Abstract
Given limited resources for motility, sperm cell activation must be precisely timed to ensure the greatest likelihood of fertilization. Like those of most species, the sperm of C. elegans become active only after encountering an external signaling molecule. Activation coincides with spermiogenesis, the final step in spermatogenesis, when the spherical spermatid undergoes wholesale reorganization to produce a pseudopod. Here, we describe a gene involved in sperm activation, spe-46. This gene was identified in a suppressor screen of spe-27(it132ts), a sperm-expressed gene whose product functions in the transduction of the spermatid activation signal. While spe-27(it132ts) worms are sterile at 25°C, the spe-46(hc197)I; spe-27(it132ts)IV double mutants regain partial fertility. Single nucleotide polymorphism mapping, whole genome sequencing, and transformation rescue were employed to identify the spe-46 coding sequence. It encodes a protein with seven predicted transmembrane domains but with no other predicted functional domains or homology outside of nematodes. Expression is limited to spermatogenic tissue, and a transcriptional GFP fusion shows expression corresponds with the onset of the pachytene stage of meiosis. The spe-46(hc197) mutation bypasses the need for the activation signal; mutant sperm activate prematurely without an activation signal in males, and mutant males are sterile. In an otherwise wild-type genome, the spe-46(hc197) mutation induces a sperm defective phenotype. In addition to premature activation, spe-46(hc197) sperm exhibit numerous defects including aneuploidy, vacuolization, protruding spikes, and precocious fusion of membranous organelles. Hemizygous worms [spe-46(hc197)/mnDf111] are effectively sterile. Thus, spe-46 appears to be involved in the regulation of spermatid activation during spermiogenesis, with the null phenotype being an absence of functional sperm and hypomorphic phenotypes being premature spermatid activation and numerous sperm cell defects.
Introduction
Sperm activation is an event that requires precise timing. Precocious activation may either result in premature consumption of stored energy or interfere with efficient fertilization. Belated activation may allow competing sperm to gain the advantage in the race to fertilize the ova. For many species, activation coincides with sperm release from the male. For instance, sea urchin sperm activate upon release into water column due to an influx of Na+ from the seawater [1]. The resulting spike in intracellular pH stimulates motility and respiration through a signal transduction pathway that includes Ca2+, cAMP, and cAMP dependent kinases. The outcome is the activation of dynein ATPase in the flagellum and increased mitochondrial activity [2]–[4]. Thus, sea urchin sperm activate precisely when motility is critical, and the cue is from the external environment.
For other species, the cue to activate is a signaling molecule. In mammals, bicarbonate is the activation cue. Bicarbonate not only induces a Ca2+ influx, cAMP accumulation, and cAMP dependent protein kinase A activity, but it also increased motility and respiration [5]–[8]. As a component of the seminal fluid, bicarbonate stimulates sperm activation precisely when the sperm enter the female reproductive tract and require motility to reach the ova and compete with rival sperm. The similarity between sea urchin and mammalian sperm activation suggests that the cellular processes are conserved across distantly related taxa.
In the nematode Caenorhabditis elegans, sperm activation occurs during the final step in sperm development, spermiogenesis. During activation, the spherical spermatids undergo wholesale cytoplasmic reorganization to form a pseudopod, which propels the sperm to the fertilization site. In C. elegans, sperm are produced by both males and hermaphrodites, each type activating at its own precise time point: (i) male sperm at ejaculation, and (ii) hermaphrodite sperm at the time they arrive at the spermatheca. Only recently has an activation signaling molecule been identified. The male spermatids activate within the uterus when exposed to seminal fluid containing the protease TRY-5 [9]. The signaling molecule for hermaphrodite sperm remains elusive. Intracellularly, the events of C. elegans spermatid activation include an influx of cations [10], a brief elevation in pH [11], the release of intracellular Ca2+ [12]–[14], polymerization of MSP, and fusion of the membranous organelles (MOs) with the plasmalemma [12].
While an external signal induces activation in C. elegans sperm, precise control over activation is brought about antagonistically: a set of inhibitory, or “brake,” proteins maintains the spermatid stage until the activation signal relieves the inhibition [15]–[16]. In hermaphrodites, the signaling cascade for activation involves a set of proteins known as the “SPE-8 group”: SPE-8 [17], SPE-12 [18]–[19], SPE-19 [20], SPE-27 [21], and SPE-29 [22]. Male spe-8 group mutants are fertile, but mutant hermaphrodites are self-sterile because their spermatids fail to activate. Notably, the arrested hermaphrodite self-spermatids can be “rescued” when exposed to seminal fluid from males, which, along with other data, suggests that SPE-8 group proteins serve specifically to amplify the activation signal for hermaphrodite self sperm [12] [19].
The brake proteins that prevent activation include SPE-6 [15] and SPE-4 [16]. These proteins have functions earlier in spermatogenesis that center on the fibrous body-membranous organelle complexes (FB-MOs). The FB-MOs act to shuttle the Major Sperm Protein (MSP) through the meiotic divisions. The most abundant protein in sperm, MSP is encoded by numerous paralogs [23], is packaged into the FBs in the primary spermatocyte and released into the cytosol of the spermatid [24]. MSP is polymerized into fibers in the pseudopod of the spermatozoon [25] thereby driving motility through treadmilling of the pseudopod membrane [26]. The SPE-4 protein localizes to the MO membrane, and null spe-4 mutant sperm have fibrous bodies (FBs) that fail to associate normally with the membranous organelles (MOs) to form normal FB-MO complexes [27]–[28]. Null spe-6 mutants fail to package the Major Sperm Protein (MSP) into the FBs [29]. However, at a later developmental stage, current models suggest that SPE-4 and SPE-6 function as brake proteins prior to entry into spermiogenesis [15]–[16]. Certain hypomorphic mutations in spe-4 and spe-6 suppress the sterility of spe-27(it132) mutants. These mutations were recovered in a suppressor screen of spe-27(it132) [17]. These spe-27 suppressor mutations not only restore fertility to other spe-8 group mutants, but they also result in premature spermatid activation. Here, we report the identification of spe-46, another gene isolated in the suppressor screen of spe-27(it132). The spe-46(hc197) mutation causes premature spermatid activation, and results in improper segregation of nuclei during meiotic divisions.
