Abstract
Background
Recently, several important polymorphisms have been identified in T-cell activation and effector pathway genes and have been reported to be associated with inter-patient variability in alloimmune responses. The present study was designed to assess the impact of these genetic variations on the outcomes of allogeneic hematopoietic stem cell transplantation.
Design and Methods
We first investigated ten single nucleotide polymorphisms in six genes, CD28, inducible co-stimulator, cytotoxic T-lymphocyte antigen 4, granzyme B, Fas and Fas ligand, in 138 pairs of patients and their unrelated donors and a second cohort of 102 pairs of patients and their HLA-identical sibling donors.
Results
We observed that patients receiving stem cells from a donor with the cytotoxic T-lymphocyte antigen 4 gene CT60 variant allele (AA genotype) had a reduced incidence of grades II–IV acute graft-versus-host disease; however, they experienced early cytomegalovirus infection and relapsed more frequently, which suggested an interaction between the donor cytotoxic T-lymphocyte antigen 4 gene CT60 AA genotype and reduced T-cell alloreactivity. Furthermore, an unrelated donor with the granzyme B +55 variant genotype (AA) was an independent risk factor for development of grades II–IV acute graft-versus-host disease (P=0.024, RR=1.811). Among patients with acute myelogenous leukemia, those with the Fas -670 TT genotype were at higher risk of relapse (P=0.003, RR=3.823). The presence of these susceptible alleles in the donor and/or patient resulted in worse overall survival (54.9% versus 69.5%, P=0.029).
Conclusions
Our data suggest that genotype analysis of T-cell activation and effector pathway genes can be used for risk assessment for patients with hematologic malignancies before hematopoietic stem cell transplantation.
Key words: CD28, inducible co-stimulator, cytotoxic T-lymphocyte antigen 4, granzyme B, Fas, Fas ligand, polymorphism, hematopoietic stem cell transplantation
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is widely performed as the only curative therapy for a variety of malignant hematologic diseases. The growing indications and improvements in transplantation technology and supportive care have led to an increased number of allo-HSCT. However, disease relapse, major infection and graft-versus-host disease (GVHD) are leading causes of death following allo-HSCT, and limit the use of this important procedure.1 Defining variables that predispose to these events is, therefore, of critical importance. Current evidence suggests that in addition to HLA matching, genetic diversity among donors and recipients, including cytokine genes, innate immunity genes and pharmacogenetic polymorphisms, affect the success of transplantation by regulating alloimmune responses.2-5
Optimal donor T lymphocyte proliferation, differentiation and acquisition of effector functions play a critical role in alloimmune reactivity, and their ability to detect non-self-antigens can lead to GVHD or contribute to the prevention of relapse and infections after transplantation. CD28, inducible co-stimulator (ICOS) and cytotoxic T-lymphocyte antigen 4 (CTLA-4) belong to the same family of T-cell co-stimulatory molecules. CD28 is the primary co-stimulatory molecule and is constitutively expressed on the majority of T cells. CD28 transduces a signal that enhances the activation and proliferation of T cells.6 Another member of the CD28 family is ICOS. Although not constitutively expressed, ICOS is rapidly up-regulated on T lymphocytes upon activation.7 CTLA-4, a structural homolog of CD28, is a key factor in regulating and maintaining self-tolerance, providing a negative signal to T-cell responses.8 After activation, three effector pathways have been described for T-cell cytotoxicity: granzyme B/perforin, Fas/Fas ligand (FasL) and secreted molecules such as tumor necrosis factor-alpha (TNF-α).9,10
Recently, several important polymorphisms have been identified in these activation and effector pathway genes and have been reported to be associated with the risk of different autoimmune diseases, some malignancies and allograft rejection in solid organ transplantation patients.11-15 These data suggest that these polymorphisms correlate with gene function and contribute to inter-patient variability in alloimmune responses. However, such information is less available in allo-HSCT. We hypothesized that gene polymorphisms in T-cell activation and effector pathways from donors or patients may affect the outcomes of allo-HSCT. We have previously reported the relationship between polymorphisms in donor and/or patient cytokine genes, such as TNF-α, TNF-β, transforming growth factor-b and interleukin-10, and the risk of acute GVHD (aGVHD).16,17 In the present study, we further investigated the influence of CD28, CTLA-4, ICOS, granzyme B, Fas and FasL gene polymorphisms on T-cell alloimmune responses after HSCT. The effect of non-HLA immunogenetics may not be comparable between the unrelated donor (URD) transplantation cohort and the HLA-identical sibling transplantation cohort, with regard to the degree of the discrepancy in genetic variations between donor and patient, significant differences in the incidence of transplantation-related complications and outcomes. However, very few studies on the effect of non-HLA immunogenetics have simultaneously been evaluated in sibling and URD transplantation cohorts. We have now confirmed the analysis in two independent cohorts. The initial cohort consisted of 138 pairs of patients and their URD. The second cohort consisted of 102 pairs of patients and their HLA-identical sibling donors. Since it is known that CD28, ICOS and CTLA-4 are specifically expressed on T cells and that alloimmune responses are mediated mainly by donor T cells after allo-HSCT, only the effect of donor CD28, ICOS and CTLA-4 gene polymorphisms were investigated.
