMed] [Google Scholar]
- Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, Gaspar AH, Swierczynski A, Swaminathan A, Dwivedi P, Das A, Ton-That H. Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in gram-positive bacteria. J Bacteriol. 2009;191:5603–5612. doi: 10.1128/JB.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hava DL, Hemsley CJ, Camilli A. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J Bacteriol. 2003;185:413–421. doi: 10.1128/JB.185.2.413-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx AP, Budzik JM, Oh SY, Schneewind O. Architects at the bacterial surface – sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- Hilleringmann M, Ringler P, Muller SA, De Angelis G, Rappuoli R, Ferlenghi I, Engel A. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 2009;28:3921–3930. doi: 10.1038/emboj.2009.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Bian Z, Fan M, Huang M, Zhang P. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch Oral Biol. 2008;53:150–154. doi: 10.1016/j.archoralbio.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Huang X, Aulabaugh A, Ding W, Kapoor B, Alksne L, Tabei K, Ellestad G. Kinetic mechanism of Staphylococcus aureus sortase SrtA. Biochemistry. 2003;42:11307–11315. doi: 10.1021/bi034391g. [DOI] [PubMed] [Google Scholar]
- Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci USA. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson IM, Mazmanian SK, Schneewind O, Bremell T, Tarkowski A. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microbes Infect. 2003;5:775–780. doi: 10.1016/s1286-4579(03)00143-6. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Baker EN. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J Biol Chem. 2009;284:20729–20737. doi: 10.1074/jbc.M109.014514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci USA. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Proft T, Baker EN. Crystal structure of Spy0129, a Streptococcus pyogenes class B sortase involved in pilus assembly. PLoS ONE. 2011;6:e15969. doi: 10.1371/journal.pone.0015969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, et al. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol. 2009;191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMieux J, Hava DL, Basset A, Camilli A. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect Immun. 2006;74:2453–2456. doi: 10.1128/IAI.74.4.2453-2456.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMieux J, Woody S, Camilli A. Roles of the sortases of Streptococcus pneumoniae in assembly of the RlrA pilus. J Bacteriol. 2008;190:6002–6013. doi: 10.1128/JB.00379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke C, Young PG, Kang HJ, Bunker RD, Middleditch MJ, Caradoc-Davies TT, et al. Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring. J Biol Chem. 2010;285:20381–20389. doi: 10.1074/jbc.M109.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zhu J, Wang Y, Umeda A, Cowmeadow RB, Lai E, et al. Staphylococcus aureus sortase A exists as a dimeric protein in vitro. Biochemistry. 2007;46:9346–9354. doi: 10.1021/bi700519w. [DOI] [PubMed] [Google Scholar]
- Lu G, Qi J, Gao F, Yan J, Tang J, Gao GF. A novel ‘open-form’ structure of sortase C from Streptococcus suis. Proteins. 2011;79:2764–2769. doi: 10.1002/prot.23093. [DOI] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008a;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci USA. 2008b;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C, Contreras-Martel C, El Mortaji L, Izore T, Fenel D, Vernet T, et al. Sortase-mediated pilus fiber biogenesis in Streptococcus pneumoniae. Structure. 2008;16:1838–1848. doi: 10.1016/j.str.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Manzano C, Izore T, Job V, Di Guilmi AM, Dessen A. Sortase activity is controlled by a flexible lid in the pilus biogenesis mechanism of gram-positive pathogens. Biochemistry. 2009;48:10549–10557. doi: 10.1021/bi901261y. [DOI] [PubMed] [Google Scholar]
- Maresso AW, Schneewind O. Iron acquisition and transport in Staphylococcus aureus. Biometals. 2006;19:193–203. doi: 10.1007/s10534-005-4863-7. [DOI] [PubMed] [Google Scholar]
- Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacol Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- Maresso AW, Chapa TJ, Schneewind O. Surface protein IsdC and Sortase B are required for heme-iron scavenging of Bacillus anthracis. J Bacteriol. 2006;188:8145–8152. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Schneewind O. Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J Biol Chem. 2005;280:16263–16271. doi: 10.1074/jbc.M500071200. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Schneewind O. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol Microbiol. 2006;62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Schneewind O. Sortase C-mediated anchoring of BasI to the cell wall envelope of Bacillus anthracis. J Bacteriol. 2007;189:6425–6436. doi: 10.1128/JB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci USA. 2002;99:2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- Mishra A, Das A, Cisar JO, Ton-That H. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol. 2007;189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci USA. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiers F, Madhurantakam C, Falker S, Manzano C, Dessen A, Normark S, et al. Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity. J Mol Biol. 2009;393:704–716. doi: 10.1016/j.jmb.2009.08.058. [DOI] [PubMed] [Google Scholar]