Results and Discussion
Identification of spe-46 Mutation, hc197
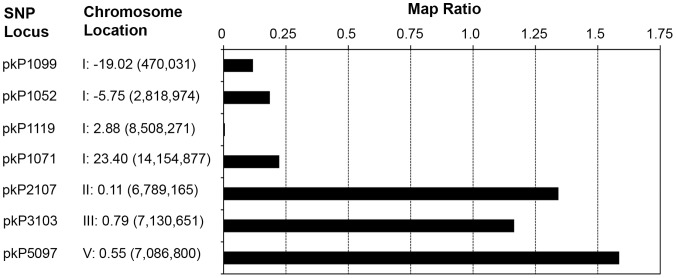
To identify the hc197 mutation, we used whole genome sequencing, which required only a rough estimate of the chromosomal location. Because hc197 segregated independently from the Chromosome IV marker, unc-22, we knew it was on another chromosome. We also ruled out the X chromosome since no sperm-expressed genes are known to reside there [30]. Single nucleotide polymorphism (SNP) mapping as described by Wicks et al. [31] was used to identify the approximate chromosomal location of hc197. Briefly, after crossing our hc197 mutant strain to the polymorphic Hawaiian strain CB4856, pooled mutant and non-mutant F2 were subjected to PCR/RFLP analysis, and map ratios were calculated for the SNP loci examined. The map ratios near 1.0 for SNPs in the middle of Chromosomes II, III, and V indicate hc197 is located elsewhere (Fig. 1). We initially analyzed pkP1119 on Chromosome I and got a map ratio near zero, indicating the SNP was linked to hc197. Our initial results were confirmed by very small map ratios from three more SNP loci across Chromosome I (Fig. 1), but pkP1119 had the smallest map ratio, suggesting it was closest to hc197.
Figure 1. SNP mapping data for hc197 on Chromosome I.
Map ratios near zero indicate linkage, while ratios near 1 indicate independent segregation. The SNP loci and chromosome locations (chromosome number, genetic map position, physical map location) are indicated.
We proceeded to sequence the genome of the hc197; spe-27(it132) unc-22(e66) strain and obtained a read depth of 62 reads per base pair. The entirety of Chromosome I was examined for SNPs that created non-synonymous alterations to coding sequences and that were present in greater than 80% of the reads. At the same time the hc197 mutant strain was sequenced, we also sequenced the genomes of six other spe-27 suppressor mutants isolated in the same screen that produced hc197. The SNPs found in hc197 were compared to those identified in the six other mutant genomes. Shared mutations were omitted from consideration, as they could not be the unique mutation responsible for the hc197 suppressor phenotypes. The remaining nine novel mutations are shown in Table 1. All nine mutations were common transition mutations induced by ethyl methanesulfonate (EMS).
Table 1. Novel1 non-synonymous mutations on Chromosome I.
| Nucleotide Position | AffectedCDS | Genetic location | Mutationtype | AA6change | fem-3/fem-1 2 | Expression3 | Phenotype4 |
| 7678275 | T24B1.1 | 2.23 | C to T | H to Y | – | Hermaphrodite; Sex enriched | – |
| 9057658 | W06D4.2 | 3.45 | C to T | Stop | 35.75 | Sperm enriched | – |
| 9142094 | ZK858.7 | 3.63 | C to T | G to E | 0.28 | Protein expression | Multiple somatic |
| 9635933 | ppfr-1 | 3.76 | C to T | A to T | – | Embryonic | Oocyte meiotic chromosome segregation5 |
| 9655212 | T05F1.11 | 3.83 | C to T | H to Y | 0.95 | Protein expression | Multiple somatic |
| 9689655 | rpl-14 | 3.85 | C to T | S to F | 0.88 | Multiple somatic | Multiple somatic |
| 11393497 | aat-9 | 7.96 | C to T | A to T | 0.54 | Germline; Oocyte; Nervous system | – |
| 11540790 | dys-1 | 9.05 | C to T | G to R | 0.89 | Muscle; Nervous system | Movement defects |
| 12905311 | car-1 | 14.18 | G to A | G to R | 0.27 | Oogenesis; Apoptosis | Germline cell death; Oocyte defects |
Because the hc197 mutation suppresses the phenotype of a sperm-expressed gene, it should be expressed in sperm and have a sperm defective, or sterile, phenotype. Wormbase (www.Wormbase.org) contains a comprehensive collection of such data for C. elegans. In Table 1, the phenotype and expression pattern for each gene identified by Illumina sequencing are presented. Phenotypic data come primarily from RNAi experiments, which have been a powerful resource for rapid gene knockdown [32], although RNAi is ineffective for sperm-expressed genes [33]. Therefore, RNAi experiments should not produce a sperm defective phenotype, but significant non-sperm phenotypes suggest expression partially or solely in somatic tissues. None of the genes presented had a sperm defective phenotype, but many did have somatic phenotypes. The Wormbase expression patterns come primarily from multiple microarray experiments, and we paid close attention to expression patterns related to sperm or the germline. Two genes are involved in oogenesis (aat-9 and car-1), but both also function in non-germline tissues [34]–[35]. Two other genes, T24B1.1 and W06D4.2, were of particular interest. T24B1.1 was annotated as sex-enriched, and W06D4.2 had a sperm-enriched expression pattern. Finally, we examined microarray data comparing transcript abundance in hermaphrodites making only sperm [fem-3(q23)] with hermaphrodites making only oocytes [fem-1(hc17)] [30]. Only W06D4.2 had a ratio indicating sperm expression, although the fem-3/fem-1 ratio for T24B1.1 was not available. The mutations identified in both genes were further confirmed by conventional sequencing. Because both W06D4.2 and T24B1.1 are located near pkP1119, the presumptive locale for hc197, we decided to pursue both genes.
