Abstract
There remains a tremendous need to develop targeted therapeutics that can both image and localize the toxic effects of chemotherapeutics and antagonists on diseased tissue while reducing adverse systemic effects. These needs have fostered the development of a nanotechnology-based approach that can combine targeting and toxicity potential. In this study, CPMV nanoparticles were chemically modified with the dye Alexa Flour 488 and were also tandemly modified with PEG1000 followed by AF488; and the derivatized nanoparticles were subsequently added to macrophages stimulated with either LPS (M1) or IL-4 (M2). Previously published studies have shown that M1/M2 macrophages are both present in an inflammatory microenvironment (such as a tumor microenvironment and atherosclerosis) and play opposing yet balancing roles; M2 macrophages have a delayed and progressive onset in the tumor microenvironment (concomitant with an immunosuppression of M1 macrophages). In this study, we show higher uptake of CPMV-AF488 and CPMV-PEG-AF488 by M2 macrophages compared to M1 macrophages. M1 macrophages showed no uptake of CPMV-PEG-AF488. More specifically, M2 macrophages are known to be up regulated in early atherosclerosis plaque. Indeed, previous work showed that M2 macrophages in plaque also correlate with CPMV internalization. These studies emphasize the potential effectiveness of CPMV as a tailored vehicle for targeting tumor macrophages involved in cancer metastasis, or vascular inflammation, and further highlight the potential of CPMV in targeted therapeutics against other diseases.
Keywords: Cowpea mosaic virus (CPMV), macrophages, M1, M2, nanoparticle, internalization, therapeutics
INTRODUCTION
Macrophages play a crucial role in innate and adaptive immunity, and are critical mediators of inflammatory processes.1 Macrophages are highly versatile cells that are involved in a range of functions including phagocytosis of pathogens, antigen presentation, removal of cellular debris and tissue remodeling, induction of immunity, thrombosis and regulation of inflammation. Macrophages are also highly adaptable cells that exhibit pro- and anti-inflammatory properties depending on the disease stage and the signals they receive. Accordingly, macrophages have been broadly classified into “classically activated macrophages” (M1), or “alternatively activated macrophages” (M2).2–4
Both populations of macrophages are present in the inflammatory environments observed in various diseases such as solid tumors, atherosclerosis, and central nervous system (CNS) inflammation. M1s and M2s are thought to be responsible for different functions in these microenvironments.5–8 In tumors for example, M2 macrophages are known to be elevated in the tumor microenvironment and are alternatively referred to as Tumor Associated Macrophages or TAMs. TAMs are notorious for their involvement in cancer metastasis by suppressing tumor immunity and promoting vascularization; thus the ability to target M2s (and/or TAMs) specifically and modify their function would be a great advantage.8–12
Macrophages may be experimentally polarized to M1 or M2 phenotype by treatment with various cytokines (SI Figure 1). Additionally, the M1 and M2 phenotypes are reversible and they can be re-differentiated by reversing the cytokine treatment.1,11,12 M1 are polarized by treatment with IFN-γ, lipopolysaccharide (LPS) and other bacterial products. M1 macrophages up regulate several pro-inflammatory cytokines and chemokines such as TNF-α, IL-12, IL-6, CCL2 and IL-1β; and also increased reactive oxygen species (ROS) and reactive nitrogen intermediates (RNI). M1 macrophages have elevated expression of inducible nitric oxide synthase (iNOS) to metabolize arginine to nitric oxide, ROIs and RNIs to enhance their killing mechanism.
At the other extreme, M2 macrophages are polarized by stimuli such as IL-4, IL-13, IL-10 or glucocorticoid hormones. M2s up regulate scavenger, mannose and galactose receptors (SI Figure 1).2–4 M2 polarization is coupled with secretion of IL-10 and TGF-β, which results in a diminution of pathological inflammation. Release of arginase, proline, polyaminases and TGF-β by the activated M2 cell is tied to wound repair and fibrosis. M2 cells induce arginase 1 to metabolize arginine to ornithine and polyamines and collagen contributing to the production of the extracellular matrix. Interestingly, M2 macrophages also secrete various tumor-associated enzymes such as matrix metalloproteases MMP-2 and MMP-9 involved in matrix degradation, and support tumor growth and angiogenesis by releasing a number of pro-angiogenic cytokines including IL-8 and IL-10 and various growth factors.13 M2s therefore have a range of functions with the capacity to affect diverse aspects of neoplastic tissues including angiogenesis and vascularisation, stroma formation and dissolution, and modulation of tumor growth (enhancement and inhibition).
The plasticity of macrophage populations and their influence on the tumor microenvironment presents an opportunity to develop therapeutics that can target the tumor microenvironment.14–17 Current treatments for cancer typically employ systemic chemotherapies that often have severe adverse effects such as nausea, hair loss, weight loss and immunosuppression. These adverse effects limit the doses that patients can tolerate and the success of treatment. In addition to classical chemotherapeutics (doxorubicin, cisplatin, etc), various ‘molecularly targeted therapeutics’ (such as growth factor receptor inhibitors, anti-angiogenic inhibitors, proteasome inhibitors) have enriched the therapeutic armory with their ability to more selectively interfere with certain cancer signatures.16 However, therapeutic strategies are gaining momentum to influence the tumor microenvironment to be more hostile to tumor cells, and the ability to develop nanotherapeutics to target tumor macrophages to achieve this goal would be an important advance.
A variety of novel nanoparticle platforms are in development for targeted therapeutic applications including dendrimers, nanocrystals, lipids, iron oxides, polymers, quantum dots, and virus and other protein-based particles (termed protein cages).18–30 Viruses are attractive scaffolds for nanoscale constructions because they are predisposed to self-assemble into highly symmetrical structures, are polyvalent, can be produced in large quantities, and are highly stable, biocompatible and bioavailable. Most importantly, the particles are programmable units that can be modified by both genetic modification and chemical bioconjugation methods.
