Abstract
This chapter describes the methods to form and optimize samples of protein–DNA complexes that are suitable for detailed structure and dynamics studies by NMR spectroscopy.
Keywords: Protein–DNA complex, NMR, Structure, Intermolecular NOEs
1. Introduction
Interactions between proteins and DNA molecules play an essential role in a wide range of important biological processes, including gene expression and genomic replication, recombination, and repair. Of particular interest are site-specific DNA-binding transcription factors, which regulate gene expression by recognizing specific nucleotide sequences. These proteins locate, and bind to, the correct site within the genome despite the presence of a vast number of competitor sites with similar geometries and electrostatic surfaces. Over the past several decades, X-ray crystallography has been used extensively to determine a large number of high-resolution structures of protein–DNA complexes (1, 2). This work has provided a wealth of detailed stereochemical information about binding site recognition, which typically is achieved through complementary hydrogen-bonding and van der Waals interactions that can be maximized by protein folding and/or DNA distortions (3, 4). However, crystallography provides only a static view of a protein–DNA complex, and thus little insight into the conformational dynamics that underpin macromolecular recognition (5).
NMR spectroscopy is a powerful tool that can be used to investigate protein–DNA recognition in the solution state. When the spectra obtained are of good quality, NMR can be used to elucidate high-resolution structures and atomic-level conformational dynamics (6, 7). NMR can also be applied to investigate other key aspects of recognition, including the basis of nonspecific binding (8), hydration lifetimes (9), on/off rates of binding, and the process by which a protein locates its binding site (10). Even when the quality of the NMR spectra are poor due to resonance line broadening, structural models of a protein–DNA complex can be generated using chemical shift mapping techniques. In recent years, the size and complexity of protein–DNA complexes amenable for NMR studies have increased due to several methodological advances, such as selective isotopic labeling, residual dipolar coupling measurements (11), paramagnetic relaxation enhancement methods (12), and transverse relaxation-optimized experiments, that exploit ultrahigh magnetic field strengths (13, 14). However, one of the largest obstacles to successfully studying a protein–DNA complex by NMR is the preparation of sufficiently stable, concentrated, and homogeneous samples of the complex.
For a protein–DNA complex to be suitable for detailed NMR studies, it typically must satisfy several criteria. The components and nature of the interaction should be well-defined. In particular, biochemical experiments should have been performed to clearly delineate the specific nucleotide sequence recognized by the protein, as well as the stoichiometry and affinity of the resulting complex. The same molecular weight limitations that hinder NMR studies of other macromolecules apply, so the final size of the complex is also an important consideration. Ideally, the complex should have a dissociation constant (Kd) in the submicromolar range, thus making it more likely that it will be in the slow-exchange regime on the chemical shift timescale. However, weaker affinity complexes that are in fast exchange have also been successfully studied. To optimize the production and spectral qualities of a complex, large quantities of DNA and isotopically labeled protein are also needed to enable different preparative procedures and conditions to be tested. Pilot studies typically make use of purified 15N-enriched protein and a range of DNA species that differ in their length and sequence. In our experience, the greatest chance of success occurs when the protein is soluble in its DNA-free state and its 1H–15N HSQC spectrum is well-resolved. However, DNA is highly soluble and as a result the aggregation behavior of the protein may be greatly reduced upon complex formation.
To create a stable complex suitable for NMR studies, the DNA molecule must have the appropriate nucleotide sequence and length to form productive contacts with the protein. For sequence-specific DNA binding proteins, this information can be obtained from previously reported biochemical studies, which should define the specificity, affinity, and stoichiometry of the complex. If the protein is known to bind to several DNA sites, then an alignment of their nucleotide sequences may reveal conserved positions essential for binding. This knowledge is helpful later in optimizing the spectra of the protein–DNA complex, since it identifies nucleotides within the DNA molecule that can presumably be altered without affecting stability. To reduce spectral overlap, the minimal DNA sequence with good binding affinity for the protein should be used to make the complex. If the structure of the protein in the DNA-free state is known, a model of it docked to B-form DNA should be constructed. This may help to determine the minimum DNA length that can be used, and whether additional nucleotides outside of the known binding site are required to form nonspecific stabilizing contacts. In practice, DNA molecules studied in our group almost always contain a G:C base pair (bp) at each end, which limits fraying by increasing the melting temperature.