Design and Methods
Characteristics of the patients, donors and transplants
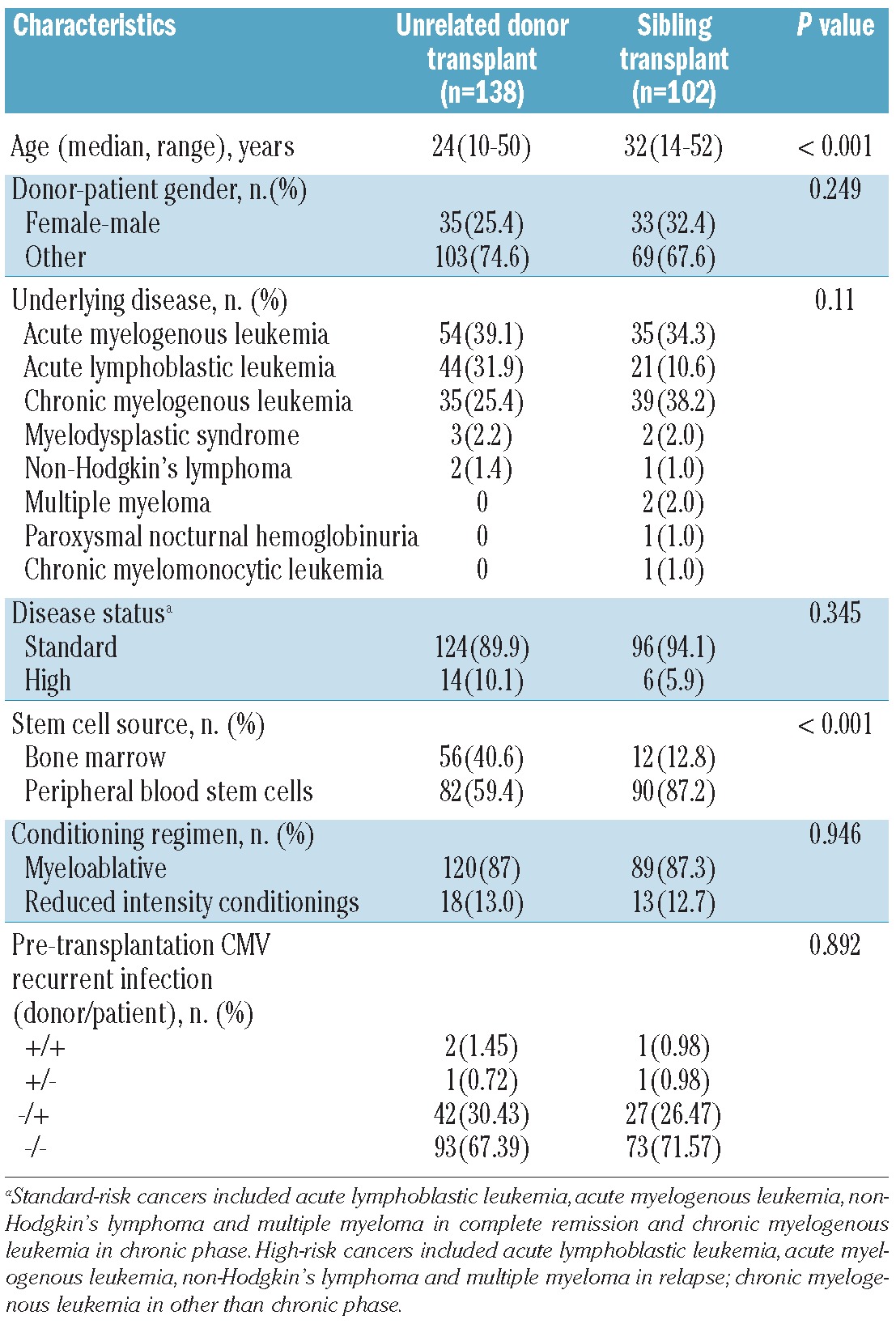
We unselectively included all patients undergoing allo-HSCT in our Bone Marrow Transplantation Center between January 2001 and March 2009 into the present study if the donors' and patients' DNA were stored in our specimen pool and clinical data were available. Divided according to donor type (URD versus sibling), there were no significant differences in terms of donor-patient gender, underlying disease, disease status before transplantation, conditioning regimen or the status of cytomegalovirus (CMV) recurrent infection in donors and patients pre-transplantation (Table 1). The only significant differences between the two cohorts were in patients' age and the source of stem cells: these variables were not identified as significant in univariate or multivariate outcome analysis. Thus the transplantation characteristics of the two cohorts were very similar and the final analysis of the clinical end-points included both cohorts.
Table 1.
Characteristics of the patients, donors and transplants in the two cohorts.
All the patients and their donors were of Chinese origin. The study was approved by the local ethics committee. All the patients and donors gave their written informed consent.
Graft-versus-host disease prophylaxis and conditioning regimen
Low-resolution HLA typing had been performed for HLA-A, -B, and -DRB1 in the sibling transplantation cohort, and high-resolution DNA typing for HLA-A, -B, -C, -DRB1, and -DQB1 in the URD transplantation cohort. The main myeloablative conditioning regimen used involved busulfan/cyclophosphamide without total body irradiation; reduced-intensity conditioning regimens were predominantly fludarabine-based combinations without total body irradiation. On the basis of clinical and animal studies, mycophenolate mofetil as an adjunct to the combination of cyclosporine A and methotrexate appeared to augment the actions of standard immunosuppressant agents without adding overlapping toxicities.18-21 Furthermore, our previous clinical data and animal studies have suggested that low-dose mycophenolate mofetil provides similar immunosuppression without adversely affecting engraftment.22,23 Following this rationale, in our center, all patients received the same GVHD prophylaxis, consisting of cyclosporine A, methotrexate and low-dose mycophenolate mofetil. Cyclosporine A was scheduled to be given intravenously at a dose of 2.5 mg/kg/day from day -1, with a target blood level of 200–300 ng/mL. The dosage was tapered during the second month after transplantation, according to chimeric status and evidence of GVHD. Mycophenolate mofetil was initiated orally at a dose of 500 mg/day on day +1, and withdrawn on day +100. Methotrexate was given at a dose of 15 mg/day on day +1 and 10 mg/day on days +3 and +6.
Genotyping single nucleotide polymorphisms in T-cell activation and effector pathway genes
Genomic DNA was extracted from peripheral blood samples obtained from recipients and donors before transplantation, using a salting-out method with a commercial DNA extraction kit (Dynal Biotech, Brown Deer, WI, USA), following the manufacturer's recommendations. DNA was quantified by spectrophotometry.
Single nucleotide polymorphism nomenclature uses a number that refers to its position in the nucleotide sequence upstream (indicated by a minus sign) or downstream (indicated by a plus sign) of the start of the transcription site, followed by two letters indicating the polymorphism, adenine (A), cytosine (C), guanine (G) or thymine (T). Ten single nucleotide polymorphsims recently described to interfere with T-cell immune responses, including -594 A/G (rs35593994) in the CD28 gene promoter region; -693 A/G (rs11883722) in the ICOS gene promoter region; -1722 A/G (rs733618) in the promoter, +49 A/G in exon 1 (rs231775), and CT60 A/G (rs3087243) in the 3-terminal untranslated region of the CTLA-4 gene; +55 A/G (rs8192917) in exon 2 and -295 C/T (rs7144366) in the promoter of the granzyme B gene; -1377 A/G (rs2234767) and -670 C/T (rs1800682) in the promoter of the Fas gene, and -844 C/T (rs763110) in the promoter of the FasL gene, were determined by multiplex SNaPshot technology, according to previously described methods,16,17 using an ABI fluorescence-based allelic discrimination method assay (Applied Biosystems, Foster City, CA, USA). Detailed information on the primers used is available in the Online Supplementary Methods (Online Supplementary Table S1).
Evaluation of clinical outcome
The end-points of T-cell alloimmune responses post-HSCT which were studied were induction of GVHD, prevention of major infection and recurrence of malignancy. Only patients with successful engraftment were included in the analysis of aGVHD. Acute GVHD was diagnosed and graded using established criteria.24 Given that grade I typically refers to patients with clinically uncertain aGVHD and most clinicians do not treat these patients systemically, only aGVHD of grades II and higher were considered for analysis. Patients who survived ≥100 days were analyzed for chronic GVHD (cGVHD).
CMV reactivation and disease continue to be important complications post-HSCT25,26 and CMV infection is of special concern in the Chinese population. The incidence of asymptomatic infection is high. The episodes of major infection were focused on early CMV infection post-HSCT. Surveillance of CMV (by pp65 antigenemia) was performed using peripheral blood samples on a weekly basis until day +100 and monthly thereafter. According to published standard criteria,27 recurrent CMV infection is defined as new detection of CMV antigenemia or CMV seropositivity [CMV-specific immune globulin M positive, CMV IgM(+)] in a patient or donor who had a previously documented infection and who had not had virus detected for an interval of at least 4 weeks during active surveillance. CMV antigenemia was defined as more than two pp65 antigen-positive cells per 50,000 leukocytes. Early CMV infection refers to antigenemia or disease occurring within day 100 after transplantation. Ganciclovir or foscarnet sodium was given for CMV prophylaxis or pre-emptive pre-transplantation treatment. Pre-emptive therapy was started whenever CMV antigenemia was detected.
