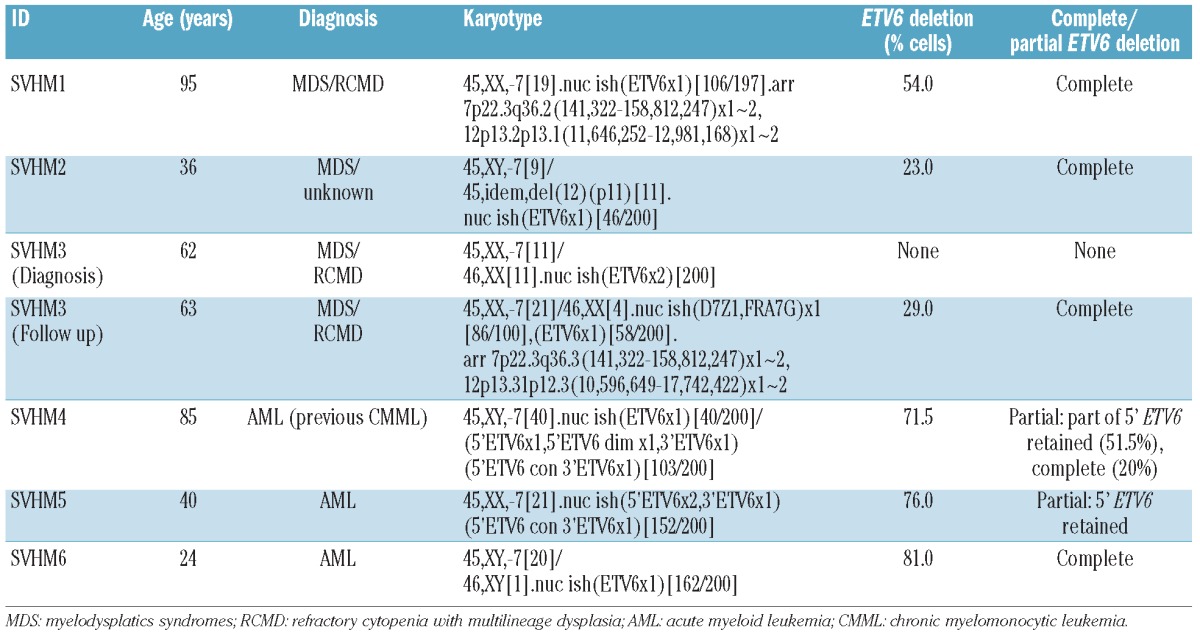
Chromosome 7 abnormalities are common in myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML). Often, there is partial loss of 7q as the result of deletion (del(7q)) or loss of one entire copy of chromosome 7 (monosomy 7 or -7). Del(7q) and -7 may occur in isolation, in simple or in complex karyotypes.1 It is assumed that the pathogenicity of -7/del(7q) is due to loss of tumor suppressor gene(s). However, the molecular mechanisms underpinning the clinical behavior of patients with 7/del(7q) MDS/AML are not well understood.
Deregulation of the ETV6 gene through translocation, deletion or somatic mutation is also recurrent in myeloid malignancies. By metaphase cytogenetics (MC), approximately 1% of patients with AML and 0.6% of patients with MDS have deletion of part of 12p.2,3 Mapping studies place ETV6 within the smallest commonly deleted region.4
It has been reported that 5% of patients with -7/del(7q) have del(12p) by MC.1 Using a single nucleotide polymorphism array (SNP-A), we identified a cryptic ETV6 deletion in a patient with -7. The poor chromosome morphology typically encountered in patients with -7 means that it may be difficult to identify subtle 12p abnormalities in metaphases. We, therefore, reasoned that the frequency of ETV6 deletion in patients with -7 may be higher than is currently appreciated. In order to test this hypothesis, we performed fluorescence in situ hybridization (FISH) using an allele-specific ETV6 probe in patients with -7.