CPMV is a member of the Comoviridae family of viruses, and is composed of 60 copies each of a large and small capsid protein to make a 31 nm diameter icosahedral particle that contains 5 surface lysine residues per capsid (300 total); and thereby displays multivalency which has potential for high cargo loading. Previous work demonstrated that fluorescent-labeled CPMV can be used in intravital imaging of healthy and tumor vasculature. Interestingly, CPMV is readily internalized in a variety of cell types including endothelial cells, fibroblasts, and macrophages, and dendritic cells, as well as in atherosclerotic plaque and CNS inflammatory lesions31. The uptake of CPMV is mediated by specific interaction with a surface-displayed form of the cytoskeletal protein vimentin. Surface vimentin expression has been detected in endothelial cells in vivo as well as in activated macrophages, and the ability of cells to internalize CPMV is correlated with the presence of surface vimentin on these cell types. CPMV also accumulates within the tumor margin in tumor on-plants grown on chick chorioallantoic membrane, a region where macrophages also home during i.v. delivery.32–34 Thus it is important to further investigate the interaction between CPMV and specialized macrophage populations that exist in tumor and other inflammatory microenvironments. Here the association of CPMV with M1 and M2 macrophages was studied using the murine macrophage cell line RAW 264.7 as an in vitro M1/M2 differentiation model. Polarization of the cells to M1 vs. M2 was studied by investigating expression of inducible nitric oxide synthase (iNOS) and Arginase 1. Vimentin expression and CPMV uptake was compared in M1 and M2 macrophages using confocal microscopy and flow cytometry.
MATERIALS AND METHODS
Propagation, extraction and purification of CPMV
CPMV was extracted and purified from infected Cowpea plants as previously described.35,36 Briefly, a homogenate of CPMV-infected leaves in potassium phosphate buffer (pH 7.0) was used as an inoculum for passage to new cowpea plants to produce stocks of virus. CPMV was subsequently extracted and purified from infected secondary leaves, and resuspended in sterile phosphate-buffered saline (PBS). Virus concentration was measured by UV-Vis spectroscopy at 260 nm (ε = 8.1 mL mg−1 cm−1). The integrity of the particles was analyzed on a native agarose gel (1.2%) and evaluated using an ÄKTA™ Superose-6 size-exclusion column (GE Healthcare, Piscataway, NJ) at 0.4 mL/min in 0.1 M potassium phosphate buffer at pH 7.0.
Synthesis and characterization of CPMV-SH, CPMV-AF488 and CPMV-PEG-AF488
The lysine residues on CPMV particles were chemically modified by SATP and deacetylated using hydroxylamine hydrochloride (Thermo Scientific, Rockford, IL) to expose free thiol residues to yield CPMV-SH according to a published procedure.37 CPMV-SH was further reacted with Alexa Fluor® 488 C5 maleimide (Life Technologies, Carlsbad, CA) to yield CPMV-AF488.37 Virus and dye concentration were measured using UV-Vis spectroscopy (εCPMV = 8.1 mL mg−1 cm−1, MWCPMV = 5.6 × 106 g/mol, εdye = 71,000 M−1 cm−1) and the number of dye molecules per CPMV particle was calculated by the molar ratio of dye to CPMV. Average labeling of 172–178 dye molecules per particle was achieved.
For tandem modification of CPMV with dye and PEG, CPMV was first reacted with mPEG-NHS, PEG succinimidyl ester, MW 1000 (NANOCS, Inc., New York, NY) in a 900:1 molar ratio of mPEG-NHS to CPMV (1 mg). The reaction was allowed to proceed for 2 h in a 500 µL solution of 1:4 DMSO and 10 mM Na2PO4 buffer and the resulting CPMV-PEG conjugate was purified via 10 kDa Molecular Weight Cut Off (MWCO) spin columns (Millipore, Billerica, MA). The CPMV-PEG conjugate was further reacted with SATP, hydroxylamine hydrochloride and Alexa Fluor® 488 C5 maleimide as described above to yield CPMV-PEG-AF488 with an average labeling of 80 – 84 dye molecules per PEGylated CPMV particle.
Cell culture and in vitro stimulation of cells with cytokines
RAW 264.7 macrophages were grown in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 100 µg/mL penicillin and 100 µg/mL streptomycin (all from Life Technologies, Carlsbad, CA). After adhering for 5 h after plating, 50 ng/mL of either LPS (Sigma-Aldrich, St. Louis, MO) or IL-4 (Peprotech, Rocky Hill, NJ) was added to the cells for an additional 24 h to polarize RAW 264.7 cells to M1 or M2 macrophages, respectively.
RNA extraction and RT-PCR
RNA was recovered from the cultured cells by the direct addition of Trizol (Life Technologies, Carlsbad, CA) into the well, after aspirating media from each well. Total RNA was subsequently extracted according to the manufacturer’s instructions. Reverse transcription-PCR was carried out using a QIAGEN OneStep RT-PCR Kit (QIAGEN, Inc., Valencia, CA) for gene expression analysis of iNOS and Arginase1(Arg1). Murine iNOS-specific primers sense, (5’-GCCTCATGCCATTGAGTTCATCAACC-3’) and antisense, (5’-GAGCTGTGAATTCCAGAGCCTGAAG-3’) were used to amplify a 372-bp product. iNOS was amplified by using 35 cycles of the following sequential steps: 96°C for 35 sec, 62°C for 2 min, and 72°C for 2 min.38 Murine Arg1-specific primers sense, (5’-CAGAAGAATGGAAGAGTCAG-3’) and antisense, (5’-CAGATATGCAGGGAGTCACC-3’) were used to amplify a 249-bp product. Arg1 was amplified by using 40 cycles of the following sequential steps: 94°C for 30 sec, 45°C for 30 sec, and 72°C for 1 min.39
Gel Electrophoresis
iNOS and Arg1 RT-PCR products, CPMV, CPMV-AF488 and CPMV-PEG-AF488 particles (15 µg per lane) were analyzed by native 1.2% agarose (in 1× TBE buffer) gel electrophoresis in 0.5× TBE running buffer. After separation the gel was analyzed on an Alpha Imager Innotech imaging system.
Phase microscopy
RAW 264.7 cells were seeded at 500,000 cells/well in 6-well plates (BD Falcon, San Jose, CA) and polarized to M1 and M2 macrophages as described above. Cells were then washed in PBS and fixed in 4% electron-microscopy grade formaldehyde in PBS for 15 min at room temperature. After fixing, cells were washed 2× in PBS, stained in hematoxylin for 4 min, washed in water 2× for 5 min, dehydrated in 50% ethanol for 2 min, dehydrated in 75% ethanol for 2 min, counterstained in eosin for 1 min, washed 2× in 75% ethanol for 2 min, dehydrated in 95% ethanol for 2 min, dehydrated 2× in 100% ethanol for 2 min, incubated in xylene 3× for 2 min and mounted with Permount (Fisher Scientific, Hampton, NH). Images were captured at 40× using a Nikon Coolpix 5400 digital camera attached to a Nikon Eclipse TS100 microscope.
