Chronic idiopathic neutropenia (CIN) is an acquired disorder of granulopoiesis characterized by prolonged, unexplained reduction in peripheral blood (PB) neutrophils due to accelerated apoptosis of the bone marrow (BM) granulocytic progenitors.1 There has been growing evidence to suggest that CIN shares common pathogenetic features with immune-mediated BM failure syndromes, such as aplastic anemia (AA) and myelodysplastic syndromes (MDS).2
Clonal hematopoiesis has been associated with the pathophysiology of the immune-mediated BM failure syndromes.3,4 The possible existence of clonal stem/progenitor cell populations in CIN using methods other than classical karyotype has not yet been investigated. The X-chromosome inactivation pattern (XCIP) method is based on the Lyon–Beutler hypothesis of the random inactivation of one of the two X-chromosomes early in embryogenesis.5,6 Normal female tissues display a mosaic expression of genes from maternal and paternal X-chromosomes whereas monoclonal populations express either maternally or paternally-derived X-linked genes.5,6 The XCIP assay has been widely used to identify clonality in MDS and AA.7-10 In the current study, we have probed the existence of clonal hematopoiesis in CIN by investigating the XCIP of the glucose-6-phosphate dehydrogenase (G6PD) and iduronate-2-sulfatase (IDS) gene polymorphisms in PB granulocytes and lymphocytes using reverse transcription polymerase chain reaction (RT-PCR).10
Genomic DNA extracted from PB samples of 134 female patients fulfilling the diagnostic criteria for CIN1 and 124 age-matched female healthy subjects, was initially screened for C/T heterozygocity at nucleotides 1311 of G6PD and 438 of IDS using a PCR-based restriction fragment length polymorphism assay.10 Total RNA isolated from PB lymphocytes and granulocytes of subjects showing heterozygocity in at least one of the two genes was transcribed into cDNA and amplified by a nested (G6PD) or conventional (IDS) PCR using previously described primers.10 Samples showing expression of only one allele at a ratio equal to or more than 9:1 compared to the other allele despite the existence of both at the genomic level, were classified as clonal. The χ2 test was used for comparisons between patients and controls.
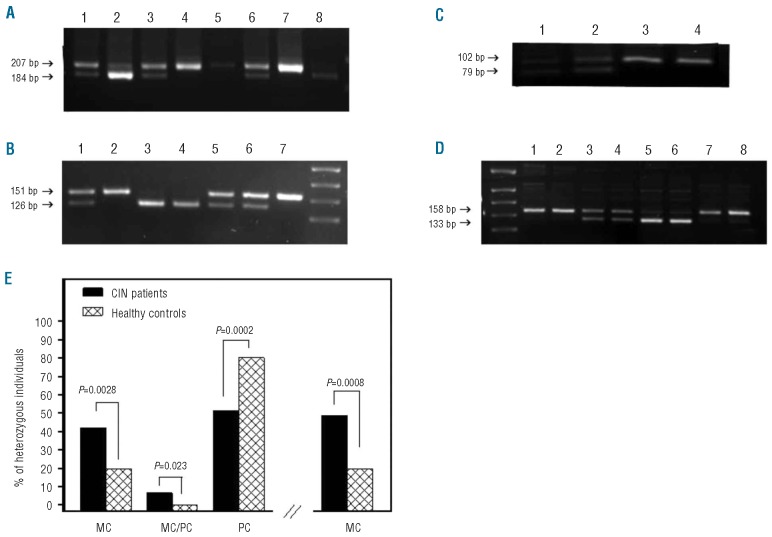
Seventy-six CIN patients (56.7%) and 76 healthy individuals (61.3%) showing heterozygocity for at least one polymorphism were assessed for clonality in PB lymphocytes and granulocytes (Figure 1A and B). Patients’ characteristics are shown in Online Supplementary Table S1. No statistically significant difference was observed in the age distribution between CIN patients and controls that were assessed for clonality. Twenty of 41 (48.78%) CIN patients heterozygous for the G6PD polymorphism showed a monoclonal pattern in at least one cell population; 16 of 41 patients (39.02%) displayed monoclonal pattern in both lymphocytes and granulocytes indicating the involvement of an early hematopoietic stem cell whereas 4 of 41 patients (9.76%) displayed monoclonal pattern in granulocytes and polyclonal in lymphocytes implying clonality within the myeloid lineage (Figure 1C). Of 28 healthy subjects showing heterozygocity for G6PD, 6 individuals (21.43%) displayed a monoclonal pattern that concerned both cell populations in all cases. This frequency is significantly lower than the respective 48.78% observed in the patients (P=0.0213).
Figure 1.