We used the Victorian Cancer Cytogenetics Service database to identify samples from patients with -7 (either as the sole autosomal abnormality or in association with an abnormality of 12p) and a diagnosis of MDS, myelodysplastic/myeloproliferative neoplasm (MDS/MPN) or AML. Thirty-eight patients referred for testing between 1st January 2000 and 1st January 2011 had residual cytogenetic suspension suitable for FISH. Two patients had a 12p abnormality. In 36 cases, -7 was the only autosomal abnormality. Twenty-one patients had a diagnosis of MDS or MDS/MPN. The remainder had a diagnosis of AML.
We identified ETV6 deletion in 6 of 38 (16%) cases. Patients with an ETV6 deletion had a median age of 51.5 years (range 24-95) (Table 1). In 5 of 6 cases, the ETV6 deletion was cytogenetically cryptic. The percentage of cells with ETV6 loss ranged from 23% to 81% and was higher in AML than in MDS (average±SEM 76.2±2.7% vs. 35.3±9.5%, P=0.02) even though the majority of metaphases in all patients were derived from the -7 clone. ETV6 deletions were heterozygous in all cases.
Characteristics and karyotypes of patients with an ETV6 deletion.
Some abnormal FISH signal patterns were consistent with complete loss of one ETV6 allele; others suggested partial loss with a breakpoint positioned within the ETV6 locus Table 1). A simple signal pattern consistent with complete deletion of one copy of ETV6 (loss of one 5’ and 3′ ETV6 signal per cell) was the only abnormality detected in 4 patients (Figure 1A). In SVHM5 there was loss of one 3′ signal with retention of the 5′ signal in all abnormal cells consistent with a partial deletion where the distal breakpoint on 12p was positioned downstream of the area recognized by the 5′ probe (Figure 1B). The remaining patient, SVHM4, had two different cell populations. A 5′ signal of diminished intensity was present together with loss of one 3′ signal per cell in 51.5% cells scored suggesting that the telomeric breakpoint of the deleted allele was located within the region recognized by the 5′ probe. In another 20% of cells, one entire copy of both the 5′ and 3′ signals was lost, indicating breakpoints located outside ETV6 (Figure 1C-E). Thus, there was heterogeneity with respect to the breakpoints relative to the position of the ETV6 locus.
Figure 1.
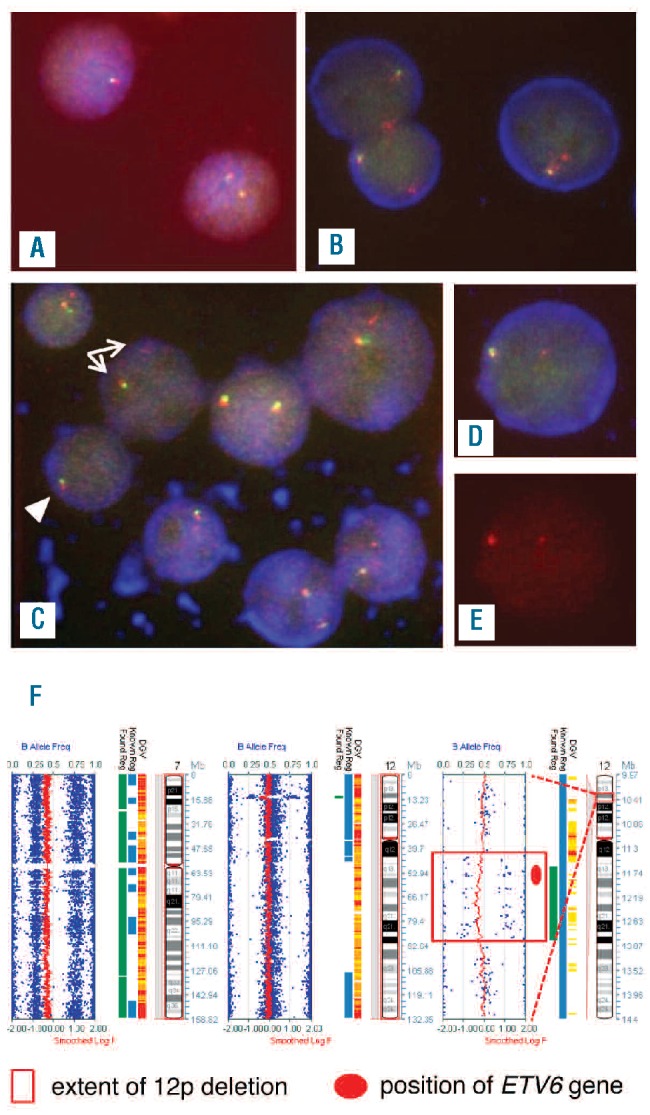
Representative FISH and SNP-A results for patients with ETV6 deletion. (A) An interphase cell with one ETV6 fusion signal (top left) consistent with complete loss of one ETV6 allele and a cell with two ETV6 fusion signals (bottom right) in SVHM3. The same signal patterns were observed in SVHM1, SVHM2 and SVHM6. (B) One ETV6 fusion signal (5’ and 3’ ETV6) and one red 5’ ETV6 signal per cell in three interphase cells from SVHM5. (C) A mixed population of cells, some with one ETV6 fusion signal and one red 5’ ETV6 signal of diminished intensity (white arrows) and one cell with a single ETV6 fusion signal only (white arrowhead) in SVHM4. (D and E) The diminished intensity of the red signal on the deleted ETV6 allele in SVHM4 was more readily apparent on