Minor changes in the DNA and protein sequences can dramatically affect the NMR spectrum of a protein–DNA complex and are therefore parameters that can be optimized. A common mistake is to choose a DNA fragment that is too long with unnecessary base pairs at either its 3′ or 5′ end. This can be problematic as longer DNA fragments can contain weaker, “cryptic” binding sites for the protein that become occupied at the high protein concentrations present in the NMR sample (typically > 0.5 mM). For example, a protein that forms numerous interactions with an A-T sequence located at the center of the primary site might also bind to a secondary A-T dinucleotide sequence present elsewhere in a longer DNA fragment. If this occurs, the multiple binding modes of the protein cause resonance line broadening. Modeling studies and a comparison of the DNA-binding sites can be used to identify potential “cryptic” sites, which can then be eliminated by altering the nucleotide sequence of the DNA molecule. A nucleotide sequence comparison can also identify dsDNA molecules that have NMR spectra that can more readily be assigned. For example, it may be preferable to maximize the number of thymine bases in the sequence, as its methyl groups are good anchor points in the assignment process. The length and sequence of the protein can also be adjusted to improve spectral quality. Typically, this involves deleting unstructured amino acids at the polypeptide termini to reduce spectral overlap. However, even subtle single amino acid changes can have a dramatic impact on spectral quality. For example, in our studies of an ARID-DNA complex, a single phenylalanine-to-leucine mutation was found to dramatically reduce line broadening, salvaging a protein–DNA complex that was originally ill suited for structural analysis by NMR (15). This biochemical approach is not a general method, but may prove useful in the spectral optimization of other protein complexes that suffer from interracial line broadening caused by dynamic changes in proximal aromatic rings.
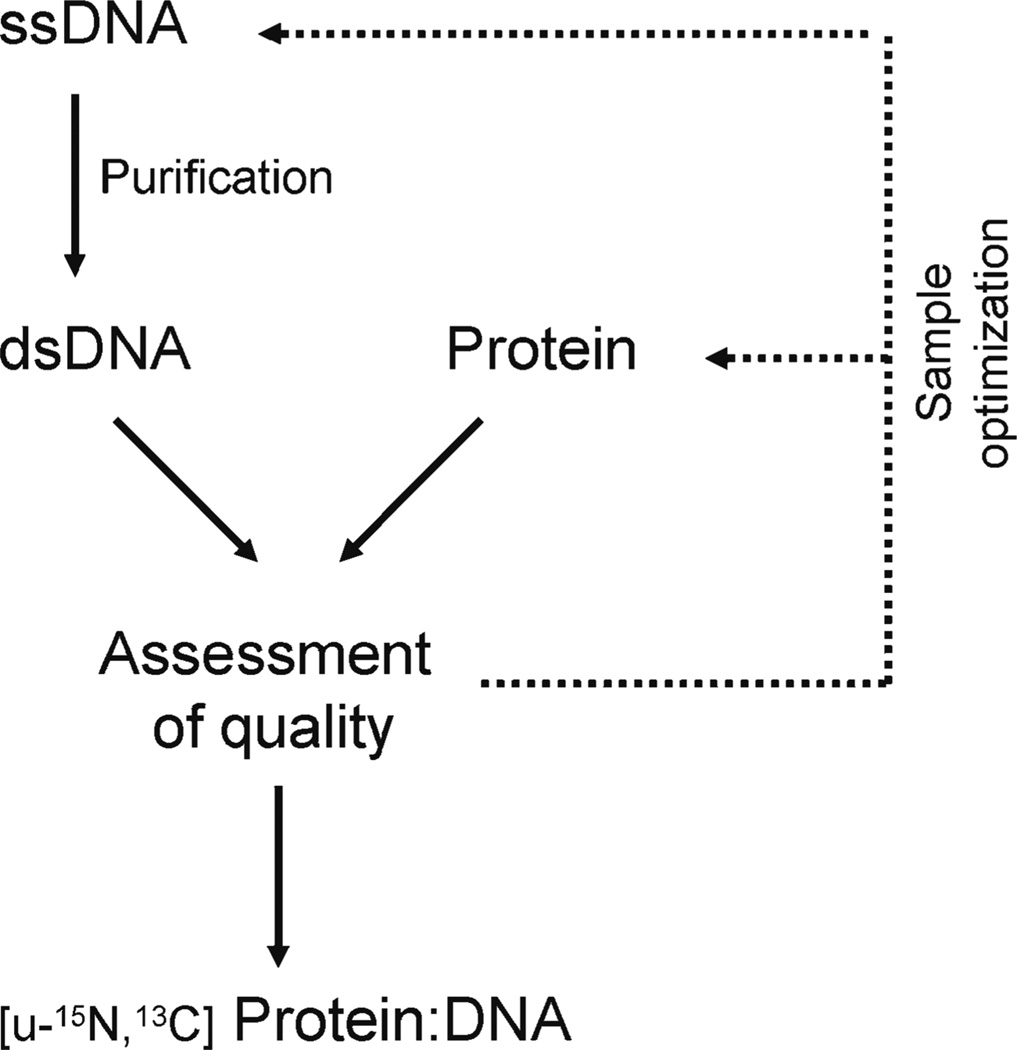
In this chapter, we outline the approaches we typically use to form protein–DNA complexes suitable for NMR studies. The overall procedure for this protocol is outlined in Fig. 1.