- Newton SM, Klebba PE, Raynaud C, Shao Y, Jiang X, Dubail I, et al. The svpA-srtB locus of Listeria monocytogenes: fur-mediated iron regulation and effect on virulence. Mol Microbiol. 2005;55:927–940. doi: 10.1111/j.1365-2958.2004.04436.x. [DOI] [PubMed] [Google Scholar]
- Nobbs AH, Rosini R, Rinaudo CD, Maione D, Grandi G, Telford JL. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect Immun. 2008;76:3550–3560. doi: 10.1128/IAI.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura M, Osaki M, Fittipaldi N, Gottschalk M, Sekizaki T, Takamatsu D. The Minor Pilin Subunit Sgp2 is Necessary for Assembly of the Pilus Encoded by the srtG Cluster of Streptococcus suis. J Bacteriol. 2011;193:822–831. doi: 10.1128/JB.01555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Lam AC, Antonio M, Dunbar K. An embarrassment of sortases – a richness of substrates? Trends Microbiol. 2001;9:97–102. doi: 10.1016/s0966-842x(01)01956-4. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Chaudhuri RR, Henderson IR. Genomic analysis of secretion systems. Curr Opin Microbiol. 2003;6:519–527. doi: 10.1016/j.mib.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Persson K. Structure of the sortase AcSrtC-1 from Actinomyces oris. Acta Crystallogr D Biol Crystallogr. 2011;67:212–217. doi: 10.1107/S0907444911004215. [DOI] [PubMed] [Google Scholar]
- Pishchany G, McCoy AL, Torres VJ, Krause JC, Crowe JE, Jr, Fabry ME, Skaar EP. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe. 2010;8:544–550. doi: 10.1016/j.chom.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Dougan SK, Chuang TY, Spooner E, Ploegh HL. Sortase-catalyzed transformations that improve the properties of cytokines. Proc Natl Acad Sci USA. 2011;108:3169–3174. doi: 10.1073/pnas.1016863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T. Sortase-mediated protein ligation: an emerging biotechnology tool for protein modification and immobilisation. Biotechnol Lett. 2010;32:1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria – structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley BR, Zahner D, Hatkoff M, Thanassi DG, Scott JR. Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Mol Microbiol. 2009;72:1379–1394. doi: 10.1111/j.1365-2958.2009.06727.x. [DOI] [PubMed] [Google Scholar]
- Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, et al. Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J Biol Chem. 2009;284:6924–6933. doi: 10.1074/jbc.M805406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz A, Fischetti VA. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc Natl Acad Sci USA. 2008;105:18549–18554. doi: 10.1073/pnas.0808301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- Scott JR, Barnett TC. Surface proteins of gram-positive bacteria and how they get there. Annu Rev Microbiol. 2006;60:397–423. doi: 10.1146/annurev.micro.60.080805.142256. [DOI] [PubMed] [Google Scholar]
- Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- Smith WD, Pointon JA, Abbot E, Kang HJ, Baker EN, Hirst BH, et al. Roles of minor pilin subunits Spy0125 and Spy0130 in the serotype M1 Streptococcus pyogenes strain SF370. J Bacteriol. 2010;192:4651–4659. doi: 10.1128/JB.00071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suree N, Jung ME, Clubb RT. Recent advances towards new anti-infective agents that inhibit cell surface protein anchoring in Staphylococcus aureus and other gram-positive pathogens. Mini Rev Med Chem. 2007;7:991–1000. doi: 10.2174/138955707782110097. [DOI] [PubMed] [Google Scholar]
- Suree N, Liew CK, Villareal VA, Thieu W, Fadeev EA, Clemens JJ, et al. The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J Biol Chem. 2009;284:24465–24477. doi: 10.1074/jbc.M109.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczynski A, Ton-That H. Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188:6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004a;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004b;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- Tsukiji S, Nagamune T. Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering. Chembiochem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- Weiner EM, Robson S, Marohn M, Clubb RT. The Sortase A enzyme that attaches proteins to the cell wall of Bacillus anthracis contains an unusual active site architecture. J Biol Chem. 2010;285:23433–23443. doi: 10.1074/jbc.M110.135434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner D, Scott JR. SipA is required for pilus formation in Streptococcus pyogenes serotype M3. J Bacteriol. 2008;190:527–535. doi: 10.1128/JB.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wu R, Joachimiak G, Mazmanian SK, Missiakas DM, Gornicki P, et al. Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site. Structure. 2004;12:1147–1156. doi: 10.1016/j.str.2004.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lu C, Standland M, Lai E, Moreno GN, Umeda A, et al. Single mutation on the surface of Staphylococcus aureus Sortase A can disrupt its dimerization. Biochemistry. 2008;47:1667–1674. doi: 10.1021/bi7014597. [DOI] [PubMed] [Google Scholar]
- Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SV. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J Biol Chem. 2004a;279:31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- Zong Y, Mazmanian SK, Schneewind O, Narayana SV. The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall. Structure. 2004b;12:105–112. doi: 10.1016/j.str.2003.11.021. [DOI] [PubMed] [Google Scholar]