Summary
Fluorescence-activated cell sorting (FACS) permits specific biologic parameters of cellular populations to be quantified in a high throughput fashion based on their unique fluorescent properties. Relative quantitation of mitochondrial-localized dyes in human cells using FACS analysis allows sensitive analysis of a variety of mitochondrial parameters including mitochondrial content, mitochondrial membrane potential, and matrix oxidant burden. Here, we describe protocols that utilize FACS analysis of human lymphoblastoid cell lines (LCL) for relative quantitation of mitochondrial-localized fluorescent dye intensity. The specific dyes described include MitoTracker Green FM to assess mitochondrial content, tetramethylrhodamine ethyl ester (TMRE) to assess mitochondrial membrane potential, and MitoSOX Red to assess mitochondrial matrix oxidant burden. Representative results of FACS-based mitochondrial analyses demonstrate the variability of these three basic mitochondrial parameters in LCLs from healthy individuals, as well as the sensitivity of applying FACS analysis of LCLs to study the effects of pharmacologic induction and scavenging of oxidant stress.
Keywords: mitochondria, MitoSOX Red, MitoTracker Green, TMRE, FACS
1. Introduction
Fluorescence-activated cell sorting (FACS) offers a high-throughput means to quantify fluorescent indicators for a variety of cell and tissue applications. It has long been used to analyze a multitude of cellular characteristics ranging from cell size to organelle abundance to specific protein levels (1). The approach first involves timed incubation of cellular suspensions with specific fluorescent dyes. Fluorescent-labeled cellular suspensions are then injected into a FACS-enabled flow cytometer. Following excitation of cells at wavelengths specific to each fluorescent dye, their emitted and scattered light is recorded as they flow individually past a detector. Dye-specific data can be plotted to permit visualization of particular cellular properties. The capacity to quickly and reproducibly generate large amounts of quantifiable data, combined with an adaptability to a wide range of tissue types and fluorescent dyes, have made FACS analysis a widely used method to probe cell biology.
FACS analysis of mitochondrial biology has been utilized in a wide range of cell types (2–3). Mitochondria-targeted fluorescent dyes are commercially available that permit targeted examination of distinct mitochondrial parameters including matrix oxidant burden (4–5), membrane potential (6–7), and mitochondria content (8) in living cells (9). Such fluorescent dyes have been increasingly utilized to interrogate mitochondria-specific biology in both in vitro systems and, more recently, in vivo using microscopic animal models (10–11).
Here, we describe methods for FACS analysis of mitochondria-localized fluorescent dyes in human lymphoblastoid cell lines (LCL). Relative quantitation is performed of mean LCL fluorescence following timed incubation with MitoTracker Green FM to assess mitochondrial content, tetramethylrhodamine ethyl ester (TMRE) to assess mitochondrial membrane potential, and MitoSOX Red to assess mitochondrial matrix oxidant burden (MitoSOX Red). We further describe the effect on relative matrix oxidant burden of antimycin A (AA)-induced mitochondrial oxidant stress, both alone and in combination with an antioxidant, N-acetyl-cysteine (NAC) (12). The methods described can be readily adapted to perform relative quantitation in LCLs of a wide range of drug or toxin effects across a range of mitochondrial parameters.
2. Materials
2.1 Cell culture and treatment
RPMI 1640 Medium: 15% fetal calf serum, 2 mmol/L L-glutamine, 100 U/mL penicillin-streptomycin
Phosphate-buffered saline (PBS) (GIBCO)
Dimethyl sulfoxide (DMSO)
5 mM MitoSOX Red stock solution: Dilute 50 μg of MitoSOX Red with 13μl of 100% DMSO.
10 μM MitoSOX Red working solution: Dilute 4 μl of 5 mM MitoSOX Red with 2 mL of RPMI 1640.
100 μM MitoTracker solution: Dilute 50 μg MitoTracker Green FM stock with 750 μl of 100% DMSO.