the single color (Spectrum Orange) image (E) than on the triple color (DAPI/FITC/Spectrum Orange) image of the same cell (D). Fluorescent signals were visualized under an Axioplan 2 (Zeiss) microscope and captured and analyzed using ISIS image analysis software (Metasystems, Altlussheim, Germany). (F) SNP-A abnormalities in SVHM1. Smoothed log R values (red) and B allele frequencies (blue) for chromosome 7 (whole chromosome view, left panel), chromosome 12 (whole chromosome view, middle panel) and the deleted region of 12p (right panel) with the extent of deletion shown by the red box and the position of the ETV6 gene indicated by the red circle. DNA (200 ng) was hybridized to CytoSNP-12 BeadChip arrays (Illumina, San Diego, CA, USA) and analysis was performed using Karyostudio v1.2 software (Illumina).
Two patients had genomic DNA collected allowing SNP-A karyotyping to be performed. In SVHM1, the SNP-A demonstrated a 1.3 Mb mono-allelic deletion encompassing ETV6 (Figure 1F and Table 1). In SVHM3, the SNP-A delineated a 7.2 Mb heterozygous deletion spanning the entire ETV6 locus (Table 1). Strikingly, in both cases, no additional cytogenetically cryptic somatic copy number changes or UPD were identified.
SVHM3 was the only case where paired diagnostic and follow-up samples were available. In this patient, ETV6 deletion was not observed at diagnosis but was detected in a sample obtained five and a half months later (Table 1). On the balance of probability, given that the -7 clone is larger and that either acquisition of the ETV6 deletion or expansion of an occult ETV6 deleted clone occurred in the months between diagnosis and reassessment, it is likely that in this case -7 preceded development of the ETV6 lesion.
Previous reports place the prevalence of ETV6 deletion in MDS/AML somewhere between 0% and 64% with the frequency heavily dependent on the characteristics of the study population. Large representative studies in MDS and AML consistently find rates of less than 5%.5-7 The highest rates occurred in cohorts enriched for patients with structural or numerical abnormalities of chromosome 12.8,9 Interestingly, in other studies where the rate of ETV6 deletion exceeded 5%, there was a strong association with complex karyotypes that frequently exhibited -7/del(7q).10-12
Outside of two cohorts with chromosome 12 abnormalities8,9 and one case series with complex karyotypes,12 our study has the highest reported frequency of ETV6 deletion. At 16%, the prevalence of ETV6 loss in -7 patients is in excess of 3 times the expected rate in unselected MDS/AML cohorts. Although the association between -7/del(7q) also holds up in complex karyotypes in published series10-12 and in our own experience (R. MacKinnon et al., unpublished data, 2012) the presence of gross chromosomal instability in that setting can make it difficult to distinguish driver from passenger lesions. Co-occurrence of just two copy number changes, at rates higher than might be expected by chance in this study is more compelling evidence in support of functional cooperation between -7 and ETV6 deletion.