Genotyping, clonality analysis and frequency of the expression pattern of G6PD and/or IDS polymorphisms in CIN patients and healthy individuals. Genomic DNA was extracted from PB samples of CIN patients and healthy controls and genotyping for polymorphisms of G6PD and IDS genes was performed by a PCR-based restriction fragment length polymorphism assay. Total RNA was isolated from lymphocytes and granulocytes of heterozygous subjects and clonality analysis was performed by RT-PCR. (A) Genotyping of G6PD. Lanes 1, 3 and 6 show heterozygous (C/T), lanes 2 and 8 homozygous (T/T), and lanes 4, 5 and 7 homozygous (C/C) subjects. (B) Genotyping of IDS. Lanes 1, 5 and 6 show heterozygous (C/T), lanes 2 and 7 homozygous (T/T), and lanes 3 and 4 homozygous (C/C) subjects. (C) Clonality analysis of G6PD RT-PCR products in CIN patients. Lanes 1 and 2 show a polyclonal pattern in granulocytes and lymphocytes, respectively, and lanes 3 and 4 show a monoclonal pattern in granulocytes and lymphocytes, respectively. (D) Clonality analysis of IDS RT-PCR products. Lanes 1 and 2 show monoclonal patterns in granulocytes and lymphocytes, respectively (CIN); lanes 3 and 4 show polyclonal patterns in granulocytes and lymphocytes, respectively (CIN); lanes 5 and 6 show monoclonal pattern in granulocytes and lymphocytes, respectively (healthy control); lanes 7 and 8 show monoclonal pattern in granulocytes and lymphocytes, respectively (healthy control). (E) The bars on the left represent the frequency of patients and healthy individuals with a monoclonal (MC) pattern of at least one gene polymorphism in both lymphocytes and granulocytes, monoclonal pattern in granulocytes and polyclonal pattern in lymphocytes (MC/PC) and polyclonal (PC) pattern in both cell populations. The bars on the right represent the overall frequency of patients and healthy individuals displaying monoclonality in at least one cell population. Comparison between patients and controls has been performed by means of the χ2 test and P values are shown.
Similarly, 23 of 50 (46%) CIN patients heterozygous for the IDS polymorphism displayed a monoclonal pattern in at least one PB cell population (Figure 1D); 20 of 50 (40%) CIN patients displayed monoclonal pattern in both cell populations and 3 of 50 patients (6%) displayed monoclonal pattern in granulocytes and polyclonal in lymphocytes suggesting the involvement of a primitive and committed stem cell, respectively. Of the 57 healthy subjects showing heterozygocity for IDS, 12 individuals (21.05%) displayed monoclonal pattern that concerned both cell populations in all cases (Figure 1D). This proportion is significantly lower than the respective 46% observed in CIN patients (P=0.0061).
Of the 76 CIN patients displaying heterozygosity in the expression of at least one gene polymorphism, 32 (42.10%) showed monoclonal pattern in both cell populations, 5 (6.58%) showed monoclonal pattern in granulocytes and polyclonal in lymphocytes, and 39 showed polyclonal pattern in both cell populations (51.32%). No statistically significant difference was obtained in the frequency of clonality between patients with mild and those with more severe neutropenia using the threshold of 1,000x106/L. Of the 76 healthy individuals who were heterozygous for the G6PD and/or IDS polymorphism, 15 (19.74%) displayed monoclonal pattern in both cell subpopulations (P=0.0028), none displayed monoclonal/polyclonal pattern in granulocytes/lymphocytes (P=0.023), and 61 of 76 (80.26%) displayed polyclonal pattern in both cell subpopulations (P=0.0002) (Figure 1E). Overall, 48.68% (37 of 76) CIN patients and 19.74% (15 of 76) healthy individuals displayed a monoclonal pattern in at least one cell population (P=0.0008). For patients (n=15) and controls (n=9) who were heterozygous for both polymorphisms, a concordance was obtained in the pattern of polymorphism expression (monoclonal/polyclonal) in both cell populations (Online Supplementary Table S1).
Our data suggest that CIN may represent a clonal disorder of hematopoiesis mostly at the pluripotent stem cell level. Although the validity of our method was screened by analyzing PB cell populations from patients with hematologic malignancies (data not shown), we recognize that interpretation of results is limited by the skewed pattern of X-chromosome inactivation that occurs in a significant proportion of females and may increase with age.11 Excessive allele skewing has been reported to be 20% in females older than 60 years but proportions ranging from 0% to 38% have also been reported, depending on the criteria for the excessive Lyonization definition, the type of XCIP, and even the tissue type. However, given that CIN patients and healthy controls are age-matched, the differences in clonality assessment obtained should be taken into account as they cannot be attributed to the influence of age-related skewing. Furthermore, no significant difference was found in age distribution between subjects (patients or controls) with clonal and those with non-clonal XCIP, suggesting that our results are unlikely to be biased by age.