4 mM Tetramethylrhodamine ethyl ester perchlorate (TMRE) stock solution: Dissolve 25 mg TMRE with 12.14 mL 100% DMSO.
20μM TMRE working solution: Dilute 4 mM TMRE stock 1:200 in DMSO to make a 20 μM TMRE working solution.
1 mM Antimycin A (AA) stock solution: Dissolve 5.4 mg AA in 10 ml of DMSO. Stored at −20°C.
100 mM N-acetyl-cysteine (NAC) stock solution: Dissolve 163.19 mg of NAC in 10 ml of distilled water, stored at 4°C.
Human lymphoblastoid cell lines (LCL)
Multi-well cell culture plate (3 ml capacity per well)
2.2 Fluorescence-activated cell sorting (FACS) flow cytometry
12 × 75 mm round bottom polystyrene tubes
-
Dual Laser Becton Dickinson Analytical FACS Calibur Flow Cytometer equipped with a 488 nm laser and the following channels:
FL1 (530/30, 560 shortpass (SP))
FL2 (585/42, 640 longpass (LP))
FL3 (670 LP)
3. Methods
3.1 LCL culture plate preparation
Collect 1 × 107 LCLs in a 15 mL conical tube for each drug and dye combination.
Pellet cells by centrifugation at 300 g for 1 min.
Discard supernatant.
Count LCLs.
Resuspend 200,000 cells per 1 mL of RPMI 1640 (see Note 1).
Plate 1 mL resuspended cells in a single well of a 24 cell culture plate.
Plate 4 replicate wells for each desired drug treatment group.
Plate 4 control wells containing untreated cells to determine baseline fluorescence for FACS analysis.
3.2 LCL incubation with Antimycin A (AA) and N-acetyl-cysteine (NAC)
3.3 LCL incubation with MitoTracker Green FM
Add 2 μL of 100 μM MitoTracker Green FM solution to 1 mL cells in the desired wells of the culture plate to achieve a final concentration of 200 nM.
Incubate cells for 20 minutes at 37°C in a 5% CO2 incubator.
Collect medium and cells in a 15 mL conical tube.
Centrifuge cells at 300 g for 1 minute. Remove supernatant.
Wash cells by resuspending them in 1 mL of PBS maintained at 37°C.
Centrifuge cells at 300 g for 1 minute. Remove supernatant.
Resuspend pelleted cells with 0.4 mL of PBS.
Incubate cells first for 10 minutes at 37°C in a 5% CO2 incubator and then for 20 minutes at room temperature (see Note 4).
3.4. LCL incubation with TMRE
Dilute 20 μM TMRE in RPMI 1640 (by adding 2 μL TMRE in 2 mL RPMI 1640) to achieve a final concentration of 20 nM TMRE in RPMI 1640.
Add 2 mL of 20 nM TMRE in RPMI 1640 solution to cells (see Note 1) in the desired wells of the culture plate to achieve a final concentration of 13.3 nM.
Incubate cells for 10 minutes at 37°C in a 5% CO2 incubator.
Collect medium and cells in a 15 mL conical tube.
Centrifuge cells at 300 g for 1 minute. Remove supernatant.
Wash cells by resuspending in 1 mL of PBS maintained at 37°C.
Centrifuge cells at 300 g for 1 min. Remove supernatant.
Repeat wash as detailed in steps 6 through 8.
Resuspend pelleted cells with 0.4 mL of PBS.
Incubate cells first for 10 minutes at 37°C in a 5% CO2 incubator, and then for 20 minutes at room temperature (see Note 4).
3.5. LCL incubation with MitoSOX Red
Add 2 mL of 10 uM MitoSOX Red working solution to cells (see Note 1) in the desired wells of the culture plate to achieve a final concentration of 6.6 μM.
Incubate cells with MitoSOX Red at 37°C for 10 min in a 5% CO2 incubator (see Note 5).
Collect medium and cells in a 15 mL conical tube.