Statistical analysis
Linkage disequilibrium analysis among pairs of alleles was performed using Fisher's exact test. Cumulative incidences using the competing risk method,28 as described by Gray,29 were studied to determine the differences in probabilities of aGVHD, cGVHD, early CMV infection and relapse in the presence of polymorphisms. Death without signs of GVHD was considered a competing risk in the analysis of GVHD incidence. The competing risks for early CMV infection and relapse were death without CMV infection and death in complete remission, respectively. Overall survival was calculated from transplantation to death from any cause and estimated using the Kaplan-Meier method. Multivariate Cox regression models using a forward stepwise procedure with the likelihood ratio criterion (inclusion/exclusion criteria: P<0.05/P>0.1 respectively) were applied to analyze the effects of these characteristics and other known clinical and biological factors on allo-HSCT outcomes. All variables in the univariate analysis with a P-value at or below 0.2 were included in the multivariate analysis.
Statistical analysis was performed using SPSS software version 16.0 and the cmprsk and CumIncidence28 packages in R (The R foundation, http://www.r-project.org). All probability values were two-sided and P values less than 0.05 were considered statistically significant. P values between 0.05 and 0.1 were considered to be indicative of a trend.
Results
Frequency of polymorphisms in donors' and patients' genes and corresponding phenotypes
The frequencies of CD28, ICOS, CTLA-4, granzyme B, Fas and FasL genotypes in 240 allo-HSCT donors and patients were in good agreement with those referenced in the GenBank NCBI SNP databases (Online Supplementary Table S2). This is the first report of the allele frequencies at CD28 -594 and Fas -1377 loci in a Chinese population. There were no significant differences in the distribution of these genotypes between the URD transplantation cohort and the HLA-identical sibling donor transplantation cohort. No significant deviations from the expected Hardy-Weinberg genotype frequencies were observed (data not shown).
Effect of genetic variations in T-cell activation and effector pathways on graft-versus-host disease: univariate and multivariate analyses in both cohorts
Analysis of the outcomes of the two separate cohorts in the present study demonstrated higher cumulative incidences of grades II–IV aGVHD (43.5% versus 12.7%, Gray's test P<0.0001) and severe aGVHD (13.2% versus 7.8%, Gray's test P=0.0022) in the URD transplantation cohort compared with the sibling donor transplantation cohort. However the incidences of limited and extensive cGVHD were comparable in the two cohorts (16.7% versus 22.5%, P=0.31; 15.9% versus 10.8%, P=0.343, respectively).
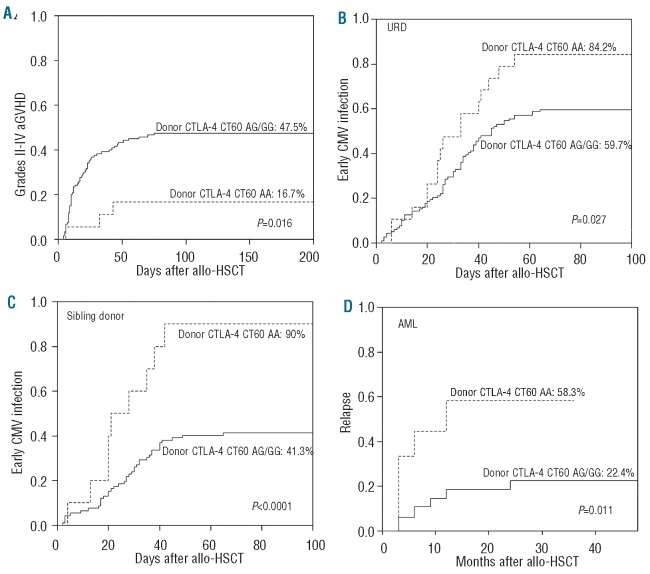
When assessing the risk of aGVHD according to genetic variations in T-cell activation and effector pathways in recipients and donors, we found that two single nucleotide polymorphisms in donors, CTLA-4 CT60 and granzyme B +55, influenced the risk of development of aGVHD. The association was particularly strong in the URD transplantation cohort. Patients receiving stem cells from an URD with the CTLA-4 CT60 variant allele (A allele) had a lower incidence of grades II–IV aGVHD than those who received stem cells from a URD with the wild-type allele (G allele) (AA: 16.7% versus AG/GG: 47.5%, Gray's test P=0.016; Figure 1A). However, URD with the granzyme B +55 variant allele (A allele) were associated with a higher incidence of grades II–IV aGVHD (AA: 54.8% versus AG/GG: 34.2%, Gray's test P=0.038; Figure 2). The same effect was observed in the cohort of sibling donors although the incidence of clinically significant aGVHD in this cohort was low overall and the association was not statistically significant (for CTLA-4 CT60: Gray's test P=0.2; for granzyme B +55: Gray's test P=0.17). The small number of patients with severe aGVHD in the two separate cohorts precluded statistical analysis of the relationship between these gene polymorphisms and severe aGVHD.
Figure 1.
The capacity for T-cell alloimmune reactivity stratified according to the donors' CTLA-4 CT60 genotype. Cumulative incidence of grades II to IV aGVHD (A), early CMV infection in the URD transplantation cohort (B), early CMV infection in the sibling transplantation cohort (C) and relapse in AML patients (D).
Figure 2.