The pathophysiological significance of the clonal populations identified in CIN patients is obscure. They might indicate a contraction of the stem cell pool resulting in support of hematopoiesis by one or a few stem cells, as previously described in aplastic anemia and hypoplastic myelodysplastic syndromes (MDS).4 Alternatively, they might imply an early event in a transformation process and although malignant evolution has been rarely reported in CIN, a thorough long-term follow up of patients with clonal populations is needed.12 The clonal XCIP in our patients should not be interpreted as indicative for MDS diagnosis because the majority of these patients had a long history of stable disease. Overall, we have provided evidence for possible clonal hematopoiesis in CIN. However, clonality data cannot be currently used as a diagnostic criterion for the disease and further studies with newer technologies, such as comparative genomic hybridization and single nucleotide polymorphism array-based karyotyping, are required to confirm clonal patterns of hematopoiesis in these patients.
Supplementary Material
Acknowledgments
Funding: this work was partially supported by the grant 09SYN-13-880 of the Greek Ministry of National Education and Religious Affairs to HAP and a grant from the Hellenic Hematology Association to SM.
Footnotes
The online version of this article has a Supplementary Appendix.
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure o Competing Interests are also available at www.haematologica.org.
References
- 1.Papadaki HA, Eliopoulos AG, Kosteas T, Gemetzi C, Damianaki A, Koutala H, et al. Impaired granulocytopoiesis in patients with chronic idiopathic neutropenia is associated with increased apoptosis of bone marrow myeloid progenitor cells. Blood. 2003;101(7):2591-600 [DOI] [PubMed] [Google Scholar]
- 2.Palmblad J, Papadaki HA. Chronic idiopathic neutropenias and severe congenital neutropenia. Curr Opin Hematol. 2008;15(1):8-14 [DOI] [PubMed] [Google Scholar]
- 3.Anan K, Ito M, Misawa M, Ohe Y, Kai S, Kohsaki M, et al. Clonal analysis of peripheral blood and haemopoietic colonies in patients with aplastic anaemia and refractory anaemia using the polymorphic short tandem repeat on the human androgen-receptor (HUMARA) gene. Br J Haematol. 1995;89(4):838-44 [DOI] [PubMed] [Google Scholar]
- 4.Tiu R, Gondek L, O'Keefe C, Maciejewski JP. Clonality of the stem cell compartment during evolution of myelodysplastic syndromes and other bone marrow failure syndromes. Leukemia. 2007;21(8):1648-57 [DOI] [PubMed] [Google Scholar]
- 5.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 1961;190:372-3 [DOI] [PubMed] [Google Scholar]
- 6.Beutler E, Yeh M, Fairbanks VF. The normal human female as a mosaic of X-chromosome activity: studies using the gene for C-6-PD-deficiency as a marker. Proc Natl Acad Sci USA. 1962;48:9-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guidetti F, Grazioli S, Capelli F, Marini C, Gallicchio M, De Micheli D, et al. Primitive hematopoietic stem cells shows a polyclonal pattern in myelodysplastic syndromes. Haematologica. 2004;89(1):21-8 [PubMed] [Google Scholar]
- 8.Cermak J, Belickova M, Krejcova H, Michalova K, Zilovcova S, Zemanova Z, et al. The presence of clonal cell subpopulations in peripheral blood and bone marrow of patients with refractory cytopenia with multilineage dysplasia but not in patients with refractory anemia may reflect a multistep pathogenesis of myelodysplasia. Leuk Res. 2005;29(4):371-9 [DOI] [PubMed] [Google Scholar]
- 9.van Kamph H, Landegent JE, Jansen RP, Willemze R, Fibbe WE. Clonal hematopoiesis in patients with acquired aplastic anemia. Blood. 1991;78(12):3209-14 [PubMed] [Google Scholar]
- 10.Mortazavi Y, Chopra R, Gordon-Smith EC, Rutherford TR. Clonal patterns of X-chromosome inactivation in female patients with aplastic anaemia studies using a novel reverse transcription polymerase chain reaction method. Eur J Haematol. 2000;64(6):385-95 [DOI] [PubMed] [Google Scholar]
- 11.Chen GL, Prchal JT. X-linked clonality testing: interpretation and limitations. Blood. 2007;110(5):1411-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papadaki HA, Pontikoglou C. Pathophysiologic mechanisms, clinical features and treatment of idiopathic neutropenia. Expert Rev Hematol. 2008;1(2):217-29 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.