To determine the gene that harbors the hc197 mutation, we performed transformation rescue. PCR amplified wild-type products of both W06D4.2 and T24B1.1 including ∼1Kbp of promoter and ∼1Kbp of downstream sequence were injected into the gonads of hermaphrodites with the hc197 mutation but an otherwise wild-type background. F2 transformants were reared at 25°C from the L4 stage onward and examined for rescue of the fertility defect associated with hc197. Transformants with wild-type T24B1.1 had the same fertility as did control worms, so T24B1.1 failed to rescue the mutant phenotype (Table 2). However, W06D4.2 did result in rescue: transgenic F2 were significantly more fertile than were controls (Table 2). These results confirm that the hc197 mutation resides in W06D4.2, which is henceforth referred to as spe-46. We confirmed the exonic arrangement of spe-46 by sequencing the cDNA derived from RT-PCR. Based upon EST sequences mapping to spe-46 in Wormbase, there is no alternative splicing for this gene. The mutation changes the last codon in the fifth exon from a glutamine to a stop (Fig. 2).
Table 2. Results from transformation rescue experiments.
| T24B1.1 * | W06D4.2 ** | |||||
| Transgenic F2 | Control F2 | Statistical Result | Transgenic F2 | Control F2 | Statistical Result | |
| Mean No. Progeny (N) | 22.0 (14) ±2.96 | 23.4 (13) ±3.91 | t = −0.30 P = 0.7661 | 210.2 (16) ±13.15 | 42.6 (15) ±3.05 | t = 12.431 P = 8×10−10 |
hc197; him-5(e1490) worms were transformed with T24B1.1.
hc197 worms with an otherwise wildtype genomic background were transformed with W06D4.2.
Sample Sizes are Listed in Parentheses, and Error is Shown as ± SEM
Figure 2. The gene structure of W06D4.2.
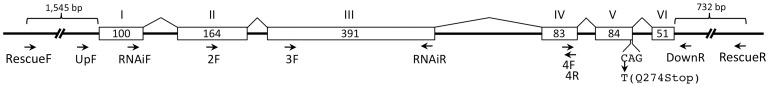
Boxes represent individual exons (I-VI). The hc197 mutation (C −>T) converts a glutamine residue to a premature stop. Horizontal arrows refer to oligonucleotide sequences. RescueF and RescueR were used to amplify the rescuing 3,626 bp fragment. RNAiF and RNAiR provided the RT-PCR amplicon (585 bp) that was used to construct an RNAi feeding clone. The UpF and DownR primers were used to amplify the cDNA sequence, which was confirmed by conventional DNA sequencing employing the remaining primers.
spe-46 Encodes a Predicted Membrane Protein with Sperm-specific Expression
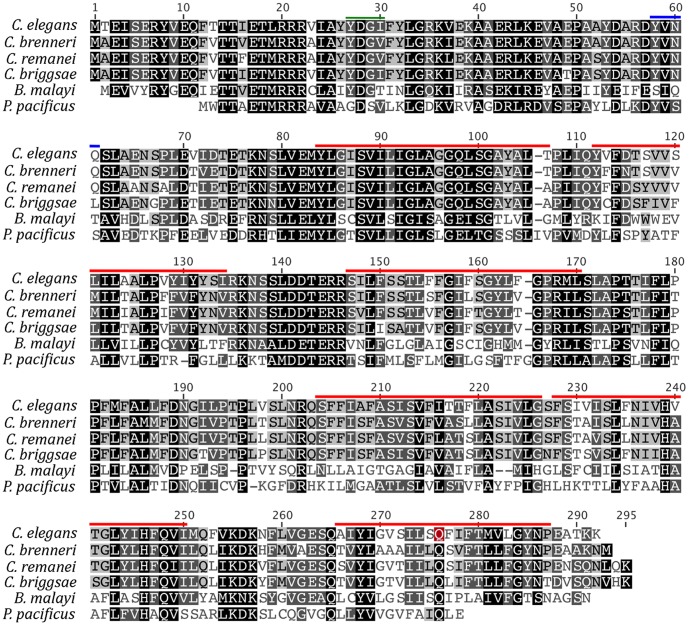
Sequence analysis with the ELM resource [36] suggests that the SPE-46 protein has six putative transmembrane domains and a Tyr-based sorting signal important in trafficking proteins to cellular compartments [37] (Fig. 3). Interestingly, both the ELM resource and the NetPhos 2.0 Server [38] predict a Tyr phosphorylation site at the same position the ELM predicts an SH2 binding domain (AA position 58–61; Fig. 3). Hypothetically, this site could interact with SPE-8 protein, a kinase with an SH2 domain and which transduces the signal to activate [17]. The NetPhos 2.0 Server also predicts numerous other Ser and Thr phosphorylation sites on the SPE-46. The premature stop caused by hc197 falls within the final transmembrane domain. SPE-46 appears exclusive to nematodes, as homologs were not found in other taxa, although the nematode sequences are highly conserved (Fig. 3).
Figure 3. An alignment of the C. elegans SPE-46 protein sequence with those identified as homologs in other nematode species.
The Geneious Alignment option with Blosum62 cost matrix generated the alignment. Red bars represent predicted transmembrane domains. The green bar represents a predicted Tyr-based sorting signal, and the blue bar represents both a predicted Tyr phosphorylation site and a predicted SH2 binding domain. The Q residue highlighted in red is transformed into a stop codon by the hc197 mutation.