Fig. 1.
Flowchart showing the procedures used to form and optimize protein–DNA complexes for NMR studies.
2. Materials
The exact reagents used to form protein–DNA complexes suitable for high-resolution solution-state NMR studies vary depending upon the specific system that is being studied. In this section, the materials used to produce the Integrase(Int)-DNA complex are described (16).
2.1. Binding Affinity Measurements
2× binding buffer: 40 mM Tris–HCl, pH 7.5, 40 mM NaCl, 40 mM KC1, 10% (w/v) glycerol, 2 mM EDTA, and 2 mM DTT.
32P-labeled DNA: Labeled at its 5′-termini with T4 polynucleotide kinase and γ-32P-ATP.
PhosphorImager (Molecular Dynamics Inc.): To quantify radioactivity in the gels.
1× TBE: For 1 L of 10× TBE, dissolve 108 g of Tris base, 55 g of boric acid, and 7.4 g of disodium EDTA salt. Dilute tenfold to get 1× TBE.
Bovine serum albumin (BSA): Stock concentration 1 µg/µL in H2O.
Poly dI/dC: Stock concentration 0.5 µ5/µL.
Protein stock solution: 20 µM (4× of the highest protein concentration used in the binding assay). For protein–DNA complexes with nanomolar affinity, a typical titration range is as follows (nM): 5,000, 1,000, 200, 100, 50, 25, 5, 1, 0.5, and 0. Protein should be dissolved in a solution that is most stable for that particular protein. This protein buffer is diluted in the binding assay and replaced with binding buffer solution.
6× DNA loading dyes: 0.03% (w/v) xylene cyanol FF and 0.03% (w/v) bromphenol blue in 10 mM Tris, pH 8, and 30% (w/v) glycerol.
1 M Tris, pH 8: Dissolve 121.1 g of Trizma (MW = 121.1 g/mol) in 800 mL of H2O, titrate with concentrated HC1 until pH 8 is achieved, and then bring final volume to 1 L.
2.2. Preparation of Purified Single-Stranded DNA
DNA oligonucleotides: 1 µmol scale synthesis (Integrated DNA Technologies, IDT) (see Note 1).
17% acrylamide–urea gel: For 400 mL total volume, mix 168.2 g of urea, 170 mL of 40% acrylamide (37.5:1), 40 mL of 10× TBE (Subheading 2.1), 60 mL of H2O, 1 mL of 10% (w/v) APS, and 100 µL of TEMED.
DNA loading buffer: 7 M urea, 50 mM Tris, pH 8.0 (using 1 M Tris, pH 8; Subheading 2.1), 5 mM EDTA (using 0.5 M EDTA, pH 8), and 10% (w/v) glycerol.
0.5 M EDTA, pH 8: Add 186.1 g of EDTA disodium salt and ~20 g of NaOH pellets to 700 mL of H2O (EDTA dissolves as it approaches pH 8), and then bring the solution to 1 L once pH 8 is achieved.
6× DNA loading dyes (see Subheading 2.1).
1× TBE (see Subheading 2.1).
FLEX TLC plates.
Electroelution chamber.
Dialysis buffer: 50 mM Tris–HCl, pH 7.5, 200 mM NaCl, and 2 mM EDTA (using 0.5 M EDTA, pH 8).
2.3. Preparation of Duplex DNA for NMR Studies
Annealing buffer: Same as dialysis buffer (Subheading 2.2).
D2O.
2.4. Preparation of the Protein–DNA Complex
High-salt protein buffer: 50 mM Hepes, pH 7.0 (using 1 M Hepes, pH 7), 500 mM NaCl, and 1 mM DTT.
1 M Hepes, pH 7: Stock solution is adjusted to pH with NaOH.
High-salt DNA buffer: 50 mM Tris, pH 7.5 (using 1 M Tris, pH 7.5, see Subheading 2.1, except adjust pH to 7.5), 500 mM NaCl, and 0.1 mM EDTA.
Low-salt buffer: 25 mM Hepes, pH 7.0 (using 1 M Hepes, pH 7), 15 mM NaCl, 2 mM DTT, 7% D2O, and 0.01% NaN3.
Centricon YM-3 centrifugal filter device (Amicon Bioseparations).
~50 µM protein solution.
~50 µM dsDNA solution.
3. Methods
Our lab has solved the structures of six protein–DNA complexes. Four have been determined by NMR spectroscopy and two have been determined by X-ray crystallography (17–22). Below, we describe the procedures we generally use to form complexes between a sequence-specific binding protein and a duplex-DNA molecule. Unless otherwise stated, the procedures described below are used to produce samples of the complex between the Int protein and its cognate DNA site (17–22). Four procedures are presented: (1) an electrophoretic mobility shift assay (EMSA) for affinity and specificity measurements, (2) methods to purify single-stranded DNA (ssDNA), (3) methods to prepare duplex DNA (dsDNA), and (4) the procedures used to assess the spectral quality of a protein–DNA complex to determine if additional NMR studies are warranted.