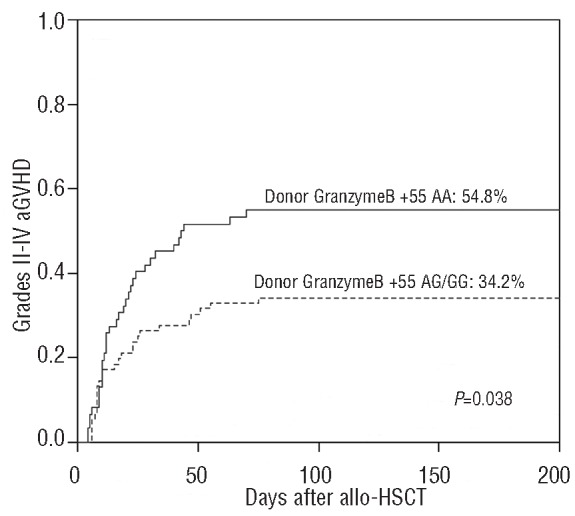
Cumulative incidence of grades II to IV aGVHD stratified according to the donors' granzyme B +55 genotypes
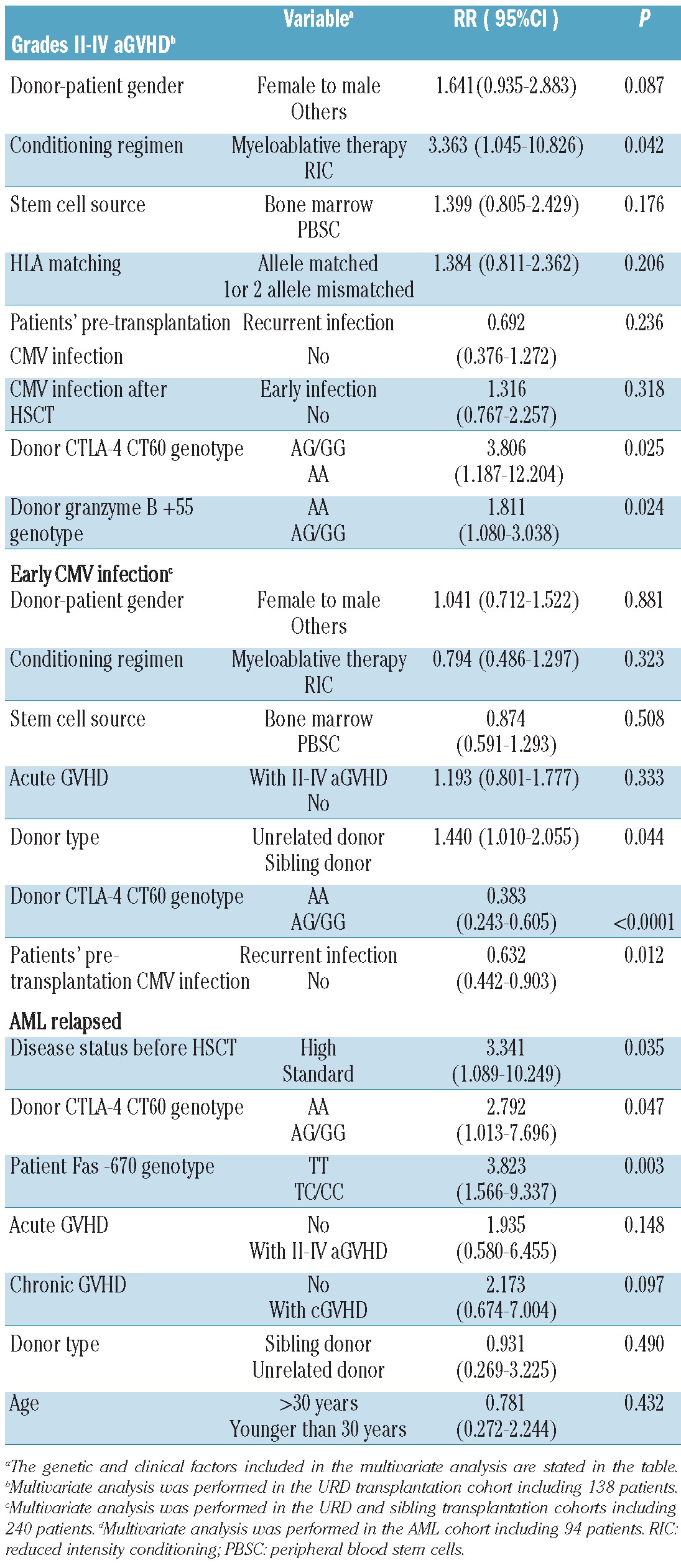
Multivariate Cox regression analysis for aGVHD in the URD transplantation cohort including donor CTLA-4 CT60 genotype, donor granzyme B +55 genotype and major known risk factors (donor-patient gender relation, conditioning regimen, stem cell source, HLA matching, and patients' pre-transplantation and post-transplantation CMV infection status) (Table 2) confirmed that an URD with the granzyme B +55 variant genotype (AA) was an independent risk factor for development of grades II–IV aGVHD (P=0.024, RR = 1.811, 95% CI, 1.080–3.038). In contrast, an URD with the CTLA-4 CT60 variant genotype (AA) was protective (P=0.025, RR=3.806, 95% CI, 1.187–12.204). Myeloablative conditioning also contributed significantly to the development of grades II–IV aGVHD (P=0.042, RR=3.363, 95% CI, 1.045–10.826). A female donor and male recipient pairing was found to be a less significant factor (P=0.087). Severe grades of aGVHD were not influenced by the CTLA-4 CT60 or granzyme B +55 genotype. The incidence of aGVHD was not affected by polymorphisms of the CD28, ICOS, Fas or FasL genes. No polymorphism was significantly associated with the risk of cGVHD.
Table 2.
Multivariate analysis for the capacity of T-cell alloimmune reactivity.
Effect of genetic variations in T-cell activation and effector pathways on early cytomegalovirus infection: univariate and multivariate analyses in both cohorts
In the present study cohorts, all patients and almost all donors were CMV seropositive [CMV-specific immune globulin G positive, CMV-IgG(+)] before HSCT and only one donor from the Taiwan Tzu Chi Stem Cells Center was CMV seronegative. All patients received CMV prophylaxis and those experiencing pre-transplantation recurrent CMV infection received pre-emptive treatment. In the URD transplantation cohort, 44 patients (31.88%) experienced recurrent CMV infection pre-transplantation, as did 28 patients (27.45%) in the sibling donor cohort (Table 1). Of the total of 240 patients, 134 (55.8%) experienced early CMV infection with a median onset at 27 days (range, 2–64) post-HSCT. Of these 134 patients, 28 (20.9%) developed CMV infection during granulocytopenia and 106 (79.1%) developed the infection after neutrophil recovery. However the different CMV serostatus in the Chinese population did not result in a significantly higher incidence of CMV disease in an allo-HSCT setting. In the present study, the majority of patients developed CMV-positive antigenemia without disease; only 9 (6.7%) patients developed CMV disease (7 patients with CMV pneumonia and 2 patients with CMV enteritis), compared with the 8%–10% described in the literature.25
Patients who received stem cells from donors with the CTLA-4 CT60 variant genotype (AA) had a higher incidence of early CMV infection than those with the wild-type genotype (AG or GG) in both the URD transplantation cohort (AA: 84.2% versus AG or GG: 59.7%, Gray's test P=0.027, Figure 1B) and the sibling transplantation cohort (AA: 90% versus AG or GG: 41.3%, Gray's test P<0.0001, Figure 1C), although a higher incidence of early CMV infection was observed in the URD transplantation cohort (63% versus 46%, Gray's test P=0.025). Donor CTLA-4 CT60 genotype, together with other clinical and biological factors (donor-patient gender relation, conditioning regimen, stem cell source, aGVHD, donor type and patients' pre-transplantation CMV infection status) known to contribute to the development of early CMV infection were subjected to multivariate analysis including the two cohorts. The results are given in Table 2. Three independent risk factors were significantly associated with early CMV infection: URD (P=0.044, RR=1.440, 95% CI, 1.010–2.055), patients experiencing recurrent CMV infection pre-transplantation (P=0.012, RR=0.632, 95% CI, 0.442–0.903) and donors with the CTLA-4 CT60 AA genotype (P<0.0001, RR=0.383, 95% CI, 0.243–0.605) all contributed to the development of early CMV infection. There was no significant difference in the impact of CD28, ICOS, granzyme B, Fas and FasL single nucleotide polymorphisms on early CMV infection.