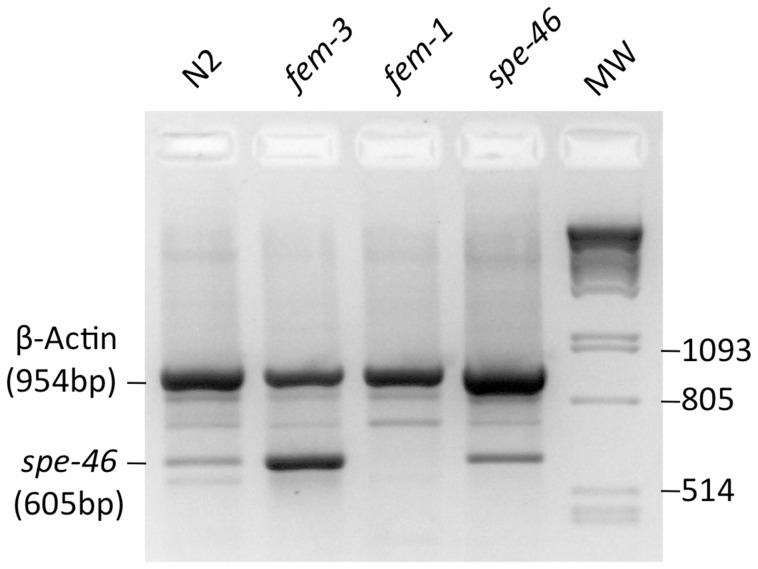
While microarray studies suggest that spe-46 is highly sperm-specific, we sought to test this conclusion by RT-PCR. Our results show that the spe-46 transcript is only present in animals making sperm cells [fem-3(q23) mutants] and not in animals making only oocytes [fem-1(hc17) mutants] (Fig. 4). The spe-46 product was amplified using primers 2F and 4R (Fig. 2), and the expression in spe-46 mutants was similar to that from wild-type worms, suggesting that the transcripts with the premature stop codon appear stable.
Figure 4. Products of RT-PCR reactions using RNA from wild-type (N2) worms, hermaphrodites that produce only sperm [fem-3(q23)], hermaphrodties that produce only oocytes [fem-1(hc17)], and spe-46(hc197) mutant hermaphrodites.
The reactions were multiplexed with primers for both the spe-46 transcript and the transcript for the C. elegans β-Actin homolog act-2. The molecular weight marker (MW) is lambda phage cut with PstI. While non-specific products were amplified, the specific amplicons for both spe-46 and act-2 were present. In particular, the spe-46 product is present in all lanes but fem-1, a strain that makes no sperm. The size of each specific amplicon is indicated.
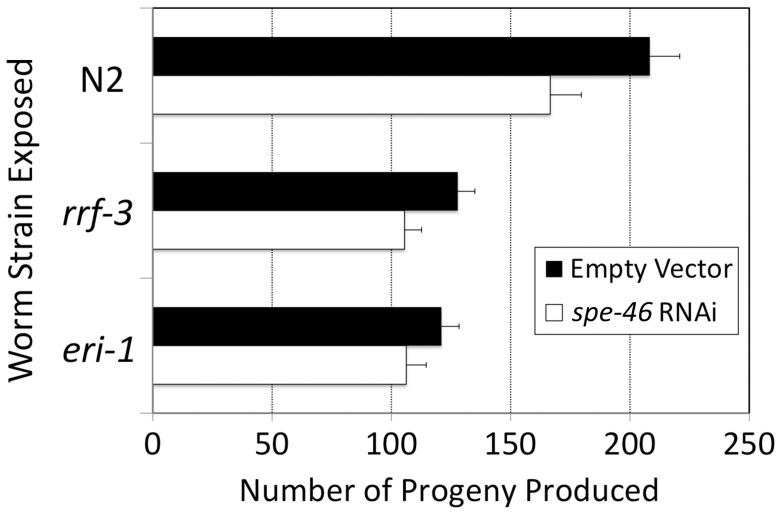
The sperm specificity of spe-46 expression was also tested by looking for somatic defects in response to RNA interference (RNAi) for spe-46. We constructed an RNAi feeding strain of E. coli HT115 expressing double stranded RNA corresponding to 585 bp of the cDNA (amplified with primers RNAiF and RNAiR – see Fig. 2). In addition to wild type strain N2, the RNAi sensitive strains rrf-3(pk1426)II and eri-1(mg366)IV were exposed to spe-46 RNAi. In no case did we observe evidence of somatic phenotypic defects such as dumpy, uncoordinated, slow growth, etc. The exposed worms appeared somatically normal, especially compared to control worms exposed to bacteria containing the RNAi empty vector. The only difference was that RNAi exposed worms produced slightly but significantly fewer progeny than did controls (Fig. 5; F1.77 = 10.554; P = 0.002). Such a result cannot necessarily be interpreted as a spe-46 specific sperm defect because sperm-expressed genes in C. elegans are typically resistant to RNAi [33]. Alternatively, it is possible that the reduction in fertility is due to a non-specific effect of titrating away limited component(s) of the RNAi machinery, which are active during spermatogenesis [39].
Figure 5. The effect of spe-46 RNAi on lifetime fecundity.
Worms were exposed to transformed E. coli strain HT115 expressing dsRNA corresponding to the spe-46 transcript (spe-46 RNAi) or to control bacteria transformed with only the RNAi vector, L4440 (empty vector). In addition to wild-type strain N2, the strains rrf-3 and eri-1 were utilized due to their increased sensitivity to RNAi. Error bars represent the SEM.
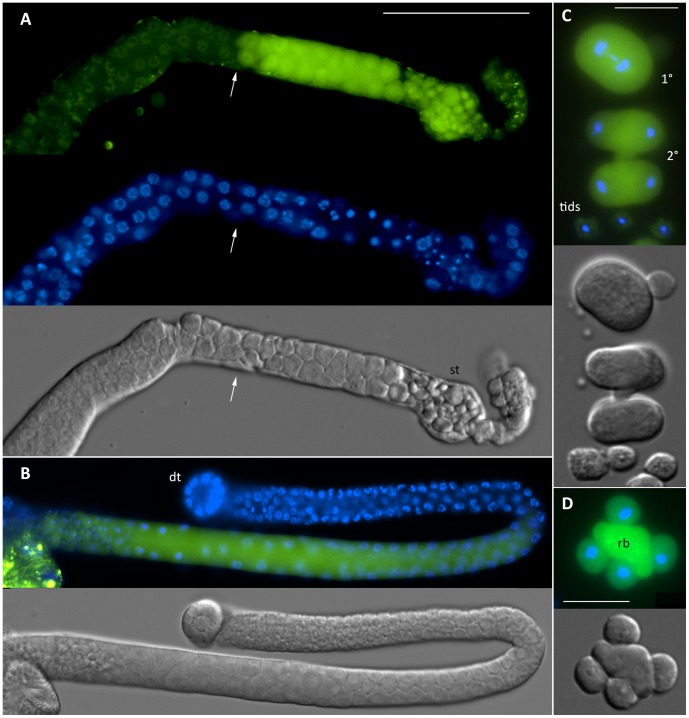
The expression of SPE-46 was investigated using a transcriptional GFP fusion. Our reporter construct included 1 Kbp of genomic sequence on either side of the spe-46 coding sequence with GFP replacing the spe-46 coding sequence. Green fluorescence was localized to the spermatogenic tissue (Fig. 6). In gonads from hermaphrodite L4 larvae, GFP is present in a very specific population of cells, likely those that will produce the sperm (Fig. 6A). In the male gonad, GFP labeling begins at approximately mid to late pachytene stage, and it is present through the appearance of the sperm (Fig. 6B). Both primary and secondary spermatocytes contain GFP, as do the sperm (Fig. 6C, D). Unfortunately, we were unsuccessful in out attempts to determine the subcellular localization of SPE-46 via construction of a translational GFP fusion. We are left to conclude that spe-46 expression occurs in the spermatogenic tissue beginning with entry into the meiotic pachytene phase.