3.1. Binding Affinity Measurements
Biochemical assays should be available to rapidly estimate the affinity of wild-type and mutant proteins for different DNA molecules. A variety of methods can be employed to measure binding, such as the EMSA, isothermal titration calorimetry (ITC), fluorescence anisotropy, surface plasmon resonance (SPR; e.g., Biacore), and fluorescence quenching (if the protein contains an appropriately positioned tryptophan). However, we favor the EMSA because it is robust and simple to perform (23). This procedure has been described in detail previously (24) and is outlined below.
Mix the following components: 12 µL of 2× binding buffer, 3 µL of 1 µg/µL BSA, 2 µL of 0.5 µg/µL Poly dI/dC (competitor DNA) for a total of 17 µL.
Add to this mixture the protein stock solution and the appropriate amount of H2O to achieve a final volume of 23 µL (see Subheading 2.1 and Note 2).
Incubate on ice for 20 min.
Add 1 µL of 32P-labeled DNA probe (~4,000 cpm/mL).
Incubate on ice for 20 min.
Prepare an 8% polyacrylamide gel and pre-electrophorese the gel by running it for 30–60 min at 10 V/cm in 1× TBE at ambient temperature or 4°C (see Note 3).
Load the reaction mixtures onto an 8% polyacrylamide/TBE gel at 4°C (see Note 4). Load 6× DNA loading dyes in a separate lane as a reference to track the migration of the free DNA. The gel run time should be optimized for each specific system so as to resolve the species of interest and to be as short as possible. A typical gel run time is ~1.5 h at constant voltage (10 V/cm), but the voltage should be reduced if the gel becomes warm during electrophoresis.
Quantify the amount of free and bound DNA in each lane by using a PhosphorImager system or the equivalent.
Determine the dissociation constant by fitting to the following equation: θ = [L]/([L] + Kd), where θ, [L], and Kd are the fraction of DNA bound, the total protein concentration in the reaction, and the dissociation constant, respectively. θ is equal to the counts present in the shifted band divided by the total counts for the DNA (free plus shifted bands).
3.2. Preparation of Purified Single-Stranded DNA
In this section, we discuss how to purify large quantities of commercially available ssDNA oligonucleotides for NMR studies. We initially purchase the ssDNA in a crude form at a cost of ~$2 per nucleotide for 1 µmole of material. ssDNA with lengths less than 20 base pairs is purified on 20% acrylamide–urea gels and with lengths longer than 20 bp is purified on 17% gels. The ssDNA is then eluted from the gel and dialyzed into native buffer for further use. The procedure we use generally yields ssDNA that is >98% pure for molecules up to 40 bp in length. For ssDNA shorter than 15 bp, more conventional approaches are sufficient to produce ssDNA suitable for NMR studies (see Note 5).
Prepare one 17% acrylamide/urea gel with a single lane (see Note 6).
Dissolve the DNA oligonucleotide (1 µmol or 3–6 mg) in 2 mL of DNA loading buffer.
Load 20 µL of 6× DNA loading dyes on the right and left edge of the gel within the single lane. The migration of the xylene cyanol FF and bromophenol blue dyes along the gel gives an estimate of how far the DNA has migrated (see Note 7).
Run the gel in 1× TBE. It typically takes ~10–13 h for ssDNA (depending on the length) to migrate ~3/4 of the entire gel length. For a single gel running at a constant 50 W, the voltage is ~600–700 V (65–80 mA) (see Note 8).
Transfer the gel onto saran wrap by removing the top gel plate and placing a layer of saran wrap directly on top of the gel. Invert the gel onto FLEX TLC plates with the saran wrap on top of the TLC plates. Remove the remaining gel plate and in a darkroom, use a handheld UV lamp (254 nm) to locate the ssDNA. Quickly excise the DNA using a razor blade. Cut the excised gel containing the desired ssDNA into ½-in. pieces to ensure efficient DNA electroelution. The purpose of the FLEX TLC plate is to enhance the DNA signals during exposure to UV lights.
Assemble an Elutrap™ electroelution device (Whatman). Membranes for trapping DNA are BT1 (14 nucleotide cutoff) and BT2 (cellulose acetate membrane). Alternatively, BT1 can be replaced with a low-molecular-weight cutoff dialysis membrane.
Elute the DNA in 1× TBE. Run the Elutrap at 150 V for 8–10 h (remove eluted DNA from the Elutrap two to three times during this period).
Dialyze extensively with dialysis buffer to remove all denaturing reagents from the ssDNA.