Effect of genetic variations in T-cell activation and effector pathways on disease relapse
There was no significant difference in the cumulative incidence of relapse between the two cohorts. Given that allo-HSCT ultimately fails in a significant fraction of patients with acute leukemia due to disease relapse, and that the relapse risk differs based on the diagnosis, further analysis of the effect of genetic variations in the activation and effector pathways on disease relapse was performed separately in 94 patients with acute myelogenous leukemia (AML) including five patients in the blast phase of chronic myelogenous leukemia, and 65 patients with acute lymphoblastic leukemia. In the AML cohort, more than half (58.3%) of patients receiving grafts from donors with the CTLA-4 CT60 variant genotype (AA) relapsed, but the incidence of relapse decreased to 22.4% in those receiving grafts from donors with the CTLA-4 CT60 wild-type allele (Gray's test P=0.011, Figure 1D). AML patients with the Fas -670 homogeneous variant allele (TT genotype) had a higher incidence of relapse compared to those with the wild-type allele (CC or CT genotype) (TT: 43.9% versus CC/CT: 16.7%, Gray's test P=0.02, Figure 3). Disease relapse was not affected by polymorphisms of the CD28, ICOS, granzyme B or FasL genes. Multivariate analysis in 94 AML patients including donor CTLA-4 CT60 genotype, patients' Fas -670 genotype and major known risk factors such as disease status before HSCT, aGVHD, cGVHD, donor type and patients' age, confirmed that donors with the CTLA-4 CT60 variant genotype (P=0.047, RR=2.792, 95% CI, 1.013–7.696), patients with the Fas -670 variant genotype (P=0.003, RR=3.823 95% CI, 1.566–9.337) and disease status before transplantation (P=0.035, RR=3.341 95% CI, 1.089–10.249) contributed to the risk of AML relapse after allo-HSCT. However, patients experiencing cGVHD were less prone to relapse (P=0.097) (Table 2). Interestingly, the effects of CTLA-4 CT60 and Fas -670 polymorphisms futures were highly significant in AML but did not show any trend in patients with acute lymphocytic leukemia (data not shown). No significant impact of CD28, ICOS, Granzyme B and FasL single nucleotide polymorphisms was observed on the incidence of relapse of either AML or acute lymphocytic leukemia.
Figure 3.
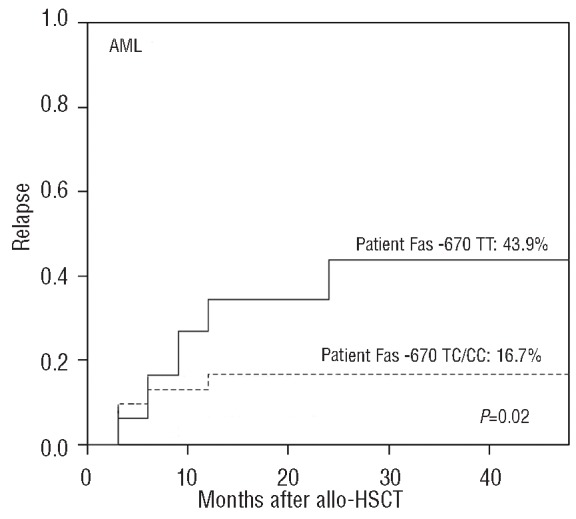
Cumulative incidence of relapse in AML patients stratified according to the patients' Fas -670 genotypes.
Risk assessment before allogeneic hematopoietic stem cell transplantation according to genetic variations and outcomes
Since donor CTLA-4 CT60 genotype, donor granzyme B +55 genotype and recipient Fas -670 genotype were significantly associated with transplant-related life-threatening complications, including aGVHD, early CMV infection and disease relapse, analysis of these genotypes can be used pre-HSCT to assess risk. When overall survival was analyzed in relation to the occurrence of isolated patient or donor variants, a significant worse overall survival was observed in AML patients with the Fas -670 homogeneous variant allele (TT genotype) than in those with the wild-type allele (CC or CT genotype) (TT: 40.8% versus CC/CT: 74.7%, generalized Wilcoxon's test P=0.004). Although a trend toward worse overall survival was observed both in patients receiving grafts from CTLA-4 CT60 AA donors and in patients receiving grafts from granzyme B +55 AA donors, the analysis did not reach true significance (data not shown). We next analyzed overall survival in relation to the occurrence of combined variants: patients were stratified into two groups with respect to the presence or absence of the susceptible alleles in the donor and/or patient. One hundred and thirty-one patient-donor pairs included at least one susceptible allele (patients with a CTLA-4 CT60 AA donor, patients with a granzyme B +55 AA donor, AML patients with a Fas -670 TT genotype or a combination of these), and 109 patient-donor pairs involved wild-type genotypes. With a median follow up of 34 months (range, 8-113), the presence of the susceptible alleles in the donors and/or patients resulted in a worse overall survival at 5 years than did the presence of the wild-type genotypes (54.9% versus 69.5%, log-rank, P=0.029, Figure 4).
Figure 4.
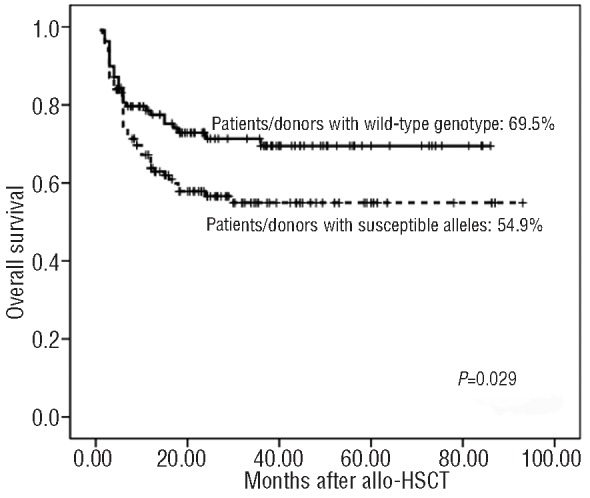
Overall survival of patients stratified according to the donors' CTLA-4 CT60 genotype, donors' granzyme B +55 genotype and patients' Fas -670 genotype.
Discussion
Transplantation outcome may be influenced not only by the ability of T cells to recognize antigens but also by their capacity to become activated or de-activated. We first examined the roles of a wide range of genetic variations in T-cell activation and effector pathways in alloimmune responses following allo-HSCT. The primary and novel findings of this study highlight the previously unrecognized effect of variations in the CTLA-4, granzyme B and Fas genes in both the donors and patients on the outcomes of allo-HSCT.