Surface and cytosolic vimentin expression in M1 and M2 macrophages
For confocal microscopy, cells were seeded at 250,000 in glass bottom 35-mm dishes (MatTek Corporation, Ashland, MA), and post adhering, were treated with LPS or IL-4 for 24 h for polarization to M1 or M2 macrophages respectively. Cells were washed 3× with PBS and fixed in 3% electron-microscopy grade formaldehyde in PBS for 15 min at room temperature. Cells were then blocked in 5% FBS for 1 h. Vimentin was stained with a polyclonal goat anti-vimentin antibody (V4630, 1:500, Sigma-Aldrich, St. Louis, MO) for 1 h followed by either chicken anti-goat Alexa Fluor 488 conjugated antibody (1:500) or donkey anti-goat Alexa Fluor 647 conjugated antibody (1:500, both from Life Technologies, Carlsbad, CA) for 1 h. Cell nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI) at 1:5000 for 10 min (all done at room temperature). Coverslips were mounted using Aqua Mount (Thermo Scientific, Waltham, MA). Confocal images were obtained at 40× on an Olympus (Center Valley, PA) FV1000 confocal laser scanning microscope. Images were processed using Olympus Confocal Microscope FluoView FV1000 software.
For fluorescence activated cell sorting (FACS), M1 and M2 macrophages were removed from the plates using enzyme-free Hank’s based Cell Dissociation Buffer (Life Technologies, Carlsbad, CA) and re-suspended in sterile PBS at 2.5 × 106 cells/mL and 200 µL of the cell suspension was added to the wells (5 × 105 cells/well) of a V-bottom 96-well plate for FACS analysis. Cells were fixed in 2% electron-microscopy grade formaldehyde in PBS for 15 min at room temperature and resuspended in FACS buffer (PBS supplemented with 1 mM EDTA, 25 mM HEPES pH 7.0 and 1% FBS). Vimentin was stained with the aforementioned primary and secondary antibodies with intermittent wash steps in FACS buffer. Cells were analyzed on a BD Canto (6 Color Analyzer).
CPMV uptake into M1 and M2 macrophages
For confocal microscopy, M1 and M2 cells were plated as described above. CPMV-AF488 and CPMV-PEG-AF488 were added to the cells at 100,000 particles/cell and incubated for 3 h at 37 °C. Cells were then washed, fixed and stained. Cell membranes were stained with Alexa Fluor 555-conjugated wheat germ agglutinin (Life Technologies, Carlsbad, CA) and cell nuclei were stained with DAPI.
For FACS analysis, M1 and M2 cells were plated in V-bottom 96-well plates as described above and incubated with CPMV-AF488 and CPMV-PEG-AF488 (at 100,000 particles/cell) for 3 h at 37 °C and analyzed on a BD Canto (6 Color Analyzer).
Data obtained from FACS was statistically analyzed using GraphPad Prism Software (La Jolla, CA). Data (n = 3) comparing M1 and M2 were analyzed using a unpaired two-tailed t-test with a 5% significance threshold.
RESULTS
Chemical and tandem modification of CPMV with Alexa Fluor 488 and mNHS-PEG
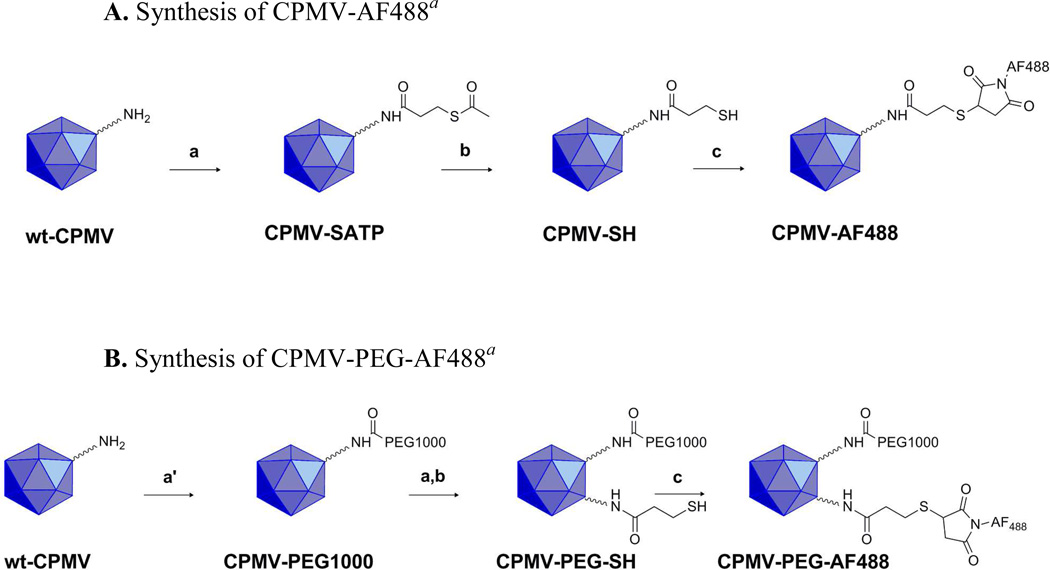
The outer capsid of CPMV is decorated with 300 exposed lysine residues that are amenable to chemical and biological modification.40 After extraction and purification, the ε-amino groups of the exposed lysine residues were reacted with different NHS ester groups as shown in Scheme 1. Scheme 1A highlights the introduction of thiol groups on the surface of CPMV via reaction with N-succinimidyl-S-acetylthiopropionate (SATP) followed by a deacetylation reaction with hydroxylamine hydrochloride to yield CPMV-SH and acetohydroxamic acid (not shown). CPMV-SH is then reacted with a thiol reactive maleimide dye of Alexa Fluor 488 to yield CPMV-AF488. Scheme 1B highlights the tandem modification of CPMV with PEG succinimidyl ester, MW 1000 followed by the SATP, hydroxylamine hydrochloride reaction to yield CPMV-PEG-SH. CPMV-PEG-SH was also reacted with Alexa Fluor® 488 C5 maleimide to yield CPMV-PEG-AF488. The stability and integrity of the dye conjugated particles were analyzed via UV-Vis spectroscopy, SEC-FPLC and native gel electrophoresis. CPMV was labeled with fluorescent dye AF488 to study the direct uptake of CPMV in vitro in M1 and M2 polarized macrophages.
Scheme 1.