Figure 6. Expression of the spe-46:GFP reporter.
Here, the worms expressed an integrated GFP flanked by the spe-46 promoter and 3′ UTR, and the tissue was labeled with the DNA dye Hoechst 33342. (A) Fluorescence was found in a specific set of cells in the gonads of L4 hermaphrodites, likely those that will produce sperm. The arrow provides reference for the region where GFP expression begins. Gamete maturation proceeds to the right, with the sperm taking up residence in the spermatheca (st). (B) In male gonads, GFP expression corresponds with the transition zone and entry into meiotic pachytene (approximately at the bend in the gonad). dt refers to the distal tip of the gonad. (C) The primary (1°) and secondary (2°) spermatocytes, and spermatids (tids) are shown. (D) The GFP was detected in the residual body (rb) during the second meiotic division. The scale bar in A is 50 µm and is the same reference for B. The bars in C and D represent 10 µm.
spe-46(hc197) is a Recessive Suppressor of the Spermiogenesis Activation Pathway and has a Conditional Spermatogenesis-defective Phenotype on its Own
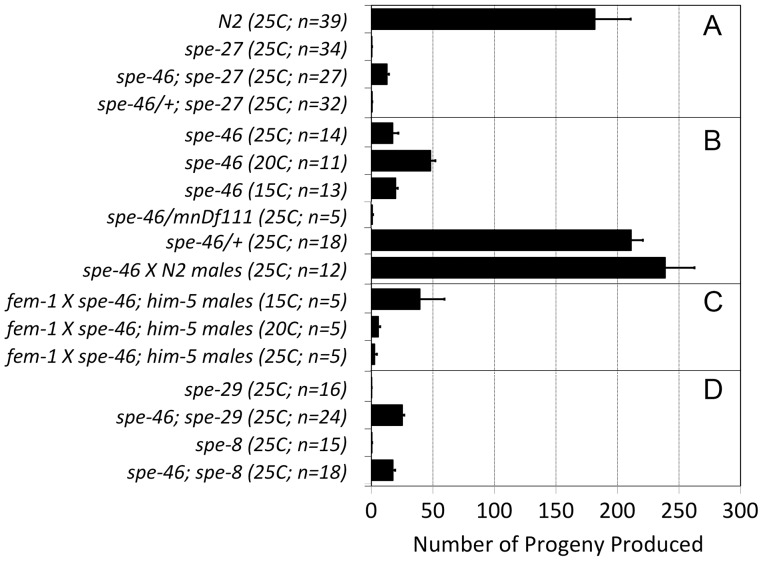
Worms homozygous for spe-27(it132ts) are sterile at 25°C, unless they are also homozygous for spe-46(hc197) (Fig. 7). This suppression of spe-27 sterility is a recessive phenotype, as spe-46(hc197)/+ are sterile in a spe-27 background. It was previously shown that the spe-4(hc196) and spe-6(hc163) suppressor alleles restore fertility not only to spe-27 mutants but also to mutants of the other spe-8 group genes [15]–[16]. Given this fact, we tested the ability of spe-46(hc197) to suppress the sterility of mutations in the spermiogenesis genes spe-8, and spe-29. In both cases, spe-46(hc197) restored fertility to the other spermiogenesis gene mutants (Fig. 7). Therefore, the suppression phenotype of spe-46(hc197) is neither allele specific nor gene specific, suggesting that it bypasses the spermiogenesis pathway in a manner similar to spe-4 and spe-6 suppressor alleles.
Figure 7. The number of progeny associated with the spe-46(hc197) mutation.
(A) Hermaphrodite self-progeny showing that spe-46(hc197) is a recessive suppressor of spe-27(it132) sterility. (B) In an otherwise wild-type background, selfing spe-46 hermaphrodites exhibit a conditional spermatogenesis defect that is rescued by mating with N2 males. Further, the spe-46 defect is recessive. (C) Male spe-46 mutant worms display a temperature-sensitive sperm defect when mating with spermless fem-1(hc17ts) hermaphrodites, being most fertile at 15°C. Finally, (D) the spe-46(hc197) mutation suppresses mutations in both spe-8 and in spe-29, restoring partial hermaphrodite self fertility. Error bars represent SEM.
Further evidence supporting a bypass hypothesis is that spe-46(hc197) causes premature spermatid activation. Virgin male him-5(e1490) worms contained only inactive spermatids when dissected into SM1 buffer, whereas nearly 30 percent of the sperm within spe-46(hc197); him-5(e1490) mutant males were active spermatozoa under the same conditions (see below). The existence of spermatozoa within virgin males shows that they activated prematurely, and they are likely the source of male infertility because crawling spermatozoa obstruct sperm transfer during copulation [40]. These results suggest that the SPE-46 protein inhibits spermiogenesis until the activation signal is received by the spermatid. Including SPE-4 [16] and SPE-6 [15], the addition of SPE-46 makes three proteins that inhibit spermiogenesis.