In the past few years, with the discovery of a large array of co-stimulatory molecules and the realization of these molecules' potential as targets to modulate clinical immune responses, interest in co-stimulatory molecules has increased remarkably. CTLA-4 is the most extensively studied inhibitory T-cell molecule, essential for T-cell homeostasis and tolerance induction. CTLA-4, a competitive antagonist for B7, has an approximately 100-fold higher affinity and avidity for B7 than has CD28. In conventional T cells, CTLA-4 efficiently competes with the positive co-stimulatory molecule CD28 and transduces negative signals. Furthermore, Foxp3+CD25+CD4+ regulatory T cells critically require CTLA-4 to suppress immune responses by down-regulation of B7 on dendritic cells and by affecting the potency of dendritic cells to activate other T cells.30 CT60 is the most widely studied disease-associated polymorphism of the CTLA-4 gene. Previous studies have indicated that the CTLA-4 CT60 AA genotype is a tolerogenic genotype associated with autoimmune diseases including systemic lupus erythematosus and Graves' disease; however, it is also a strong candidate susceptibility gene in several malignancies, which suggests an interaction between CTLA-4 CT60 AA genotype and lower T-cell immunological responses.31,32 However, the results of recent studies on the impact of the CTLA-4 CT60 genotype on the outcomes of allo-HSCT have been inconsistent. Our data suggest that the A-to-G transition at CT60 of the CTLA-4 gene has a profound effect on the capacity of T cells to mount alloimmune responses after allo-HSCT. Patients receiving stem cells from a donor with the CTLA-4 CT60 variant allele (AA genotype) had a lower incidence of grades II–IV aGVHD; however, they experienced early CMV infection and relapsed more frequently than those who received grafts from donors with wild-type alleles (AG/GG genotypes). Bosch-Vizcaya A et al. suggested that the donor CTLA-4 CT60 AA genotype was also associated with lower overall survival and disease-free survival, this being related to an increased risk of relapse.33 These observations and our data all indicate that the polymorphic feature of CTLA-4 CT60 influences T-cell activation and that the AA genotype is associated with a reduced capacity for T-cell activation. In contrast, Orru et al. reported that thalassemia patients with the CT60 AA genotype experienced grades II–IV aGVHD more frequently than patients with other genotypes after unrelated donor allo-HSCT.34 Perez-Garcia et al.35 reported that the donor CT60 AA genotype was associated with an increased risk of grades II–IV aGVHD and a lower risk of relapse in a HLA-identical sibling transplantation cohort. Murase et al. found that the donor CT60 A allele did not affect the incidence of GVHD, but was associated with a significantly lower risk of relapse and a trend toward higher overall survival after HLA-identical sibling transplantation in a Japanese population.36 Metaxas et al. further evaluated the CTLA-4 CT60 genotype with the outcomes of donor lymphocyte infusion in 79 patients after allo-HSCT and observed that patients receiving lymphocytes from a donor with the CTLA-4 CT60 A-allele had a better chance of achieving sustained complete donor chimerism and had better overall survival.37 Taken together, these observations indicate that the CTLA-4 CT60 AA genotype is associated with a higher capacity for T-cell activation. Intriguingly, Perez-Garcia's group investigated the association between CTLA4 CT60 genotype and the incidence of leukemic relapse in adult AML patients after chemotherapy and found that the CT60 AA genotype was associated with a higher rate of leukemic relapse,38 which seemed to contradict their own previous results. The rational explanation for those discrepancies resides in the full understanding of the exact influence of this polymorphism on CTLA-4 function and expression.
The CTLA-4 gene consists of four exons and is translated into two protein isoforms: a full-length isoform (flCTLA-4) and a soluble form (sCTLA-4) that lacks exon 3, which encodes the transmembrane domain. CT60 maps to the 3' untranslated region of CTLA-4 and plays a role in the efficiency of the splicing and production of sCTLA-4. The A allele at CT60 produced more sCTLA-4 than did the G allele both in patients with autoimmune diseases and in healthy controls.11,35 Regarding the function of sCTLA-4, primary investigations indicated that this molecule, lacking a transmembrane domain, is able to bind B7 with a similar affinity to flCTLA-4 and is functionally active with an inhibitory effect on the mixed leukocyte response in vitro.39 Thus our results may be explained by greater production of sCTLA-4 in patients receiving stem cells from a donor with the CTLA4 CT60 AA genotype, which contributes to suppression of T-cell alloimmune responses post-HSCT.
These data also indicated a previously unrecognized role for variations of the T-cell effector pathway genes in the outcomes of allo-HSCT. Granzyme B, a unique effector protease for immune defense, is used by cytotoxic T lymphocytes and natural killer cells as a molecular weapon in their defense against alloantigens, virus-infected and malignantly transformed host cells.40 The importance of the granzyme B/perforin pathway in the pathogenesis of aGVHD is well established.41,42 Increased gene expression of granzyme B was detected in patients with aGVHD43 and an elevated serum level of granzyme B post-HSCT was highly predictive of the risk and severity of aGVHD.44 However, inter-individual variability in the baseline expression and activity of granzyme B may result from variations in the granzyme B gene. Espinoza et al. reported that donors and/or patients with the granzyme B +55 G- allele positive genotype were associated with improved overall survival after unrelated HLA-fully matched T-cell-replete bone marrow transplantation, but failed to find any significant association with aGVHD.45 We identified for the first time that polymorphism of granzyme B +55 influenced the risk of aGVHD and that the AA genotype was a strong candidate for the susceptibility genotype. The non-synonymous single nucleotide polymorphism +55 A/G, located in exon 2, is responsible for the amino acid substitution of a glutamine (CAA) to arginine (CGA) (numbering with reference to the human chymotrypsinogen A sequence). Previous studies showed that the granzyme B +55 AA genotype is associated with increased expression of granzyme B, and that the G- allele positive genotype was incapable of inducing apoptosis,46,47 a finding which is in good agreement with our results.
Fas is another effector molecule and Fas-mediated apoptotic death is an important pathway in alloimmune responses. The Fas-mediated apoptotic pathway has been found to be dysregulated in several immune system malignancies, resulting in down-regulation of apoptosis and subsequent persistence of the malignant clone.48 Fas -670 C/T, one of the most important promoter polymorphisms, which occurs within the signal transducers and activators of transcription 1 (STAT1) transcription factor binding site, may dysregulate promoter activity and affect Fas gene expression.49 The Fas -670 C-allele positive, wild-type allele with higher Fas expression,49 reduced the risk of AML sixfold compared to the risk in subjects with the -670 variant allele (T-allele)50 and the same effects were observed in adult T cell leukemia51 and invasive squamous cell carcinomas.52 Furthermore, our study proves that AML patients with the Fas -670 C-allele had a significantly lower risk of relapse following allo-HSCT. This suggests that polymorphism of Fas -670 may contribute to apoptosis-resistant clones in AML. However, this strong association was only observed in AML patients but not in patients with other myelogenous or lymphoid malignancies. A possible explanation for this discrepancy may be that the mechanisms and/or threshold for the graft-versus-leukemia effect differ between acute myelogenous malignancies, chronic myelogenous malignancies and lymphoid malignancies.
In addition to HLA matching, huge advances in human genomics have identified many non-HLA polymorphisms that have been found to influence the outcomes of allo-HSCT. Our present study suggests that a donor's CTLA-4 CT60 genotype, donor's granzyme B +55 genotype and a patient's Fas -670 genotype are significantly associated with the outcomes of allo-HSCT, and that the presence of these susceptible alleles in the donor and/or patient results in worse overall survival. Analysis of the donor's and/or patient's genotype for T-cell activation and effector pathway genes could, therefore, be used for pre-HSCT risk assessment for patients with hematologic malignancies. The compelling urgency is to construct a risk “index” of clinical and genetic variables that would not only enable selection of an optimal unrelated donor out of multiple potential donors, but that could also be used to divide patients into genetically low- and high-risk subjects. Transplant recipients would thus benefit from accurate risk assessment and individualization of immunosuppressive therapy, taking into account the donor's and patient's genetic data, as well as other risk factors.
This study is the first report of the relationship between genetic variations of the activation and effector pathway molecules in transplant donors and recipients and the capacity for T-cell alloimmune responses after allo-HSCT. According to our results, special targeting of the CTLA-4, granzyme B and Fas pathways may be a promising therapeutic strategy in patients with aGVHD, CMV infection and leukemia relapse after allo-HSCT.
Supplementary Material
Acknowledgments
we would like to thank Shanghai Genesky Bio-Tech Genetic Core Laboratory for their excellent technical assistance with sequencing analysis. We also thank the HLA Typing Laboratory of the Blood Center of Zhejiang Province for assistance.