A. Synthesis of CPMV-AF488a
B. Synthesis of CPMV-PEG-AF488a
aReagents and conditions: (a) N-succinimidyl-S-acetylthiopropionate (SATP), 1:4 DMSO/10 mM Na2PO4 buffer, pH 7.5, 2 h, room temperature; (a′) 900:1 mPEG-NHS, PEG succinimidyl ester, MW 1000/CPMV (1 mg), 1:4 DMSO/10 mM Na2PO4 buffer, pH 7.5, 2 h, room temperature; (b) 0.5 M NH2OH.HCl, 25 mM EDTA/PBS, pH 7.25, 2 h, room temperature; (c) AF488 maleimide, 1:4 DMSO/10 mM Na2PO4 buffer, pH 7.5, overnight, 4 °C.
Polarizing macrophages in vitro to M1 and M2
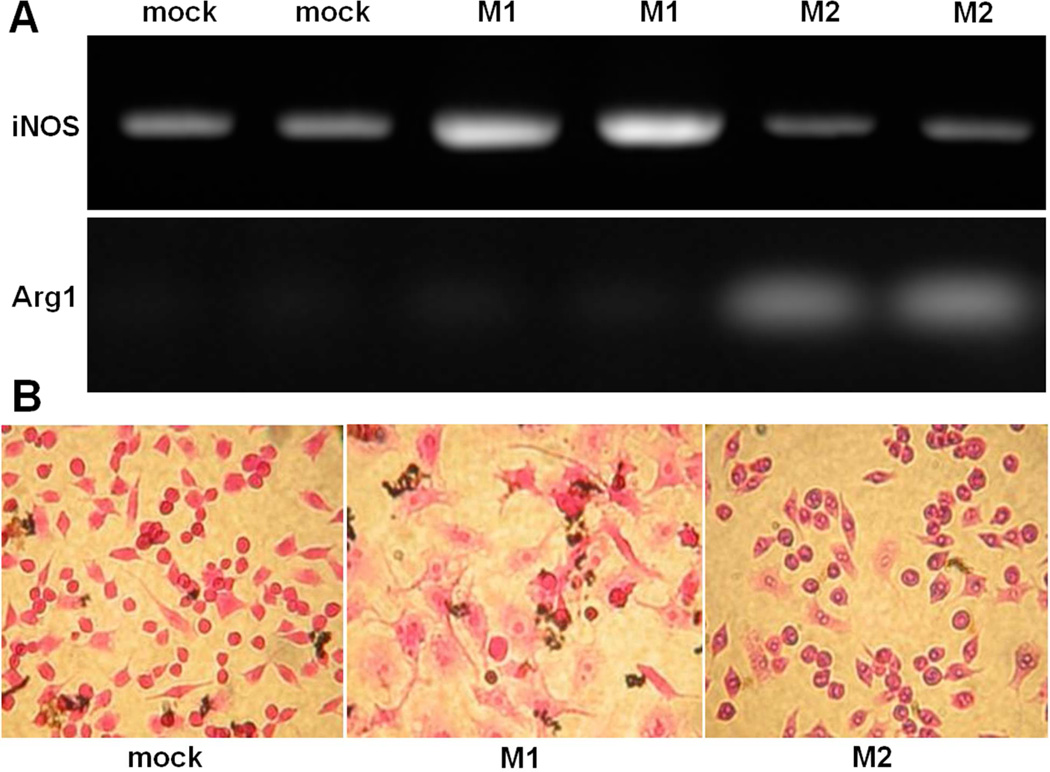
Monocytes recruited from the circulation can be broadly educated by their biological milieu into “classically activated” M1 macrophages or “alternatively activated” M2 macrophages. M1 and M2 polarized macrophages display some distinct features, including specific cytokine signatures (SI Figure 1). In order to mimic the dichotomy of macrophages present in circulation, the murine macrophage cell line RAW 264.7 cells were treated with either 50 ng/mL of LPS or 50 ng/mL of IL-4 to polarize the cells to M1 or M2 macrophages respectively. Untreated RAW 264.7 cells were used for comparison and are referred to as “mock”-treated cells. In addition to the differences between polarizing stimuli and cytokine production in M1 and M2 macrophages, the metabolism of arginine is significantly different between the macrophage subsets. M1 macrophages have elevated levels of iNOS to metabolize arginine to nitric oxide whereas M2 macrophages express arginase 1 to metabolize arginine to ornithine and various polyamines. Figure 1A shows the results of the RT-PCR for iNOS and Arg1, performed on RNA extracted from mock, LPS treated and IL-4 treated macrophages, which confirms polarization of M1 and M2 macrophages. iNOS levels are notably elevated in M1 macrophages compared to mock and M2s and further, expression of Arg1 is only present in the lanes for M2. These genotypic differences observed between M1 and M2 macrophages are accompanied by phenotypic ones as well. The phase microscopy images shown in Figure 1B are evidence of larger and more granular M1 cells compared to M2 and mock cells. The M2 cells were smaller and more proliferative than M1 cells (M2 cell count was consistently 1.5 – 3.5-fold more than the M1 cell count, despite plating the same number of cells before cytokine treatment,) and M2 cells were more multinucleated than M1 or mock cells.
Figure 1.
(A) Assessment of iNOS (top) and Arg1 (bottom) expression. RAW 264.7 cells were stimulated for 24 h using 50 ng/mL of LPS (M1) and 50 ng/mL of IL-4 (M2) in comparison to untreated RAW 264.7 murine macrophage cell line (mock). The expected RT-PCR products of miNOS and mArg1 are 372 bp and 249 bp, respectively. (B) Phase contrast microscopy images (stained with hematoxylin and eosin) showing differences in granularity and nucleation between M1 and M2 macrophages compared to the unstimulated RAW 264.7 (mock-treated) cells.