As we enter the era of molecularly targeted therapy, it is important to understand the consequences of genetic events that are critical for maintenance and progression of malignant phenotypes. Our results suggest that -7 and ETV6 loss drive complementary steps during leukemogenesis.
Supplementary Material
Acknowledgments:
the authors wish to thank staff members of the VCCS for sample set up, processing and karyotype analysis, and Brett Kennedy, Vanessa Tyrell and Derek Campbell from Illumina for training and provision of SNP-A equipment.
Funding: MW was supported by the Victorian Cancer Agency. MW and RNM were supported by the James and Vera Lawson Philanthropic Trust (ANZ Trustees).
Footnotes
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Haase D. Cytogenetic features in myelodysplastic syndromes. Ann Hematol. 2008;87(7):515-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354-65 [DOI] [PubMed] [Google Scholar]
- 3.Schanz J, Tuchler H, Sole F, Mallo M, Luno E, Cervera J, et al. New comprehensive cytogenetic scoring system for primary myelodysplastic syndromes (MDS) and oligoblastic acute myeloid leukemia after MDS derived from an international database merge. J Clin Oncol. 2012;30(8):820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato Y, Suto Y, Pietenpol J, Golub TR, Gilliland DG, Davis EM, et al. TEL and KIP1 define the smallest region of deletions on 12p13 in hematopoietic malignancies. Blood. 1995;86(4):1525-33 [PubMed] [Google Scholar]
- 5.Suela J, Alvarez S, Cifuentes F, Largo C, Ferreira BI, Blesa D, et al. DNA profiling analysis of 100 consecutive de novo acute myeloid leukemia cases reveals patterns of genomic instability that affect all cytogenetic risk groups. Leukemia. 2007;21(6):1224-31 [DOI] [PubMed] [Google Scholar]
- 6.Tiu RV, Gondek LP, O’Keefe CL, Huh J, Sekeres MA, Elson P, et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol. 2009;27(31):5219-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA. 2009;106(31):12950-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajaj R, Xu F, Xiang B, Wilcox K, Diadamo AJ, Kumar R, et al. Evidence-based genomic diagnosis characterized chromosomal and cryptic imbalances in 30 elderly patients with myelodysplastic syndrome and acute myeloid leukemia. Mol Cytogenet. 2011;4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Streubel B, Sauerland C, Heil G, Freund M, Bartels H, Lengfelder E, et al. Correlation of cytogenetic, molecular cytogenetic, and clinical findings in 59 patients with ANLL or MDS and abnormalities of the short arm of chromosome 12. Br J Haematol. 1998;100(3):521-33 [DOI] [PubMed] [Google Scholar]
- 10.Heinrichs S, Kulkarni RV, Bueso-Ramos CE, Levine RL, Loh ML, Li C, et al. Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics.Leukemia.2009;239:1605-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolquist KA, Schultz RA, Furrow A, Brown TC, Han JY, Campbell LJ, et al. Microarray-based comparative genomic hybridization of cancer targets reveals novel, recurrent genetic aberrations in the myelodysplastic syndromes. Cancer Genet. 2011;204(11):603-28 [DOI] [PubMed] [Google Scholar]
- 12.Rucker FG, Bullinger L, Schwaenen C, Lipka DB, Wessendorf S, Frohling S, et al. Disclosure of candidate genes in acute myeloid leukemia with complex karyotypes using microarray-based molecular characterization. J Clin Oncol. 2006;24(24):3887-94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