Centrifuge cells at 300 g for 1 minute. Remove supernatant.
Wash cells by resuspending them in 1 mL of PBS maintained at 37°C.
Centrifuge cells at 300 g for 1 minute. Remove supernatant.
Resuspend pelleted cells with 0.4 mL of PBS.
Incubate cells first for 10 minutes at 37°C in a 5% CO2 incubator, followed by 20 minutes at room temperature (see Note 4).
3.6 LCL imaging by fluorescence microscopy
Visualization of cell fluorescence is recommended prior to proceeding with FACS analysis to verify mitochondrial localization of fluorescence signal (Fig. 1). Higher dye concentrations and longer incubation times result in non-specific nuclear labeling, which is most easily visualized in fibroblast cell lines (Fig. 2). Thus, FACS analysis of cells having nuclear fluorescence should be avoided as they would not be informative for mitochondria-specific analyses (see Note 5).
Figure 1. Human LCL fluorescence localization with MitoTracker Green FM or MitoSOX Red.
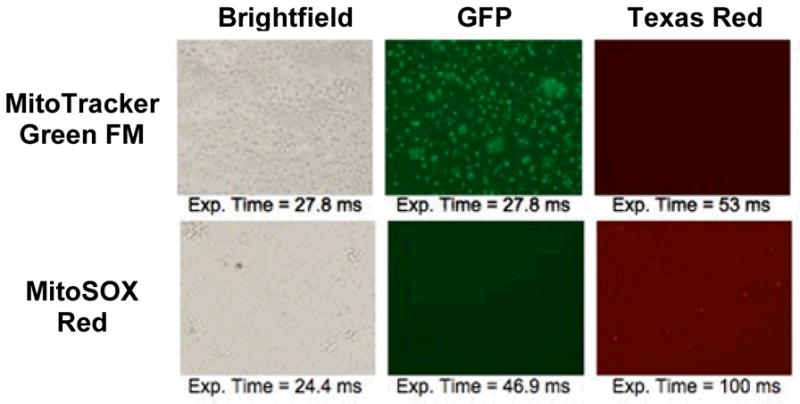
Following incubation with either MitoTracker Green or MitoSOX Red, LCLs were visualized using an Olympus phase microscope under brightfield, a GFP filter, and a Texas Red filter. Cells showed cytoplasmic labeling and nuclear sparing, as was consistent with mitochondrial localization.
Figure 2. Nuclear localization of fluorescent signal occurs following higher dye concentrations and/or longer exposure times.
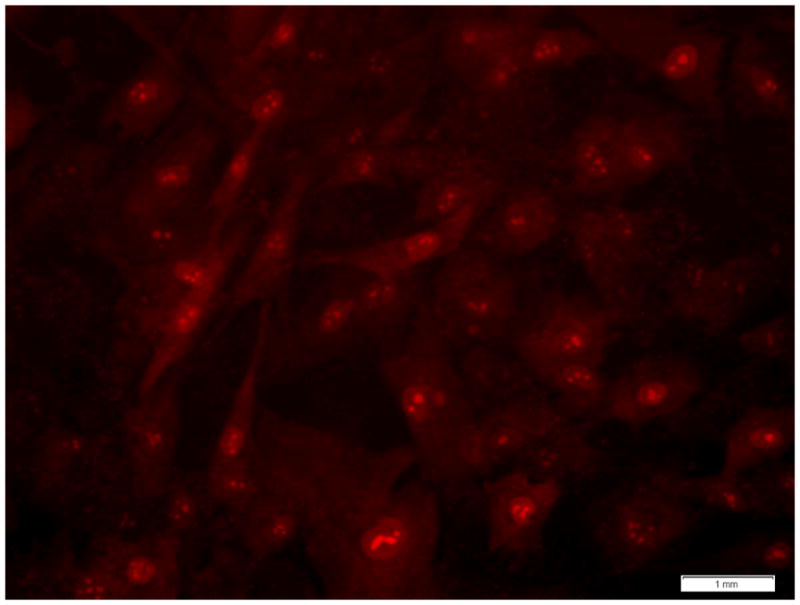
At higher dye concentrations and longer exposure times to MitoSOX Red, fluorescence was observed not only in the cytoplasm but also within the nucleus. This was evident in control fibroblast cell lines incubated with 10 uM MitoSOX Red for 60 minutes.