In an otherwise wild-type background, spe-46(hc197) worms have reduced fertility that is influenced by temperature: they produce more progeny at 20°C than they do at 25°C or 15°C. This fertility deficit is due to a sperm defect, because the fertility of spe-46(hc197) hermaphrodites increases after wild type males supply sperm at mating (Fig. 7). Furthermore, the spe-46(hc197) phenotype is recessive; spe-46(hc197)/+ worms have wild-type fertility. The fertility deficit is also evidenced in males. When paired with spermless fem-1(hc17) hermaphrodites at 25°C, male spe-46(hc197); him-5(e1490) sired very few offspring (Fig. 7), but the number of cross progeny increased with lowered temperature (Fig. 7), indicating that the male phenotype is also temperature sensitive. To understand the nature of the spe-46(hc197) mutation, we placed it over the mnDf111, a large deficiency that removes spe-46 entirely. The hemizygous hermaphrodites were essentially self-sterile (Fig. 7; control worms with wild-type spe-46/mnDf111 were fertile), suggesting that the null phenotype of spe-46 is complete sterility.
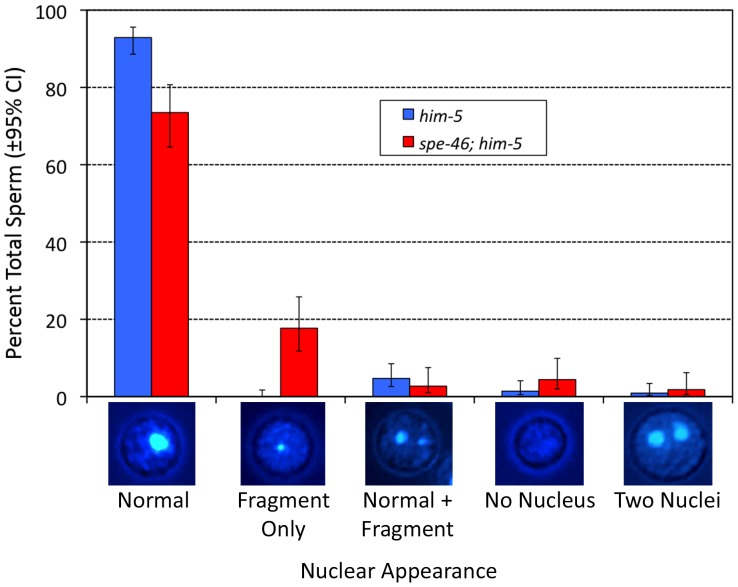
While much of the self-fertility defect in homozygous spe-46(hc197) hermaphrodites is due to a lack of fertilization, we also found a defect in embryogenesis, but only at 25°C. Of 89 fertilized eggs laid by spe-46(hc197) hermaphrodites at 25°C, only 41 hatched, giving an embryonic failure rate of 53.9%. The embryonic failure rate was 0% at 20°C (of 82 eggs) and 1.6% at 15°C (of 125 eggs). Every hatchling at all three temperatures survived to adulthood. This embryonic failure rate at 25°C was rescued in transgenic worms: all 128 eggs laid were fertilized, and 116 hatched, for a failure rate of only 9.4%. Given the sperm specificity of spe-46 expression, the observed embryonic failure is likely due to sperm that fertilized oocytes but were incapable of producing viable embryos. We found that nearly 20% of the sperm from spe-46(hc197); him-5(e1490) males appeared to have only a small fragment of the normal sperm nucleus (Fig. 8). This aneuploidy defect was almost never observed in control him-5(e1490) male sperm (Fig. 8), so we attribute this defect to spe-46(hc197), although we cannot rule out a synergistic effect between spe-46 and him-5. Other defects, including sperm with no nucleus, two nuclei, and a normal nucleus plus a fragment, were observed at very low levels in sperm from both strains (Fig. 8) Sadler and Shakes [41] showed that even anucleate sperm can fertilize oocytes and initiate early embryo development. We suspect that some nuclei that appeared normal were actually aneuploid, having excess numbers of chromosomes that were all packaged together. It is likely that self-sperm from spe-46(hc197) hermaphrodites are similarly afflicted by aneuploidy, since SPE-46 is expressed in both male and hermaphrodite spermatogenesis. While our data cannot rule out mitotic chromosomal non-disjunction, it is likely that aneuploidy occurs as a result of meiotic non-disjunction, given that our GFP reporter is expressed during meiosis and not earlier during the mitotic division (Fig. 6). These results lead to the conclusion that the SPE-46 protein participates in the meiotic divisions, in addition to its role in maintaining the spermatid stage. Such multiple roles are not unexpected, given that both SPE-4 and SPE-6 have roles early in sperm development and later in spermiogenesis inhibition [15]–[16].
Figure 8. The appearance of nuclei in spermatids labeled with Hoechst 33342.
Sperm were observed under epifluorescence, and multiple images were taken at various focal planes to ensure all nuclei were observed. The focal planes were combined to produce the images below the graph. The sperm phenotypes are expressed as percent total sperm, and the confidence interval (CI) is indicated.