Surface vimentin expression is higher in M1 macrophages
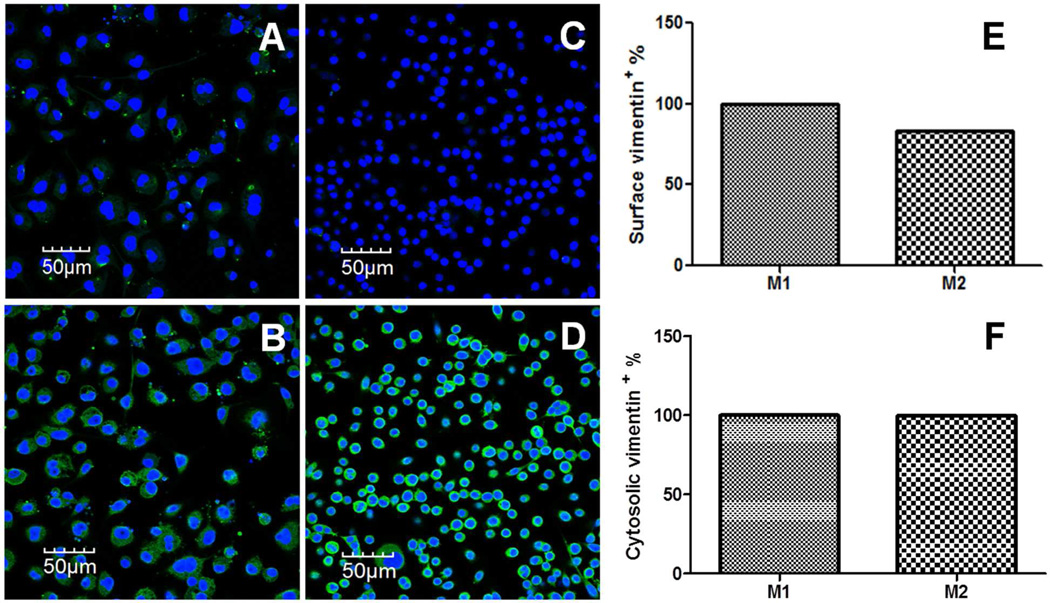
Previous studies have shown that internalization of CPMV by different cell types is mediated by surface vimentin.41,42 Confocal fluorescence microscopy and flow cytometry were used to visualize and quantify the amount of surface and cytosolic vimentin present in M1 and M2 macrophages. As seen in the confocal images in Figure 2, surface vimentin appears as a punctate staining in M1 macrophages (Figure 2A) where as surface vimentin staining in M2 macrophages is negligible (Figure 2C). On the other hand, cytosolic vimentin is broadly expressed in both types of macrophages, with a more diffuse pattern observed in M1s (Figure 2B) and a more perinuclear accumulation in M2s (Figure 2D). The flow cytometry data in Figures 2E and 2F corroborate the vimentin expression seen in confocal images of M1 and M2 cells. Figure 2E shows that surface vimentin is elevated in M1 macrophages compared to M2 macrophages (by 17%) and that the difference in expression is significant (p < 0.001, unpaired two-tailed t test). Figure 2F confirms that the levels of cytosolic vimentin are similar in both types of macrophages.
Figure 2.
Comparing surface and cytosolic vimentin expression in M1 versus M2 macrophages. (A) Confocal image of surface vimentin (green) in M1 macrophages. (B) Confocal image of cytosolic vimentin (green) in M1 macrophages. (C) Confocal image of surface vimentin (green) in M2 macrophages. (D) Confocal image of cytosolic vimentin (green) in M2 macrophages. All confocal images were obtained at 40× and the nucleus was stained with 4′,6-diamidino-2-phenylindole (blue). (E) FACS analysis of surface vimentin in M1 and M2 macrophages (p = 0.001, unpaired two-tailed t test, significant; Each column represents the mean +/− S.D. for triplicate samples, where the error bars represent approximately +/− 0.05%. (F) FACS analysis of surface vimentin in M1 and M2 macrophages (p = 0.31, unpaired two-tailed t test, not significant). Each column represents the mean +/− S.D. for triplicate samples
Enhanced CPMV uptake in M2 macrophages
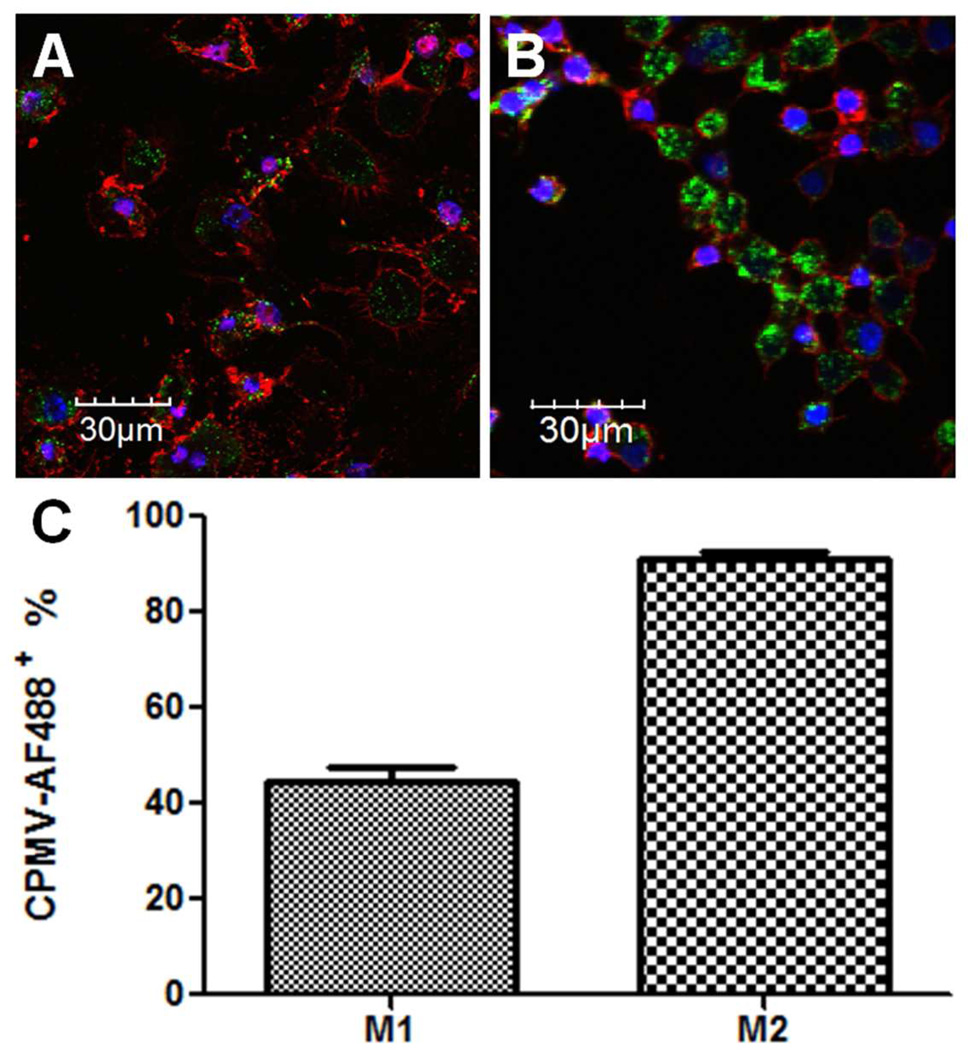
To investigate and correlate surface vimentin with CPMV uptake in M1 and M2 cells, fluorescently labeled CPMV (Scheme 1) was incubated with polarized M1 and M2 macrophages. Figures 3A and 3B illustrate elevated uptake of CPMV in M2 macrophages compared to M1. On the other hand, M1 macrophages displayed a more robust staining of the cellular membrane via Alexa Fluor 555 conjugated wheat germ agglutinin (WGA-555, staining in red), correlating with their changes in membrane morphology (Figure 1). CPMV uptake was further supported by the flow cytometry data in Figure 3C that quantified the amount of fluorescently labeled CPMV internalized by M2 macrophages compared to M1. The 46% increase in M2 uptake of CPMV is significant (p < 0.001, unpaired two-tailed t test).