Funding: this work was funded in part by the Key Project of the National Natural Science Foundation of China (81230014), National High Technology Research and Development Program of China (2012AA020905), and the National Natural Science Foundation of China (81100387, 81170501, 81170526, 81070443).
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures: The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.van den Brink MR, Porter DL, Giralt S, Lu SX, Jenq RR, Hanash A, et al. Relapse after allogeneic hematopoietic cell therapy. Biol Blood Marrow Transplant. 2010;16(1Suppl):S138-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Penack O, Holler E, van den Brink., MR Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood. 2010;115(10):1865-72 [DOI] [PubMed] [Google Scholar]
- 3.Rocha V, Porcher R, Fernandes JF, Filion A, Bittencourt H, Silva W, Jr, et al. Association of drug metabolism gene polymorphisms with toxicities, graft-versus-host disease and survival after HLA-identical sibling hematopoietic stem cell transplantation for patients with leukemia. Leukemia. 2009;23(3):545-56 [DOI] [PubMed] [Google Scholar]
- 4.Mayor NP, Shaw BE, Hughes DA, Maldonado-Torres H, Madrigal JA, Keshav S, et al. Single nucleotide polymorphisms in the NOD2/CARD15 gene are associated with an increased risk of relapse and death for patients with acute leukemia after hematopoietic stem-cell transplantation with unrelated donors. J Clin Oncol. 2007;25(27):4262-9 [DOI] [PubMed] [Google Scholar]
- 5.Espinoza JL, Takami A, Onizuka M, Sao H, Akiyama H, Miyamura K, et al. NKG2D gene polymorphism has a significant impact on transplant outcomes after HLA-fullymatched unrelated bone marrow transplantation for standard risk hematologic malignancies. Haematologica. 2009;94(10):1427-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annu Rev Immunol. 1996;14:233-58 [DOI] [PubMed] [Google Scholar]
- 7.Hutloff A, Dittrich AM, Beier KC, Eljaschewitsch B, Kraft R, Anagnostopoulos I, et al. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;397(6716):263-6 [DOI] [PubMed] [Google Scholar]
- 8.Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol. 2006;24:65-97 [DOI] [PubMed] [Google Scholar]
- 9.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, et al. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science 1994;265(5171):528-30 [DOI] [PubMed] [Google Scholar]
- 10.Lowin B, Hahne M, Mattmann C, Tschopp J. Cytolytic T-cell cytotoxicity is mediated through perforin and Fas lytic pathways. Nature. 1994;370(6491):650-2 [DOI] [PubMed] [Google Scholar]
- 11.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423(6939):506-11 [DOI] [PubMed] [Google Scholar]
- 12.Suwalska K, Pawlak E, Karabon L, Tomkiewicz A, Dobosz T, Urbaniak-Kujda D, et al. Association studies of CTLA-4, CD28, and ICOS gene polymorphisms with B-cell chronic lymphocytic leukemia in the Polish population. Hum Immuno. 2008;69(3):193-201 [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Wen QJ, Yin Y, Lu XT, Pu SH, Tian HP, et al. FASLG polymorphism is associated with cancer risk. Eur J Cancer 2009;45(14):2574-8 [DOI] [PubMed] [Google Scholar]
- 14.Marin LA, Muro M, Moya-Quiles MR, Miras M, Minguela A, Bermejo J, et al. Study of Fas (CD95) and FasL (CD178) polymorphisms in liver transplant recipients. Tissue Antigens. 2006;67(2):117-26 [DOI] [PubMed] [Google Scholar]
- 15.Haimila K, Turpeinen H, Alakulppi NS, Kyllonen LE, Salmela KT, Partanen J. Association of genetic variation in inducible costimulator gene with outcome of kidney transplantation. Transplantation. 2009;87(3):393-6 [DOI] [PubMed] [Google Scholar]
- 16.Xiao HW, Lai XY, Luo Y, Shi JM, Tan YM, He JS, et al. Relationship between TNFA, TNFB and TNFRII gene polymorphisms and outcome after unrelated hematopoietic cell transplantation in a Chinese population. Bone Marrow Transplant. 2011;46(3):400-7 [DOI] [PubMed] [Google Scholar]
- 17.Xiao H, Cao W, Lai X, Luo Y, Shi J, Tan Y, et al. Immunosuppressive cytokine gene polymorphisms and outcome after related and unrelated hematopoietic cell transplantation in a chinese population. Biol Blood Marrow Transplant. 2011;17(4):542-9 [DOI] [PubMed] [Google Scholar]
- 18.Vogelsang GB, Arai S. Mycophenolate mofetil for the prevention and treatment of graft-versus-host disease following stem cell transplantation: preliminary findings. Bone Marrow Transplant. 2001;27(12):1255-62 [DOI] [PubMed] [Google Scholar]
- 19.Neumann F, Graef T, Tapprich C, Vaupel M, Steidl U, Germing U, et al. Cyclosporine A and mycophenolate mofetil vs cyclosporine A and methotrexate for graft-versus-host disease prophylaxis after stem cell transplantation from HLA-identical siblings. Bone Marrow Transplant. 2005;35(11):1089-93 [DOI] [PubMed] [Google Scholar]
- 20.Ostronoff F, Ostronoff M, Souto-Maior AP, Domingues M, Sucupira A, Manso DA, et al. Prospective trial of mycophenolate mofetil-cyclosporine A prophylaxis for acute GVHD after G-CSF stimulated allogeneic bone marrow transplantation with HLA-identical sibling donors in patients with severe aplastic anemia and hematological malignancies. Clin Transplant. 2009;23(1):33-8 [DOI] [PubMed] [Google Scholar]
- 21.Pinana JL, Valcarcel D, Fernandez-Aviles F, Martino R, Rovira M, Barba P, et al. MTX or mycophenolate mofetil with CsA as GVHD prophylaxis after reduced-intensity conditioning PBSCT from HLA-identical siblings. Bone Marrow Transplant. 2010;45(9):1449-56 [DOI] [PubMed] [Google Scholar]
- 22.Huang H, Lin M, Meng H, Qian W, Jin J, Huang J, et al. [Combination of mycophenolate mofetil with cyclosporine A and methotrexate as acute GVHD prophylaxis after unrelated donor allogeneic bone marrow transplantation]. Zhonghua Xue Ye Xue Za Zhi. 2001;22(2):76-8 [PubMed] [Google Scholar]
- 23.Huang H, Zheng W, Lin M, Fu J, Zhang R. [Prophylaxis of acute graft-versus-host disease after allogeneic bone marrow transplantation by mycophenolate mofetil in a murine model]. Zhonghua Xue Ye Xue Za Zhi. 2002;23(4):191-3 [PubMed] [Google Scholar]
- 24.Przepiorka D, Weisdorf D, Martin P, Klingemann HG, Beatty P, Hows J, et al. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995;15(6):825-8 [PubMed] [Google Scholar]
- 25.Boeckh M, Ljungman P. How we treat cytomegalovirus in hematopoietic cell transplant recipients. Blood. 2009;113(23):5711-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeckh M, Nichols WG. The impact of cytomegalovirus serostatus of donor and recipient before hematopoietic stem cell transplantation in the era of antiviral prophylaxis and preemptive therapy. Blood. 2004;103(6):2003-8 [DOI] [PubMed] [Google Scholar]