Determine the DNA concentration from a UV absorbance reading at 260 nm (A260). The extinction coefficient may be calculated online using a program provided by IDT (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/).
3.3. Preparation of Duplex DNA for NMR Studies
Complementary purified ssDNA molecules are annealed to produce the appropriate DNA duplex. NMR and/or chromatographic approaches are then used to verify duplex formation prior to forming the protein–DNA complex. The steps used to generate dsDNA are outlined below.
Dissolve complementary ssDNA molecules in annealing buffer to a final concentration of ~100 µM. The samples should be free of urea and EDTA.
Heat the sample to ~95–100°C for 10 min and slowly cool to ambient temperature in the heat block. A water bath may be used in place of a heating block for larger annealing volumes.
Perform NMR experiments to ensure that the sample has properly annealed and that no excess ssDNA is present. Add an appropriate amount of D2O to maintain the field lock, and acquire an 1H 2D TOCSY spectrum (mixing time ~40 ms). Acquire spectra of the ssDNA components of the duplex as well. Compare the three spectra; signals from the ssDNA spectra appearing in the duplex spectrum indicate ssDNA excess. Typically, the H5-H6 cross peaks of the cytosine bases are used to discriminate between the single-stranded and double-stranded forms of DNA (see Note 9).
3.4. Preparation of the Protein–DNA Complex
Our group follows a conservative approach when forming protein–DNA complexes to minimize sample loss due to precipitation (17–22) (see Note 10). In this procedure, dilute concentrations of the components are mixed in the presence of high salt, followed by concentration and removal of the salt to form the final NMR sample. Typically, the starting concentration of the dsDNA is ~50–100 µM and it is dissolved in at least 150 mM NaCl, near physiological pH (see Note 11).
Pilot studies should be performed prior to embarking on a large-scale production of the complex as we have found that the order of addition of the protein and DNA components can have a dramatic impact on the results obtained. To test for precipitation of the sample upon component mixing, slowly titrate the protein into a ~20 µL sample of the DNA (the final volume of the fully formed complex should be ~40 µL). The concentration of the components is as described above and enough protein should be added to generate a 1:1 complex. The reverse titration in which dsDNA is titrated into a sample of the protein should also be performed. In both cases, carefully observe if any precipitation occurs during the mixing procedure and, more importantly, if any precipitation remains after the titration is complete. A light microscope can provide an easy way to assay for the presence of precipitation. As already mentioned, the order of addition can be critical. For example, in our studies of the Excisionase(Xis)-DNA complex, the addition of dsDNA to a solution of Xis resulted in irreversible precipitation, whereas the reverse titration, titrating Xis into a solution of dsDNA, yielded a soluble complex after mixing (20).
To obtain an NMR sample once the protocol for complex formation has been optimized, we typically mix the DNA and protein components such that the final volume of the complex is ~10 mL (~25 µM of the complex). The salt and unwanted buffer components are then removed by dialysis or using a centrifugal filter unit. This step is important for complex stability as the presence of salt tends to destabilize the protein–DNA complex by shielding electrostatic interactions. Generally, to obtain a sample with good NMR spectral properties, several variants of the complex are studied that differ in their pH, complex concentration, and ionic strength. Initial screening is typically performed using ~200–500 µM samples of the complex, which are then concentrated further to construct the final sample once the best conditions are discovered (see Note 12).
Prepare ~10 mL of 50 µM Int protein solution dissolved in high-salt protein buffer. Prepare ~10 mL of 50 µM dsDNA solution dissolved in high-salt DNA buffer (see Note 11).
Prepare the final NMR sample by slowly titrating 10 mL of 50 µM Int into 10 mL of 50 µM dsDNA. Add ~0.5 mL of protein, mix, and repeat until the titration is complete. The specific order of addition and the conditions used should have been optimized as described above.
Exchange the final sample into low-salt buffer using a protein concentrator (see Note 12). Concentrate the sample to a volume suitable for NMR experiments. Monitor the total amount of complex at each step of this process by measuring the A260 (see Note 13).
3.5. Assessing the Quality of the Protein–DNA Complex
To assess the quality of the DNA spectrum, record ID 1H spectra using a 1331 pulse adjusted to maximally excite the imino protons (25).
Compare the spectra of the complex and the isolated dsDNA molecule to assess protein binding.