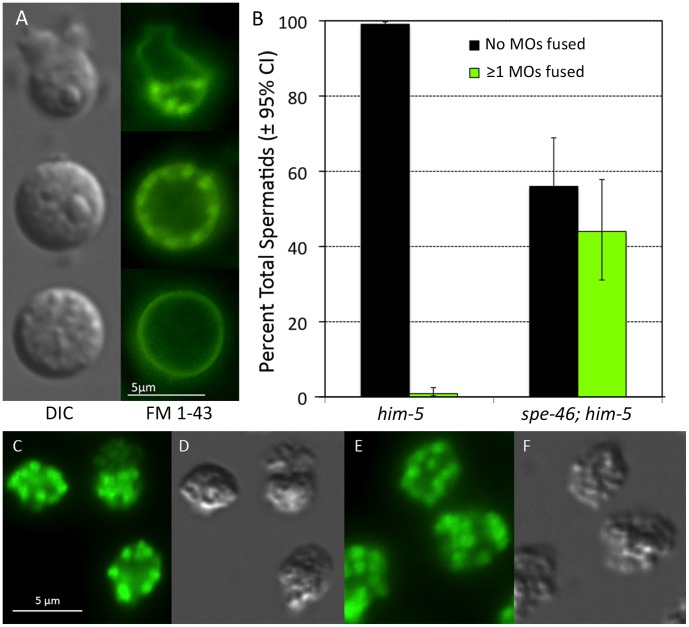
Furthermore, mutant males harbored spermatids with numerous defects. First, a large fraction of spermatids had prematurely fused MOs (Fig. 9A). Normally, the MOs fuse with the cell membrane only during spermatid activation, creating a pore in the cell membrane, which becomes contiguous with the interior of the MO. The lipid dye FM®1–43 (Life Technologies™) labels the cell membrane, and if any MOs have fused, they become labeled by FM®1–43 as well resulting in fluorescent foci abutting the cell membrane [14]. When exposed to FM®1–43, 44% of spermatids from spe-46(hc197); him-5(e1490) males had fused MOs, yet these cells showed no sign of activating (Fig. 9B). Only rarely did spermatids from him-5(e1490) have anything resembling a fused MO (Fig. 9A). Even though many MOs fused abnormally in spe-46(hc197); him-5(e1490) sperm, they segregated normally during the post-meiotic budding division. We examined >100 sperm from mutant males, and they contained a normal complement of MOs in a visual comparison to those from him-5(e1490) males (Fig. 9C-F). Therefore, spe-46(hc197) sperm have normal a number of MOs, but many of those MOs fuse prematurely with the cell membrane.
Figure 9. The status of the membranous organelles (MOs) in sperm.
A) When labeled with the membrane dye FM® 1–43 (Life Technologies™), fused MOs are visible as bright spots just inside the cell membrane, but unfused MOs are not labeled. Here, the sperm are shown in both DIC illumination and epifluorescence illumination. Top panel: a spermatozoon with an obvious pseudopod and the bright foci in the cell body indicating fused MOs. Middle panel: an abnormal spermatid with fused MOs. Bottom panel: a normal spermatid with no fused MOs. B) The percentage of normal spermatids and those that had at least a single obvious fused MO from him-5 and spe-46; him-5 male worms. Error bars represent SEM. In C-F, the sperm have been fixed, permeabilized, and exposed to an Alexa Fluor® 594 conjugate of wheat germ agglutinin (WGA; Life Technologies™), which labels all MOs, including those that have not fused with the cell membrane (original red fluorescence false colored green). C) Sperm from a spe-46(hc197); him-5(e1490) male. D) The same cells as is C, visualized in DIC optics. E) Sperm from a him-5(e1490) male labeled as in C. F) DIC image of the cells in E.
Many other morphological defects were obvious in mutant sperm. Some had large vacuoles, while others had prominent protruding spikes, and yet others were swollen (Fig. 10). The vacuolated and swollen defects were never observed in him-5(e1490) male sperm under identical conditions (Fig. 10). Swollen sperm likely had been active spermatozoa; they typically had fused MOs, and normal sperm activated in vitro sometimes take on the swollen phenotype after an extended period (CWL, personal observations).
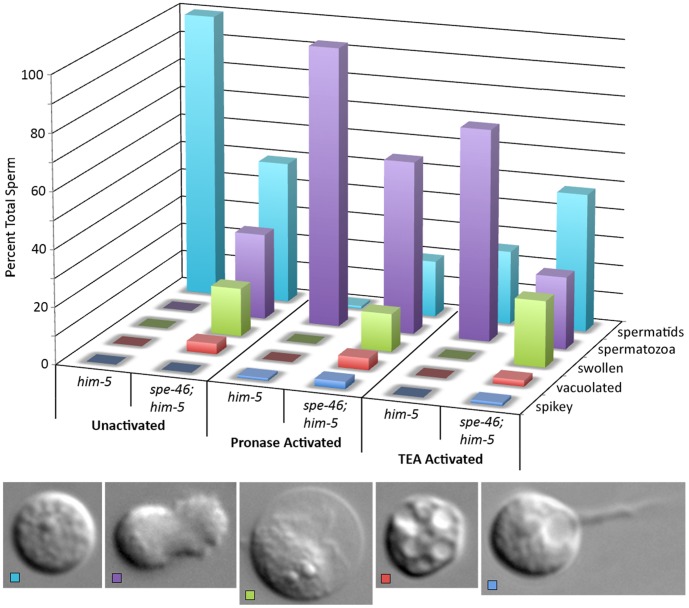
Figure 10. The sperm phenotypes from spe-46(hc197); him-5(e1490) males.
Males were dissected in SM1 buffer (unactivated), in SM1 plus Pronase (200 µg/ml) (pronase activated), and in SM1 plus 70 mM Triethanolamine (TEA) (TEA activated). Sperm phenotypes are categorized into: (i) spermatid (blue), (ii) spermatozoa (purple), (iii) sperm that appeared swollen (green), (iv) sperm that had large vacuoles (red), and (v) sperm with extended spikes (blue). The number is expressed as percent total sperm. DIC image of sperm phenotypes are shown at the bottom of the graph. Scale bar is 5 µm.
We also examined the ability of the sperm to activate in response to in-vitro activating chemicals [11]. Sperm from him-5(e1490) males behaved normally, with 98% activating to spermatozoa after exposure to Pronase (Fig. 10). Sperm from spe-46(hc197); him-5(e1490) did not respond so completely to Pronase, with nearly 20% remaining as spermatids (Fig. 10). Exposure to triethanolamine (TEA) caused more than 75% of sperm from him-5(e1490) males to activate, but spe-46(hc197); him-5(e1490) sperm did not appear to respond, with nearly identical proportions of spermatids and spermatozoa compared to sperm exposed just to SM1 buffer (Fig. 10). These results demonstrate that some spermatids from spe-46(hc197) mutant males are incapable of activating in response to in-vitro activators, suggesting that they cannot activate in-vivo.