Transfer an aliquot of washed LCLs following fluorescent dye incubation to a glass slide and cover with a coverslip.
Visualize LCL fluorescence following MitoSOX Red incubation with a Texas Red filter (Excitation: 560/40x, Emission: 630/75).
Visualize LCL fluorescence following MitoTracker Green FM incubation with a FITC/Cy2 filter (excitation: 470/40x, emission: 525/50).
3.7 FACS analysis of LCL fluorescence intensity
Transfer all LCLs that remain after washing for each sample to a 12 × 75 mm round bottom polystyrene tube.
Load cells into a Dual Laser Becton Dickinson Analytical FACS Calibur Flow Cytometer.
Obtain data using the FL1 channel for MitoTracker Green FM, the FL2 channel for TMRE, and the FL3 channel for MitoSOX Red (see Note 6).
Collect a total of 10,000 data events (cells) per sample (see Note 7).
-
Analyze data with desired flow cytometry analysis software. We used Cell Quest (Fig. 3) (BD Biosciences, San Jose, CA) and FlowJo Software (Tree Star Inc., Ashland OR).
Figure 3. Representative histograms of LCL fluorescence data viewed in the histogram format.
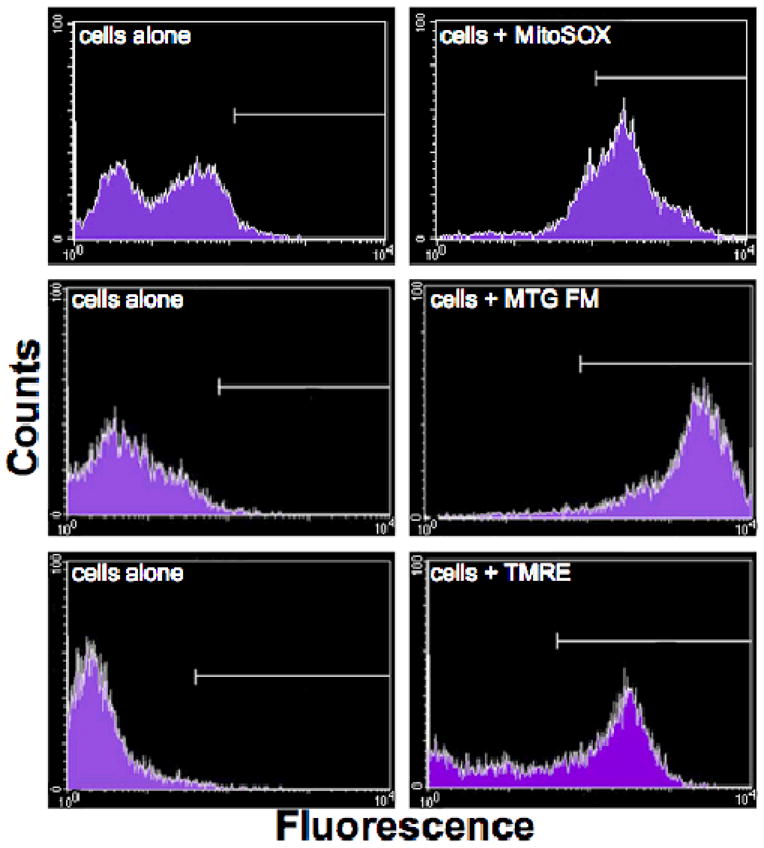
Histograms of counts versus fluorescence intensity show the clear separation in fluorescence signal between untreated LCLs and LCLs treated individually with MitoSOX Red, Mitotracker Green FM (MTG FM), or TMRE. Most control cells, which were not treated with a fluorescent dye, were excluded from the gated region. Most fluorescent dye-treated cells were captured in the gated region.