Figure 3.
Uptake of CPMV-AF488 nanoparticles in M1 versus M2 macrophages. M1 and M2 macrophages were incubated with 100,000 particles/cell for 3 h at 37 °C. (A) Confocal image of CPMV-AF488 (green) uptake in M1 macrophages (B) Confocal image of CPMV-AF488 (green) uptake in M2 macrophages. All confocal images were obtained at 40×, the nucleus was stained with 4′,6-diamidino-2-phenylindole (blue) and the cell membrane with wheat germ agglutinin (red). (C) FACS analysis of CPMV uptake in M1 and M2 macrophages (p < 0.001, unpaired two-tailed t test, significant). Each column represents the mean ± SD of triplicate samples.
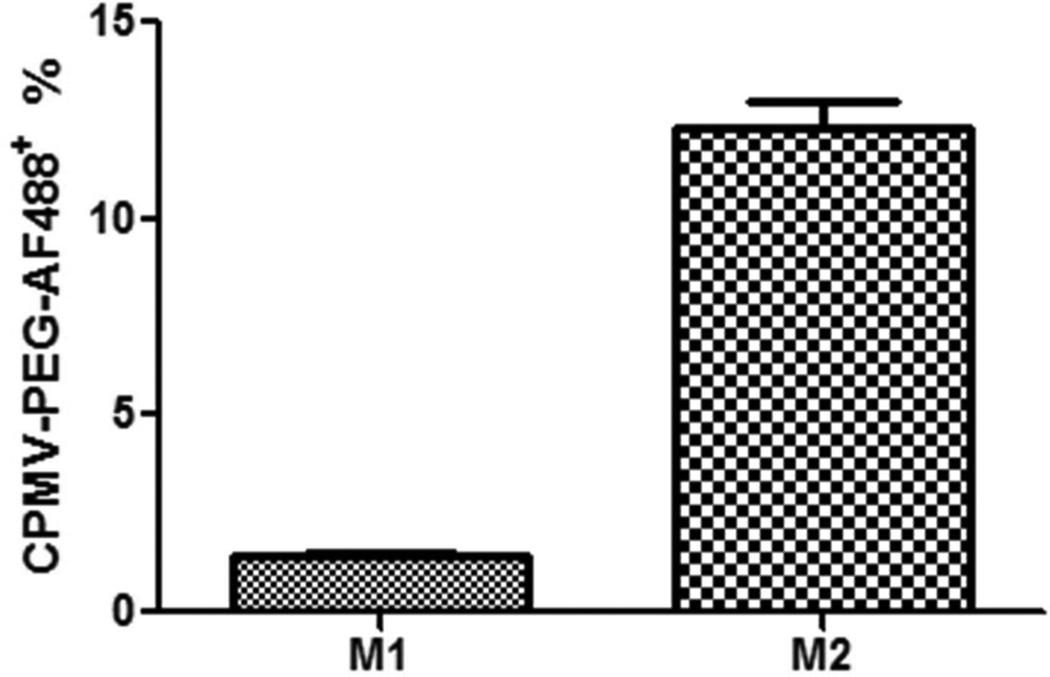
PEGylated CPMV-AF488 (CPMV-PEG-AF488) escapes internalization by M1
PEGylation of CPMV nanoparticles has been previously shown to abrogate its endogenous interaction with vimentin.40,43,44 In order to study the effect of PEGylating CPMV on uptake in M1 and M2 macrophages, CPMV was tandemly labeled with PEG1000 and Alexa Fluor 488 (Scheme 1B). Figure 4 shows the results of flow cytometry data performed on CPMV-PEG-AF488 uptake in M1 and M2 macrophages. Interestingly, M2 macrophages still showed significant internalization of PEGylated CPMV whereas in M1 macrophages the internalization was reduced to background. PEGylating CPMV inhibits interaction with surface vimentin, suggesting that there may be an alternative pathway used in internalizing CPMV in M2 macrophages. This observation is also supported by flow cytometry data shown in SI Figure 2. Quadrant 4 (SI Figure 2B) shows that 33% of the cells are CPMV positive but lack surface vimentin. When looking at the same data for M1 macrophages (SI Figure 2A), all the CPMV positive cells are also vimentin positive.
Figure 4.
Uptake of CPMV-PEG-AF488 nanoparticles in M1 versus M2 macrophages. M1 and M2 macrophages were incubated with 100,000 particles/cell for 3 h at 37 °C. FACS analysis of CPMV-PEG-AF488 uptake in M1 and M2 macrophages (p = 0.001, unpaired two-tailed t test, significant). Each column represents the mean ± SD of triplicate samples.
DISCUSSION
This study reveals that differentiated M1 and M2 macrophage populations show differential uptake of the plant virus nanoparticle Cowpea mosaic virus (CPMV). Previous work has demonstrated that CPMV uptake in different cells correlates with surface vimentin expression41,42 and FACS experiments showed that CPMV uptake in M1 macrophages indeed correlated with surface vimentin expression. Additionally, M1 macrophages showed higher surface vimentin levels than M2 macrophages. However, FACS analysis of CPMV uptake in M2 cells showed that 33% of the cells were positive for CPMV in the absence of surface vimentin, and M2 macrophages internalized more CPMV than M1 macrophages. These studies suggest that an alternative, vimentin-independent CPMV uptake pathway may exist in M2 macrophages.