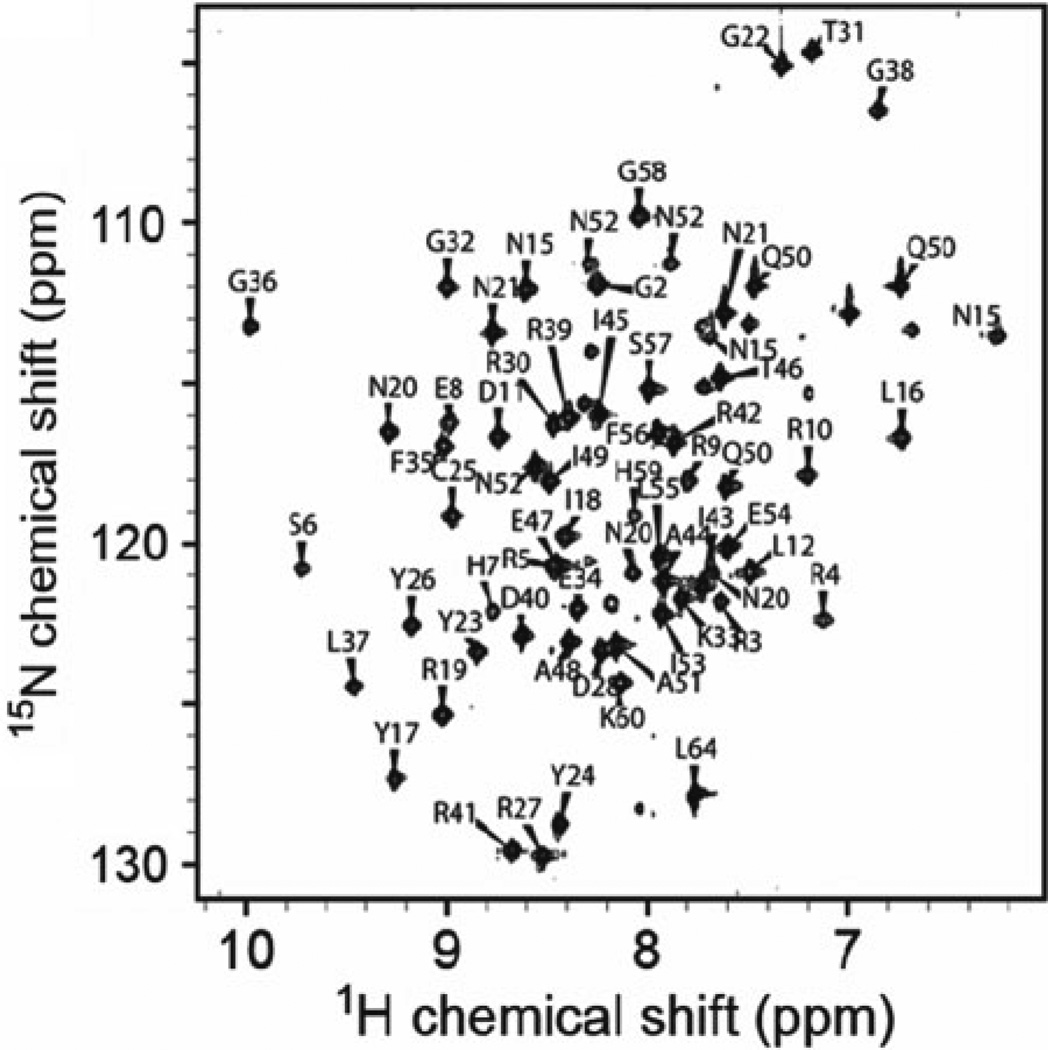
If sufficient material is present, use a similar excitation scheme to record a 2D 1H NOESY spectrum, which reveals whether the appropriate imino–imino cross peaks are present. Ascertain the quality of the protein spectrum by comparing the 1H–15N HSQC spectra of the free and DNA-bound forms of the protein. The spectrum of the complex should be well-resolved, exhibit uniform signal intensities, and differ substantially from the NMR spectrum of the DNA-free protein (Fig. 2).
Fig. 2.
The 1H–15N HSQC spectrum of the Int-DNA complex, which exhibits good dispersion and uniform line widths. Data for this complex were suitable to determine a high-resolution NMR structure ( 17–22).
In favorable cases, it may be possible to observe intermolecular NOEs using protein–DNA complexes containing only 15N-labeled protein. Depending on the structure of the complex, intermolecular NOE cross peaks are sometimes observed in the 2D 1H NOESY spectrum between the imino protons of the DNA and protein protons that resonate upfield of 1.2 ppm. This is because the most upfield resonances in the 1H spectrum of DNA are the thymine H5 methyl groups (1.2 and 1.6 ppm). Intermolecular NOEs between the protein amide and DNA imino protons can sometimes also be seen in the 3D 15N-edited NOESY spectrum of the 15N-labeled complex. However, a full assessment of whether a complex is suitable for structure determination by NMR requires the acquisition of the appropriate 2D and 3D edited and filtered NOESY experiments using samples in which the protein is labeled with 13C and 15N.
Acknowledgments
We thank Dr. Evgeny Fadeev for making Fig. 2. This work was supported by a grant from the National Institutes of Health to R.T.C. (R01 AI52217).