Figure 4. Control LCL variation in mitochondria-targeted fluorescence dyes.
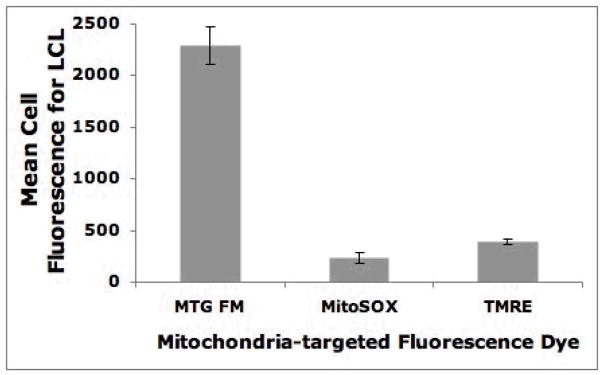
LCLs from four control subjects were labeled with MitoTracker Green FM, MitoSOX Red, or TMRE and analyzed by FACS using the appropriate channel. Bar height and error bars indicate mean and standard deviation of fluorescence intensity for each mitochondria-targeted dye among all four LCLs.
Figure 5. Mitochondrial matrix oxidant burden of MitoSOX labeled LCLs following incubation with antimycin A and/or N-acetyl-cysteine.
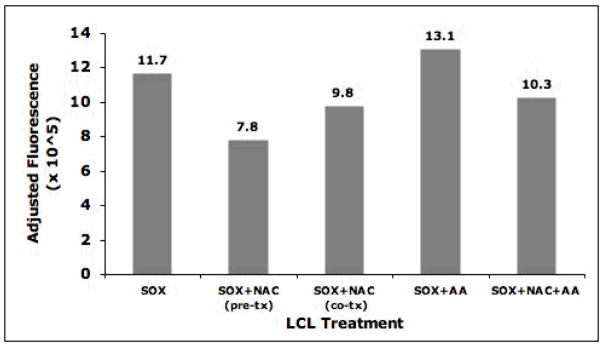
The baseline mitochondrial matrix oxidant burden as indicated by adjusted cell fluorescence (mean MitoSOX Red fluorescence intensity multiplied by number of cells in gated region) of LCLs that were incubated for 10 minutes with 6.6μM (final concentration) MitoSOX Red was decreased with 5 mM NAC treatment for either 1 hour before (pre-incubation) or concurrently with (co-incubation) fluorescence dye exposure. Mitochondrial matrix oxidant burden increased when LCLs were concurrently treated with 5 μM AA at the time of fluorescence dye exposure. However, when LCLs were treated concurrently with both 5 μM AA and 5 mM NAC, MitoSOX fluorescence intensity was reduced to levels below baseline.
Acknowledgments
This work was funded in part by grant from the National Institutes of Health (R03-DK082446) to M.J.F. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- FACS
fluorescence-activated cell sorting
- TMRE
Tetramethylrhodamine ethyl ester
- FM
MitoTracker Green
- LCL
lymphoblastoid cell lines
- NAC
N-acetyl-cysteine
- AA
antimycin A
- DMSO
Dimethyl sufoxide
Footnotes
Cell concentration was always diluted to 200,000 cells per mL so that the volume of dye and/or drug could be kept constant. Each well always contained 200,000 cells in 1 mL for our experiments.
Pre-treatment of control LCLs with AA for 6 or 24 hours prior to incubation with fluorescent dyes resulted in a progressively greater increase in MitoSOX Red fluorescence intensity than did concurrent AA treatment only during fluorescence dye incubation (data not shown).
No difference was seen when cells were incubated with NAC for 1 hour prior to the addition of fluorescent dyes or when NAC incubation was begun concurrently with fluorescent dye incubation (Fig. 5). AA and NAC can be added together to the same culture plate well, if desired (Fig. 5).