To investigate the differences in uptake observed for M2 macrophages, PEG polymers were conjugated to the particle surface followed by reaction with the same fluorophore AF488 for consistency, to generate CPMV-PEG-AF488. Previous work has demonstrated that PEGylating CPMV inhibits uptake and the natural interaction with surface vimentin on a variety of cell types.40,43,44 At the particle: cell ratios used, CPMV-PEG-AF488 completely escaped internalization by M1 macrophages, but 12% of these PEGylated nanoparticles were internalized by M2 macrophages. These data confirm our hypothesis that CPMV is internalized by M1s via the characterized vimentin-dependent route only, whereas M2s likely internalize CPMV via a vimentin-dependent as well as a vimentin-independent route. M2-specific cell surface receptors may account for the alternative uptake of CPMV, and future studies will focus on identifying these specific receptors, following procedures previously established in the lab.41,42
It will be interesting to identify the additional mechanisms of CPMV internalization in M2s, as such pathways may be useful for specifically targeting this class of macrophages. M2s have the ability to affect diverse aspects of neoplastic tissues including angiogenesis and vascularisation, stroma formation and dissolution, leading to tumor enhancement and metastasis. Since M2 macrophages are elevated in the tumor microenvironment and influence tumor immunity and vascularization, the ability of CPMV to target them specifically would be a great advantage. These observations also inform the use of PEGylated CPMV with appended ligands as targeted therapeutics. Indeed, the PEGylated nanoparticle (CPMV-PEG-AF488) was not taken up by M1 macrophages but was internalized by M2 macrophages. Recent studies showed that tumor penetration by PEGylated CPMV was enhanced compared to unmodified CPMV, which may involve the M2 population.45 Together these studies provide evidence that CPMV is being internalized via a vimentin independent pathway in M2 cells, which warrants further investigation.
Although CPMV only replicates in plants, the plant virus induces immunogenicity in mammals. In fact, this has led to the successful development of a polyclonal antibody against CPMV which allows for the detection of wild type CPMV both in vitro and in vivo. Even though repeated administration of CPMV can lead to particle accumulation in the reticuloendothelial system, the bioavailability can be improved by PEGylating the nanoparticle, which increases solubility, retention and specific uptake by M2 macrophages (vide supra).
Recent studies from our lab demonstrated that CPMV is internalized in atherosclerotic lesions in mice, noting more CPMV accumulation during the early stages of atherosclerosis than the later stage.31 Atherosclerotic lesions harbor large numbers of macrophages, which migrate to the lesion and differentiate further to foam cells that internalize lipid. Using a similar model of atherosclerosis in mice, Jamila and co-workers showed that M2 macrophages are the primary macrophage type present in early lesions, with a later rise in the number of M1 macrophages.5 This is further corroborated by studies performed by Paula and co-workers in human macrophages incubated with oxLDL that show that macrophages assume an inflammatory phenotype with progression of atherosclerotic lesions.46 Taken together one can infer from these data that the higher uptake of CPMV in early atherosclerotic lesions may be attributed to the higher population of M2 macrophages and the decline in uptake of CPMV in late atherosclerotic lesions may be attributed to the surge in M1 macrophages.
Studies of unpolarized RAW 264.7 cells and HeLa cells showed that CPMV uptake occurred via a combination of macropinocytosis and caveolar endocytosis.47 The current study suggests that the mechanism of uptake in M1 cells is likely to be similar to that observed in RAW 264.7 cells. M2s are considered to be more phagocytic than M1 cells, though preliminary phagocytosis experiments using FITC-dextran (data not shown) performed on both types of macrophages did not reveal significant differences. In summary, the different macrophage populations M1 and M2 play opposing yet balancing roles in the inflammatory microenvironments of tumors and atherosclerotic plaque. In this study, we demonstrated that CPMV internalization is enhanced by M2 macrophages. Recent work from our lab suggested the use of CPMV as a means of detecting early versus late atherosclerotic lesions.31 In a tumor setting, CPMV could be used not only as an imaging molecule to diagnose tumor progression but also as a multivalent vehicle to deliver a cargo of appended therapeutics to selectively kill off M2 cells or re-educate the microenvironment towards the production of M1 cells to stimulate anti-tumor immunity.
CONCLUSION
The goal of this project was to develop virus-based nanoparticles for targeted therapy against macrophages in the inflammatory microenvironment. Current therapies against various cancers are associated with several adverse effects. There is accumulating evidence that associates TAMs with immunosuppression of the tumor microenvironment and stimulation of angiogenesis in progressively growing solid tumors. The multi-valency and robustness of CPMV can be exploited to attach homing peptides to target specific cell types, especially macrophages. Future studies to characterize the macrophage specificity of natural and targeted interactions of CPMV will establish mechanisms to control the targeting and polarization of TAMs that will in turn enhance the efficacy of conventional chemotherapeutic strategies. In the past decade, the natural interaction between CPMV and vascular and inflammatory cells has shown immense promise for targeted therapeutics. The results obtained in this study clearly highlight the differential uptake of CPMV by M1 and M2 macrophages, and suggest that these differences in uptake of CPMV can also be exploited for targeted therapeutics against the different macrophage populations in the inflammatory microenvironment.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by NIH R01CA112075 (M.M.), AHA 12GRNT12040467 (M.M.) and the UCSD Skaggs School of Pharmacy and Pharmaceutical Sciences. The authors would also like to acknowledge the support of the Flow Cytometry Core at the UC San Diego Center for AIDS Research (AI36214), the VA San Diego Health Care System, and the San Diego Veterans Medical Research Foundation; and the support of the UCSD Neuroscience Microscopy Core Facility funded by NIH P30 NS047101.
