Methylation of DNA in CpG-rich islands is a key event in the regulation of tissue- and context-specific gene expression. Thereby, DNA methyltransferase 3A (DNMT3A) plays a pivotal role by converting cytosine to 5-methylcytosine.1 Recently, DNMT3A mutations have been described at high frequency in acute myeloid leukemia (AML), particularly with a monocytic pheno- type.2-4 In in vitro assays, mutated DNMT3A induced aberrant DNA methylation and promoted cellular proliferation. RAF kinase inhibitor protein (RKIP) negatively regulates the RAS-mitogen activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway.5 Several solid neoplasms show decreased or absent expression of RKIP and its role as a metastasis suppressor has been firmly established in animal models and studies of human tumors.5 We recently demonstrated that RKIP acts as a tumor suppressor in hematopoietic cells and that loss of RKIP expression occurs frequently in AML.6,7 Similar to mutated DNMT3A, RKIP loss is linked to AMLs with a monocytic phenotype. We, therefore, asked whether loss of RKIP and mutations in DNMT3A correlate in this particular subtype.
For PCR and direct sequencing of DNMT3A, its coding sequences and splice sites comprising 22 exons were analyzed as previously described3 in leukemic specimens of 36 patients diagnosed with acute monocytic leukemia; patients were classified in subgroups M4 and M5 according to the French-American-British (FAB) classification. Cytogenetic information was available in 32 patients with 4 of 32 (12.5%) karyotypes conferring good, 21 of 32 (65.5%) intermediate, and 7 of 32 (22%) adverse risk. Median age at diagnosis was 55.5 years (range 18-80 years), median white blood cell (WBC) count 55.5x109/L (range 5-445x109/L). This cohort has also been characterized for RKIP expression, showing its loss in 17 of 36 (47%) patients as determined at protein and mRNA level using Western blot and quantitative real-time polymerase chain reaction (PCR), respectively.7 Screening for mutations in NRAS and KRAS (codons 12, 13 and 61) as well as in NPM1 (exon 12) has been performed as previously described.7,8 Detection of FLT3 internal tandem duplications (ITD) and tyrosine kinase domain (TKD) mutations was performed using the FLT3 Mutation Assay (InVivoScribe Technologies, San Diego, CA, USA) according to the manufacturer's protocol. Informed consent was obtained from all individuals and the study was approved by the institutional review board of the Medical University of Graz, Austria. Statistical correlations were calculated by Fisher's exact (for correlation of DNMT3A with RKIP, FLT3 and NPM1) and by Mann-Whitney- Wilcoxon test (for correlation of DNMT3A with cytogenetics, WBC count and age at diagnosis), using R 2.15.1 (http://www.r-project.org).
We detected mutations in DNMT3A in 16 of 36 (44%) patients with monocytic AML. This high frequency is consistent with previous reports on DNMT3A mutations in this AML subtype.2,4 Fifteen of these mutations constituted the recently described hot spot mutations R882H and R882C, respectively.2,4 In one sample, we detected an R676W substitution, a DNMT3A mutation that has not yet been described. Its somatic origin could be proven by analysis of constitutional material (Figure 1). This substitution was predicted to be “disease causing” by MutationTaster9 and “damaging” by Sorting Intolerant From Tolerant (SIFT).10 WBC counts at diagnosis were significantly higher in patients with mutant as compared to wild-type DNMT3A (77x109/L vs. 51x109/L; P=0.040), whereas no significant difference could be observed for cytogenetic risk groups and patients age at diagnosis (data not shown). In the cohort investigated, DNMT3A mutations were associated with alterations of NPM1 as 11 of 14 (79%) DNMT3A mutated patients also harbored a NPM1 mutation compared to 6 of 20 (30%) DNMT3A wild-type patients (P=0.014). The rates of FLT3-ITD and mutations in the FLT3-TKD were found to be similar in DNMT3A mutated and DNMT3A wildtype cases. Notably, only 3 of 34 (9%) patients demonstrated mutations in NRAS or KRAS, which precluded statistical testing (Figure 2).
Figure1.