- 27.Ljungman P, Griffiths P, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002;34(8):1094-7 [DOI] [PubMed] [Google Scholar]
- 28.Scrucca L, Santucci A, Aversa F. Competing risk analysis using R: an easy guide for clinicians. Bone Marrow Transplant. 2007;40(4):381-7 [DOI] [PubMed] [Google Scholar]
- 29.Gooley TA, Leisenring W, Crowley J, Storer BE. Estimation of failure probabilities in the presence of competing risks: new representations of old estimators. Stat Med. 1999;18(6):695-706 [DOI] [PubMed] [Google Scholar]
- 30.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322(5899):271-5 [DOI] [PubMed] [Google Scholar]
- 31.Kouki T, Sawai Y, Gardine CA, Fisfalen ME, Alegre ML, DeGroot LJ. CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves' disease. J Immunol. 2000;165(11):6606-11 [DOI] [PubMed] [Google Scholar]
- 32.Parks CG, Hudson LL, Cooper GS, Dooley MA, Treadwell EL, St Clair EW, et al. CTLA-4 gene polymorphisms and systemic lupus erythematosus in a population-based study of whites and African-Americans in the southeastern United States. Lupus. 2004;13(10):784-91 [DOI] [PubMed] [Google Scholar]
- 33.Bosch-Vizcaya A, Perez-Garcia A, Brunet S, Solano C, Buno I, Guillem V, et al. Donor CTLA-4 genotype influences clinical outcome after T cell-depleted allogeneic hematopoietic stem cell transplantation from HLA-identical sibling donors. Biol Blood Marrow Transplant. 2012;18(1):100-5 [DOI] [PubMed] [Google Scholar]
- 34.Orru S, Orru N, Manolakos E, Littera R, Caocci G, Giorgiani G, et al. Recipient CTLA-4*CT60-AA genotype is a prognostic factor for acute graft-versus-host disease in hematopoietic stem cell transplantation for thalassemia. Hum Immunol. 2012;73(3):282-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Garcia A, De la Camara R, Roman-Gomez J, Jimenez-Velasco A, Encuentra M, Nieto JB, et al. CTLA-4 polymorphisms and clinical outcome after allogeneic stem cell transplantation from HLA-identical sibling donors. Blood. 2007;110(1):461-7 [DOI] [PubMed] [Google Scholar]
- 36.Murase M, Nishida T, Onizuka M, Inamoto Y, Sugimoto K, Imahashi N, et al. Cytotoxic T-lymphocyte antigen 4 haplotype correlates with relapse and survival after allogeneic hematopoietic SCT. Bone Marrow Transplant. 2011;46(11):1444-9 [DOI] [PubMed] [Google Scholar]
- 37.Metaxas Y, Bertz H, Spyridonidis A, Spyroupoulou-Vlachou M, Porzelius C, Finke J. CT60 single-nucleotide polymorphism as a surrogate marker for donor lymphocyte infusion outcome after allogeneic cell transplantation for acute leukemia. Bone Marrow Transplant. 2012;47(3):411-5 [DOI] [PubMed] [Google Scholar]
- 38.Perez-Garcia A, Brunet S, Berlanga JJ, Tormo M, Nomdedeu J, Guardia R, et al. CTLA-4 genotype and relapse incidence in patients with acute myeloid leukemia in first complete remission after induction chemotherapy. Leukemia. 2009;23(3):486-91 [DOI] [PubMed] [Google Scholar]
- 39.Huurman VA, Unger WW, Koeleman BP, Oaks MK, Chandraker AK, Terpstra OT, et al. Differential inhibition of autoreactive memory- and alloreactive naive T cell responses by soluble cytotoxic T lymphocyte antigen 4 (sCTLA4), CTLA4Ig and LEA29Y. Clin Exp Immunol. 2007;150(3):487-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurschus FC, Jenne DE. Delivery and therapeutic potential of human granzyme B. Immunol Rev. 2010;235(1):159-71 [DOI] [PubMed] [Google Scholar]
- 41.Schmaltz C, Alpdogan O, Horndasch KJ, Muriglan SJ, Kappel BJ, Teshima T, et al. Differential use of Fas ligand and perforin cytotoxic pathways by donor T cells in graft-versus-host disease and graft-versus-leukemia effect. Blood. 2001;97(9):2886-95 [DOI] [PubMed] [Google Scholar]
- 42.Higaki Y, Yamada O, Okamura T, Mizoguchi H, Kawashima M. Granzyme-B-containing lymphocyte involvement in epidermal injury in graft-versus-host disease. Dermatology. 2001;202(2):94-8 [DOI] [PubMed] [Google Scholar]
- 43.Jaksch M, Remberger M, Mattsson J. Increased immune transcript levels are correlated with acute graft-versus-host disease and cytomegalovirus response after allogeneic stem cell transplantation. Transplantation. 2004;77(2):195-200 [DOI] [PubMed] [Google Scholar]
- 44.Kircher B, Schumacher P, Nachbaur D. Granzymes A and B serum levels in allo-SCT. Bone Marrow Transplant. 2009;43(10):787-91 [DOI] [PubMed] [Google Scholar]
- 45.Espinoza LJ, Takami A, Nakata K, Yamada K, Onizuka M, Kawase T, et al. Genetic variants of human granzyme B predict transplant outcomes after HLA matched unrelated bone marrow transplantation for myeloid malignancies. PLoS One. 2011;6(8):e23827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McIlroy D, Cartron PF, Tuffery P, Dudoit Y, Samri A, Autran B, et al. A triple-mutated allele of granzyme B incapable of inducing apoptosis. Proc Natl Acad Sci USA. 2003;100(5):2562-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Girnita DM, Webber SA, Brooks MM, Ferrell R, Girnita AL, Burckart GJ, et al. Genotypic variation and phenotypic characterization of granzyme B gene polymorphisms. Transplantation. 2009;87(12):1801-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muschen M, Warskulat U, Beckmann MW. Defining CD95 as a tumor suppressor gene. J Mol Med (Berl). 2000;78(6):312-25 [DOI] [PubMed] [Google Scholar]
- 49.Kanemitsu S, Ihara K, Saifddin A, Otsuka T, Takeuchi T, Nagayama J, et al. A functional polymorphism in fas (CD95/APO-1) gene promoter associated with systemic lupus erythematosus. J Rheumatol. 2002;29(6):1183-8 [PubMed] [Google Scholar]
- 50.Sibley K, Rollinson S, Allan JM, Smith AG, Law GR, Roddam PL, et al. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res. 2003;63(15):4327-30 [PubMed] [Google Scholar]
- 51.Farre L, Bittencourt AL, Silva-Santos G, Almeida A, Silva AC, Decanine D, et al. Fas 670 promoter polymorphism is associated to susceptibility, clinical presentation, and survival in adult T cell leukemia. J Leukoc Biol. 2008;83(1):220-2 [DOI] [PubMed] [Google Scholar]
- 52.Lai HC, Lin WY, Lin YW, Chang CC, Yu MH, Chen CC, et al. Genetic polymorphisms of FAS and FASL (CD95/CD95L) genes in cervical carcinogenesis: an analysis of haplotype and gene-gene interaction. Gynecol Oncol. 2005;99(1):113-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.