Footnotes
SUPPORTING INFORMATION AVAILABLE
The Supporting Information contains a schematic describing the polarization of M1 and M2 macrophages, and additional FACS data showing the enhanced uptake of CPMV in M2 macrophages. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Porcheray F, Viaud S, Rimaniol A-C, Léne C, Samah B, Dereuddre-Bosquet N, Dormont D, Gras G. Clin. Exp. Immunol. 2005;142:481–489. doi: 10.1111/j.1365-2249.2005.02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantovani A. Blood. 2006;108:408–409. [Google Scholar]
- 3.Mantovani A, Sica A, Locati M. Eur. J. Immunol. 2007;37:14–16. doi: 10.1002/eji.200636910. [DOI] [PubMed] [Google Scholar]
- 4.Mosser DM, Edwards JP. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khallou-Laschet J, Varthaman A, Fornasa G, Compain C, Gaston AT, Clement M, Dussiot M, Levillain O, Graff-Dubois S, Nicoletti A, Caligiuri G. PLoS One. 2010;5:e8852. doi: 10.1371/journal.pone.0008852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David S, Kroner A. Nat. Rev. Neurosci. 2011;12:388–399. doi: 10.1038/nrn3053. [DOI] [PubMed] [Google Scholar]
- 7.Tabas I. Nat. Rev. Immunol. 2010;10:36–46. doi: 10.1038/nri2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, Rimoldi M, Biswas SK, Allavena P, Mantovani A. Semin. Cancer Biol. 2008;18:349–355. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Allavena P, Sica A, Solinas G, Porta C, Mantovani A. Crit. Rev. Oncol. Hematol. 2008;66:1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Qian BZ, Pollard JW. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelegrin P, Surprenant A. EMBO J. 2009;28:2114–2127. doi: 10.1038/emboj.2009.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sica A, Mantovani A. J. Clin. Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coussens LM, Tinkle CL, Hanahan D, Werb Z. Cell. 2000;103:481–490. doi: 10.1016/s0092-8674(00)00139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farokhzad OC, Langer R. Adv Drug Deliv Rev. 2006;58:1456–1459. doi: 10.1016/j.addr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Farokhzad OC, Langer R. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 16.Lammers T, Hennink WE, Storm G. Br. J. Cancer. 2008;99:392–397. doi: 10.1038/sj.bjc.6604483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maham A, Tang Z, Wu H, Wang J, Lin Y. Small. Vol. 5. Germany: Weinheim an der Bergstrasse; 2009. pp. 1706–1721. [DOI] [PubMed] [Google Scholar]
- 18.Akerman ME, Chan WCW, Laakkonen P, Bhatia SN, Rouslahti E. Proc Natl Acad Sci USA. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portney NG, Singh K, Chaudhary S, Destito G, Schneemann A, Manchester M, Ozkan M. Langmuir. 2005;21:2098–2103. doi: 10.1021/la047525f. [DOI] [PubMed] [Google Scholar]
- 20.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Nat. Med. 2004;10:993–998. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan DC, Ferrari M. Mol. Imaging. 2004;3:364–369. doi: 10.1162/15353500200404139. [DOI] [PubMed] [Google Scholar]
- 22.Feng SS, Mu L, Win KY, Huang G. Curr. Med. Chem. 2004;11:413–424. doi: 10.2174/0929867043455909. [DOI] [PubMed] [Google Scholar]
- 23.Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 24.Guccione S, Li KC, Bednarski MD. Methods Enzymol. 2004;386:219–236. doi: 10.1016/S0076-6879(04)86010-5. [DOI] [PubMed] [Google Scholar]
- 25.Guccione S, Li KC, Bednarski MD. IEEE Eng. Med. Biol. Mag. 2004;23:50–56. doi: 10.1109/memb.2004.1360408. [DOI] [PubMed] [Google Scholar]
- 26.Quintana A, Raczka E, Piehler L, Lee I, Myc A, Majoros I, Patri AK, Thomas T, Mulé J, Baker JRJ. Pharm. Res. 2002;19:1310–1316. doi: 10.1023/a:1020398624602. [DOI] [PubMed] [Google Scholar]
- 27.Bawarski WE, Chidlowsky E, Bharali DJ, Mousa SA. Nanomedicine. 4:273–282. doi: 10.1016/j.nano.2008.06.002. 23008. [DOI] [PubMed] [Google Scholar]
- 28.Majoros IJ, Williams CR, Baker JRJ. Curr. Top. Med. Chem. 2008;8:1165–1179. doi: 10.2174/156802608785849049. [DOI] [PubMed] [Google Scholar]
- 29.Manchester M, Singh P. Adv Drug Deliv Rev. 2006;58:1505–1522. doi: 10.1016/j.addr.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Soussan E, Cassel S, Blanzat M, Rico-Lattes I. Angew Chem. Int. Ed. Engl. 2009;48:274–288. doi: 10.1002/anie.200802453. [DOI] [PubMed] [Google Scholar]
- 31.Plummer EM, Thomas D, Destito G, Shriver LP, Manchester M. Nanomedicine. 2012;7:877–888. doi: 10.2217/nnm.11.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green CE, Liu T, Montel V, Hsiao G, Lester RD, Subramaniam S, Gonias SL, Klemke RL. PLoS One. 2009;4:e6713. doi: 10.1371/journal.pone.0006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leong HS, Steinmetz NF, Ablack A, Destito G, Zijlstra A, Stuhlmann H, Manchester M, Lewis JD. Nat. Protoc. 2010;5:1406–1417. doi: 10.1038/nprot.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho CF, Ablack A, Leong HS, Zijlstra A, Lewis JD. J. Vis. Exp. 2011:52. doi: 10.3791/2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rae CS, Khor IW, Wang Q, Destito G, Gonzalez MJ, Singh P, Thomas DM, Estrada MN, Powell E, Finn MG, Manchester M. Virology. 2005;343:224–235. doi: 10.1016/j.virol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Wellink J. Methods Mol. Biol. 1998;81:205–209. doi: 10.1385/0-89603-385-6:205. [DOI] [PubMed] [Google Scholar]
- 37.Steinmetz NF, Evans DJ, Lomonossoff GP. ChemBioChem. 2007;8:1131–1136. doi: 10.1002/cbic.200700126. [DOI] [PubMed] [Google Scholar]
- 38.Jenkins DC, Charles IG, Thomsen LL, Moss DW, Holmes LS, Baylis SA, Rhodes P, Westmore K, Emson PC, Moncada S. Proc Natl Acad Sci U.S.A. 1995;92:4392–4396. doi: 10.1073/pnas.92.10.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair MG, Cochrane DW, Allen JE. Immunol. Lett. 2003;85:173–180. doi: 10.1016/s0165-2478(02)00225-0. [DOI] [PubMed] [Google Scholar]
- 40.Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. Nat. Med. 2006;12:354–360. doi: 10.1038/nm1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koudelka KJ, Destito G, Plummer EM, Trauger SA, Siuzdak G, Manchester M. PLoS One. 2009;5 doi: 10.1371/journal.ppat.1000417. e1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koudelka KJ, Rae CS, Gonzalez MJ, Manchester M. J. Virol. 2007;81:1632–1640. doi: 10.1128/JVI.00960-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Destito G, Yeh R, Rae CS, Finn MG, Manchester M. Chem. Biol. 2007;14:1152–1162. doi: 10.1016/j.chembiol.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinmetz NF, Manchester M. Biomacromolecules. 2009;10:784–792. doi: 10.1021/bm8012742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinmetz NF, Maurer J, Sheng H, Bensussan A, Maricic I, Kumar V, Braciak TA. Cancers. 2011;3:2870–2885. doi: 10.3390/cancers3032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martín-Fuentes P, Civeira F, Recalde D, García-Otín AL, Jarauta E, Marzo I, Cenarro A. J. Immunol. 2007;179:3242–3248. doi: 10.4049/jimmunol.179.5.3242. [DOI] [PubMed] [Google Scholar]
- 47.Plummer EM, Manchester M. Mol. Pharm. 2012 doi: 10.1021/mp300238w. manuscript in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.