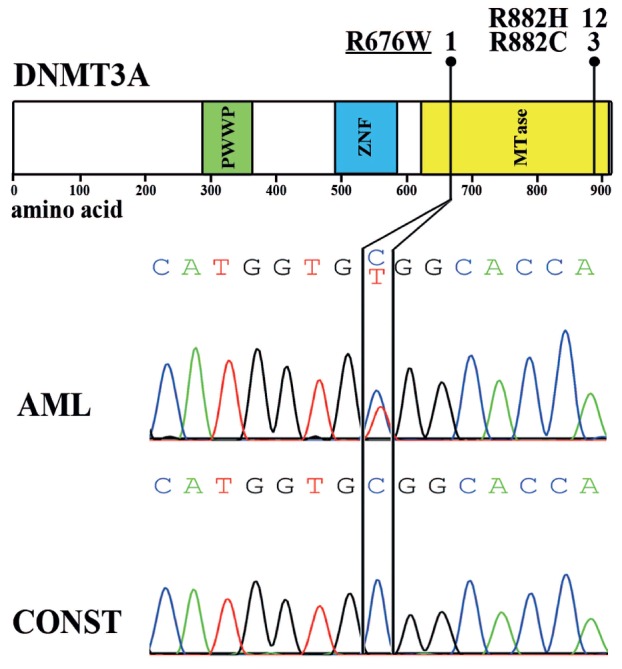
R676W is a novel somatic DNMT3A mutation. Number of distinct mutations detected in this study and their locations within the DNMT3A protein. The novel R676W substitution is caused by the heterozygous C2026T substitution as highlighted with black bars in the electropherogram of leukemic DNA. Absence of this mutation in constitutional material proves its somatic origin. PWWP: proline-tryptophan-tryptophan-proline domain; ZNF: zinc finger domain; MTase: methyltransferase domain; CONST: constitutional material.
Figure 2.
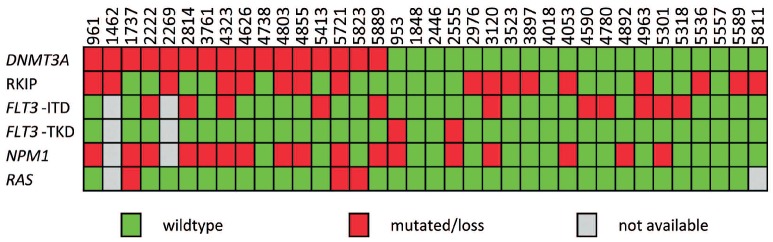
Molecular correlations of DNMT3A mutations in monocytic AML. Heatmap showing the occurrence of DNMT3A, NPM1, FLT3 and RAS mutations as well as RKIP loss in 36 patients with monocytic AML. Exon 12 alterations in NPM1 were significantly enriched in patients with mutant DNMT3A (P=0.014), whereas RKIP loss and aberrations in FLT3 did not correlate with DNTM3A mutations. Note that RAS status was excluded from statistical testing due to the low number of mutated cases.
Importantly, mutations in DNMT3A did not correlate with loss of the RKIP protein (P=0.99) as calculated by Fisher's exact test (Figure 2). This finding is of interest as the causative mechanisms of RKIP silencing have not yet been clarified. We previously screened for mutations and copy number variations in the RKIP gene as well as for promoter methylation, but did not find any abnormalities.7 Interestingly, DNMT3A has been shown to induce both hyper- and hypomethylation at distinct loci including non-promoter regions resulting in direct and indirect alteration of gene expression profiles.11,12 However, the lack of correlation between these two aberrations observed in this study suggests that RKIP silencing is not mediated by mutations in DNMT3A.
Taken together, we confirmed a high frequency of DNMT3A mutations in monocytic AML and described one novel somatic substitution. Mutant DNMT3A occurred independently of the RKIP expression status suggesting other, hitherto unknown mechanisms responsible for loss of RKIP in AML.
Supplementary Material
Acknowledgments
Funding: this study was supported by Leukamiehilfe Steiermark. Armin Zebisch was further supported by the START-Funding- Program of the Medical University of Graz, Graz, Austria.
Footnotes
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell Mol Life Sci. 2002;59(2):241-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fried I, Bodner C, Pichler MM, Lind K, Beham-Schmid C, Quehenberger F, et al. Frequency, onset and clinical impact of somatic DNMT3A mutations in therapy-related and secondary acute myeloid leukemia. Haematologica. 2012;97(2):246-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43(4):309-15 [DOI] [PubMed] [Google Scholar]
- 5.Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, et al. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2(3):232-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zebisch A, Haller M, Hiden K, Goebel T, Hoefler G, Troppmair J, et al. Loss of RAF kinase inhibitor protein is a somatic event in the pathogenesis of therapy-related acute myeloid leukemias with C- RAF germline mutations. Leukemia. 2009;23(6):1049-53 [DOI] [PubMed] [Google Scholar]
- 7.Zebisch A, Wolfler A, Fried I, Wolf O, Lind K, Bodner C, et al. Frequent loss of RAF kinase inhibitor protein expression in acute myeloid leukemia. Leukemia. 2012;26(8):1842-9 [DOI] [PubMed] [Google Scholar]
- 8.Pasqualucci L, Liso A, Martelli MP, Bolli N, Pacini R, Tabarrini A, et al. Mutated nucleophosmin detects clonal multilineage involvement in acute myeloid leukemia: Impact on WHO classification. Blood. 2006;108(13):4146-55 [DOI] [PubMed] [Google Scholar]
- 9.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575-6 [DOI] [PubMed] [Google Scholar]
- 10.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073-81 [DOI] [PubMed] [Google Scholar]
- 11.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44(1):23-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329(5990):444-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