LCLs should be incubated at room temperature with each fluorescent dye for 20 to 60 minutes. However, cell death will result if the incubation time exceeds 1 hour.
LCLs labeled with MitoSOX Red should be imaged prior to FACS analysis to confirm diffuse cytoplasmic labeling with nuclear sparing consistent with mitochondrial localization. Cells having bright punctate nuclear labeling should not be used for FACS analysis of mitochondrial-specific parameters.
The FL3 channel can also be used for FACS analysis of LCLs labeled with TMRE. However, we observed better separation of labeled and unlabeled cell populations when analyzing TMRE-labeled cells in the FL2 channel.
Voltage compensation may need to be adjusted to obtain a scatter of data points across the y-axis. However, voltage should not be strong enough to compress treated cells close to the y-axis, which would result in data points being insufficiently separated to distinguish untreated control LCLs from fluorescent-dye treated LCLs.
If the observed data peak is not tight, the number of gated events can be lowered to 90%. However, it is important to keep the percentage of gated events consistent between untreated control LCLs and dye- and/or drug-treated LCLs.
References
- 1.Martinez AO, Vigil A, Vila JC. Flow-cytometric analysis of mitochondria-associated fluorescence in young and old human fibroblasts. Exp Cell Res. 1986;164:551–555. doi: 10.1016/0014-4827(86)90053-4. [DOI] [PubMed] [Google Scholar]
- 2.Yu D, Carroll M, Thomas-Tikhonenko A. p53 status dictates responses of B lymphomas to monotherapy with proteasome inhibitors. Blood. 2007;109:4936–4943. doi: 10.1182/blood-2006-10-050294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Pastor F, Mata-Campuzano M, Alvarez-Rodriguez M, Alvarez M, Anel L, de Paz P. Probes and techniques for sperm evaluation by flow cytometry. Reprod Domest Anim. 2010;45(Suppl 2):67–78. doi: 10.1111/j.1439-0531.2010.01622.x. [DOI] [PubMed] [Google Scholar]
- 4.Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci U S A. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes A, Fernandes E, Lima JL. Fluorescence probes used for detection of reactive oxygen species. J Biochem Biophys Methods. 2005;65:45–80. doi: 10.1016/j.jbbm.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Ward MW. Quantitative analysis of membrane potentials. Methods Mol Biol. 2010;591:335–351. doi: 10.1007/978-1-60761-404-3_20. [DOI] [PubMed] [Google Scholar]
- 7.O’Reilly CM, Fogarty KE, Drummond RM, Tuft RA, Walsh JV., Jr Quantitative analysis of spontaneous mitochondrial depolarizations. Biophys J. 2003;85:3350–3357. doi: 10.1016/S0006-3495(03)74754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Enriquez S, Kai Y, Maldonado E, Currin RT, Lemasters JJ. Roles of mitophagy and the mitochondrial permeability transition in remodeling of cultured rat hepatocytes. Autophagy. 2009;5:1099–1106. doi: 10.4161/auto.5.8.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkuri KR, Cowan TM, Kwan T, Ng A, Herzenberg LA, Enns GM. Inherited disorders affecting mitochondrial function are associated with glutathione deficiency and hypocitrullinemia. Proc Natl Acad Sci U S A. 2009;106:3941–3945. doi: 10.1073/pnas.0813409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingley S, Polyak E, Lightfoot R, Ostrovsky J, Rao M, Greco T, Ischiropoulos H, Falk MJ. Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion. 2010;10:125–136. doi: 10.1016/j.mito.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estes S, Coleman-Hulbert AL, Hicks KA, de Haan G, Martha SR, Knapp JB, Smith SW, Stein KC, Denver DR. Natural variation in life history and aging phenotypes is associated with mitochondrial DNA deletion frequency in Caenorhabditis briggsae. BMC Evol Biol. 2011;11:11. doi: 10.1186/1471-2148-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]