Footnotes
A variety of companies sell pure or partially purified single-stranded oligonucleotides that are synthesized using phosphoramidite chemistry (e.g., IDT, Invitrogen, Sigma, and Applied Biosystems). To save money, we typically obtain standard, desalted, unpurified oligonucleotides, which are then further purified in-house. Olignonucleotides greater than 60 nt in length can be produced via enzymatic reactions, which may also be applied to produce DNA molecules enriched with 13C and 15N (26, 27).
In this procedure, binding isotherms are generated by varying the protein concentration (0, 0.5, 1, 5, 25, 50, 100, 200, 1,000, 5,000 nM protein).
Alternatively, Tris–acetate–EDTA (TAE) may be used for the gel casting mixture and running buffer.
A 5–15% gel could be used depending on the respective sizes of the individual components and formed complex.
A classical and more straightforward method to purify ssDNA from a crude synthesis is to use an HPLC reverse-phase column (C4 to C18; porous hydrocarbon silica gel) with 0.1 M triethylammonium acetate (TEAA; mobile phase A) and ace-tonitrile (mobile phase B). A complete purification protocol and a column selection guide are described in Current Protocols Nucleic Acid Chemistry by Andrus et al. (28). A Mono Q column (GE, Mono Q HR5/5) on an FPLC can be used to purify ssDNA under denaturing conditions. Buffers for this purification scheme are as follows: (a) Buffer A: 50 mM Tris, pH 7.5, 6 M urea, and (b) Buffer B: 50 mM Tris, pH 7.5, 6 M urea, 1.5 M NaCl. No more than 1.5 µmol of ssDNA should be loaded onto this column for good separation. A 25 mL gradient (flow rate = 1 mL/min) from 10 to 30% buffer B should be sufficient to ensure good separation of ssDNA from impurities. It should be noted that all buffers and resuspended DNA should be filtered through a 0.2-µm filter before applying to the column.
If the DNA is longer than ~20 nucleotides, a 20% acrylamide gel should be used instead.
On a 10% gel, xylene cyanol migrates equivalent to a 55-nucleotide ssDNA molecule and bromophenol blue migrates equivalent to a 12-nucleotide ssDNA molecule. On a 20% gel, xylene cyanol and bromophenol blue migrate equivalent to 28- and 8-nucleotide ssDNA molecules, respectively.
Power settings for a single 20% acrylamide/7 M urea gel should be set to 30 W and maximum voltage and current. For a single gel running at a constant 30 W, the voltage is ~600–650 V and the current is 45–50 mA.
An alternative approach to ensure that solutions of dsDNA do not contain excess ssDNA is to provide a 10% excess of one of the DNA strands in the annealing reaction. After annealing, a Mono Q column is then used to separate the dsDNA molecule from excess ssDNA under native conditions. The following buffers can be used: Buffer A: 50 mM Tris, pH 7.5, and 1 mM EDTA; Buffer B: 50 mM Tris, pH 7.5, 1 mM EDTA, and 1.5 M NaCl. For good separation of the two DNA species, no more than 2 mg of DNA should be loaded onto a 1 mL Mono Q column. ssDNA typically elutes around 25% buffer B (0.375 M NaCl) and dsDNA typically elutes at ~35% buffer B (0.525 M NaCl). A 25 mL gradient from 10 to 45% NaCl running at 1 mL/min works well for oligos between 10 and 30 bp.
A variety of approaches can be used to successfully make protein–DNA complexes for NMR studies. A strategy commonly used in the literature is to titrate a ~1 mM 15N-labeled sample of the protein with a concentrated stock solution of the DNA molecule. 1H–15N HSQC spectra are recorded at various points during the titration until the desired stoichiometry is reached. Although the titration method is simple, mixing concentrated protein and DNA samples can cause the resulting complex to precipitate.
These conditions prevent a shift in the equilibrium from dsDNA to ssDNA that occurs at low-salt concentrations. A similar protein concentration is used when forming the complex; however, the salt and pH in the protein solution can vary and are chosen to maximize the stability and solubility of the free protein.
Frequently, the components of the complex are precious and therefore methods that minimize sample loss during concentration are desired. One trick to concentrate small-volume samples (< 2 mL) is to partially evaporate the complex. In this procedure, a weak flow of nitrogen gas is blown over the sample while it rests in the NMR tube. This can be accomplished by passing the gas through drawn-out pipette that is in turn inserted into the NMR tube. The buffer components are also concentrated during this process, and therefore the initial buffer conditions must be chosen accordingly.
A rough estimate of the concentration of the complex can be obtained by measuring its A260 immediately after mixing the components. The amount of the material present at this point in the preparation procedure is known. Therefore, it is possible to estimate the extinction coefficient of the complex by determining the optical absorbance of the complex at 260 and 280 nm. Once estimated, the extinction coefficient enables the concentration of the complex to be readily determined as it is concentrated to a volume suitable for NMR. It also enables the yield of the concentration procedure to be determined as the total amount of complex before and after concentrating can be determined.
References
- 1.Pabo CO, Sauer RT. Transcription Factors-Structural Families and Principles of DNA Recognition. Annu. Rev. Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 2.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol. Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 3.Nadassy K, Wodak SJ, Janin J. Structural features of protein-nucleic acid recognition sites. Biochemistry. 1999;38:1999–2017. doi: 10.1021/bi982362d. [DOI] [PubMed] [Google Scholar]
- 4.Jen-Jacobson L. Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers. 1997;44:153–180. doi: 10.1002/(SICI)1097-0282(1997)44:2<153::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 5.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billeter M, Qian YQ, Otting G, Muller M, Gehring W, Wuthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J. Mol. Biol. 1993;234:1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- 7.Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 8.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 9.Qian YQ, Otting G, Wuthrich K. NMR detection of hydration water in the intermolecular interface of a protein-DNA complex. J. Am. Chem. Soc. 1993;115:1189–1190. [Google Scholar]
- 10.Iwahara J, Clore GM. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 11.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 12.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavanagh J, Fairbrother WJ, Palmer AGI, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles & Practice. 2nd ed. San Diego: Academic Press; 2006. [Google Scholar]
- 15.Iwahara J, Wojciak JM, Clubb RT. Improved NMR spectra of a protein-DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered NOESY experiment. J. Biomol. NMR. 2001;19:231–241. doi: 10.1023/a:1011296112710. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Abbani MA, Papagiannis CV, Sam MD, Cascio D, Johnson RC, Clubb RT. Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly. Proc. Natl. Acad. Sci. USA. 2007;104:2109–2114. doi: 10.1073/pnas.0607820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwahara J, Iwahara M, Daughdrill GW, Ford J, Clubb RT. The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA. EMBO J. 2002;21:1197–1209. doi: 10.1093/emboj/21.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fadeev EA, Sam MD, Clubb RT. NMR structure of the amino-terminal domain of the lambda integrase protein in complex with DNA: immobilization of a flexible tail facilitates beta-sheet recognition of the major groove. J. Mol. Biol. 2009;388:682–690. doi: 10.1016/j.jmb.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sam MD, Cascio D, Johnson RC, Clubb RT. Crystal structure of the excisionase-DNA complex from bacteriophage lambda. J. Mol. Biol. 2004;338:229–240. doi: 10.1016/j.jmb.2004.02.053. [DOI] [PubMed] [Google Scholar]
- 21.Wojciak JM, Connolly KM, Clubb RT. NMR structure of the Tn916 integrase-DNA complex. Nature Struct. Biol. 1999;6:366–373. doi: 10.1038/7603. [DOI] [PubMed] [Google Scholar]
- 22.Wojciak JM, Iwahara J, Clubb RT. The Mu repressor-DNA complex contains an immobilized “wing” within the minor groove. Nature Struct. Biol. 2001;8:84–90. doi: 10.1038/83103. [DOI] [PubMed] [Google Scholar]
- 23.Buratowski S, Chodosh LA. Mobility shift DNA-binding assay using gel electrophoresis. Curr. Protoc. Mol. Biol., Chapter 12. 2001 doi: 10.1002/0471142727.mb1202s36. Unit 12 2. [DOI] [PubMed] [Google Scholar]
- 24.Taylor JD, Ackroyd AJ, Halford SE. The gel shift assay for the analysis of DNA-protein interactions. In: Kneale GG, editor. DNA-protein interactions, principles and protocols. Totowa, NJ: Humana Press; 1994. [DOI] [PubMed] [Google Scholar]
- 25.Hore PJ. A new method for water suppression in the proton NMR spectra of aqueous solutions. J. Magn. Reson. 1983;54:539–542. [Google Scholar]
- 26.Louis JM, Martin RG, Clore GM, Gronenborn AM. Preparation of uniformly isotope-labeled DNA oligonucleotides for NMR spectroscopy. J. Biol. Chem. 1998;273:2374–2378. doi: 10.1074/jbc.273.4.2374. [DOI] [PubMed] [Google Scholar]
- 27.Xiong AS, Yao QH, Peng RH, Duan H, Li X, Fan HQ, Cheng ZM, Li Y. PCR-based accurate synthesis of long DNA sequences. Nat Protoc. 2006;1:791–797. doi: 10.1038/nprot.2006.103. [DOI] [PubMed] [Google Scholar]
- 28.Andrus A, Kuimelis RG. Analysis and purification of synthetic nucleic acids using HPLC. Curr. Protoc. Nucleic Acid Chem., Chapter 10. 2001 doi: 10.1002/0471142700.nc1005s01. Unit 10 5. [DOI] [PubMed] [Google Scholar]