Conclusions
Our results support the idea that spermatid activation requires proper timing. Spermatids from spe-46(hc197) worms activate prematurely to spermatozoa, and mutant males exhibit a near complete loss of fertility. Prematurely activated spermatozoa are known to compromise male fertility as they obstruct passage of sperm from male to hermaphrodite at mating [9]. Premature activation associated with the spe-46(hc197) mutation also bypasses the need for an activation signal and its transduction through the spe-8 group gene products. Not only is a fraction of the sperm prematurely activated, a large portion of the inactive spermatids have prematurely fused MOs. These results support the hypothesis that SPE-46 acts as a brake protein to inhibit spermiogenesis in the manner of SPE-4 and SPE-6 [15]–[16]. Under this hypothesis, SPE-46 has a role in preventing spermiogenesis until it is down-regulated by an incoming activation signal or by the hc197 mutation. We also show that SPE-46 expression begins when the developing gamete nuclei reach the late pachytene stage, and that SPE-46 has a role in the meiotic process. These two roles are evidenced by the degree of loss of SPE-46 function: worms with two mutated alleles exhibit a host of defects including premature spermatid activation, while hemizygous worms which have only a single mutated allele are without competent sperm altogether.
SPE-46 is predicted to reside in a membrane, but its subcellular location is as yet undetermined. The spe-4 and spe-6 genes, both of which can be mutated to cause premature spermatid activation, also have functions earlier in spermatogenesis associated with assembly of the FB-MO complex. Mutations in both genes affect FB-MO function [27] [29], and the SPE-4 protein is located in the MO membrane [27]. Recently, spe-44 has been identified as a master transcriptional regulator of a large subset of sperm-expressed genes, but it does not directly regulate expression of spe-4, spe-6, or spe-46 [42]. Given that SPE-4 and SPE-6 are associated with the MO, it is a likely destination for the SPE-46 protein. However, given the premature fusion of MOs (Fig. 8), the cell membrane is also a possible location. Resolution of this question is the subject of our future investigations.
SPE-46 is highly conserved in nematodes, but it is not present in other organisms. Given the intense scrutiny of Chromosome I sperm-expressed genes via traditional mutagenesis [43], it is surprising to discover new Chromosome I sperm-expressed genes by mutagenesis. However, we feel that the spe-27(it132) suppressor screen that identified spe-46 provides an increased level of sensitivity for the identification of genes involved in spermiogenesis. We have at least two more spe-27(it132) suppressor mutations that map to Chromosome I, so this chromosome is not exhausted as a source of new spermatogenesis genes. Further, spermatogenesis is a process that requires an inordinately large percentage of the genome. Well over 5% of the genome is enriched or expressed solely during spermatogenesis [44] [30], which probably reflects the unique nature of the sperm cell. However, fewer than 100 genes that have been ascribed a sperm function through experimental studies. Therefore, we understand only a small fraction of the mechanistic processes governing spermatogenesis; many more await investigation.
Methods
Worm Strains and Culture
All C. elegans strains were maintained on Escherichia coli OP50-seeded Nematode Growth Media (NGM) agar plates and manipulated as described by Brenner [45]. The Caenorhabditis Genetic Center kindly provided the following strains: N2, CB4856, spe-27(it132ts)IV, spe-27(it132ts) unc-22(e66)IV, spe-29(it129) dpy-20(e1282ts)IV, him-5(e1490)V, spe-27(it132ts)IV; him-5(e1490)V, fem-1(hc17ts)IV, fem-3(q23ts)IV, rrf-3(pk1426)II, eri-1(mg366 IV and mnDf111/unc-13(e1091) lin-11(n566)I. The strain bearing the hc197 mutation was isolated in a suppressor screen of spe-27(it132ts); unc-22(e66) [17] [15]. We backcrossed the hc197; spe-27(it132ts) unc-22(e66) line six times with spe-27(it132ts), each time recovering unc-22 F2 that were fertile at 25°C. Worms were mated in 35 mm petri dishes at a ratio of 12 males to four hermaphrodites unless otherwise noted. In creating the spe-46(hc197)/mnDf111 hemizygotes, spe-46(hc197)/+; him-5 males were crossed to mnDf111/unc-13(e1091) lin-11(n566) hermaphrodites. The F1 were allowed to lay eggs for three days, after which their DNA was extracted, the region containing the hc197 mutation was amplified by PCR, and the DNA was sequenced to confirm the genotype.
Single Nucleotide Polymorphism Mapping of hc197
The hc197 mutation was mapped using its spe-27(it132ts) suppression phenotype. We constructed a mapping strain which had spe-27(it132ts) backcrossed into the Hawaiian strain CB4856 six times (henceforth HA-spe-27). In the mapping cross, hc197; spe-27(it132ts) unc-22(e66) hermaphrodites were mated to HA-spe-27 males. Virgin F1 progeny were allowed to self-fertilize at 15°C, where spe-27(it132ts) worms are fertile. A total of 120 F2 worms were isolated as L4 larvae and allowed to reproduce at 25°C, where only hc197 homozygotes are self-fertile in a spe-27 background. Twenty-seven of the F2 were identified as fertile. These fertile worms were combined for bulk DNA extraction, as were another 27 sterile F2 worms chosen arbitrarily, essentially as described [31]. The results from this approach to SNP mapping are in the form of map ratios, where ratios near 1.0 indicate an unlinked state, and map ratios close to 0.0 indicate linkage. We initially tested the following SNP loci for linkage to hc197 (Chromosome: nucleotide position): pKP1119 (Chr I), pkP2107 (Chr II), pkP3103 (Chr III), pkP5097 (Chr V) (see Fig. 1 for chromosomal locations). We did not analyze SNPs on chromosome IV because preliminary data indicated hc197 was unlinked to spe-27 on chromosome IV. Only the Chromosome I SNP had a map ratio indicating linkage (Fig. 1), so we subsequently analyzed three more SNP loci across Chromosome I (Fig. 1). All SNP information is based upon WormBase release WS230.
Sequencing and Analysis of the hc197 Genome
Genomic DNA was extracted from ∼1 ml of worms from the hc197; spe-27(it132ts) unc-22(e66) strain using standard methods. Briefly, we froze ∼0.5 ml of worms in 4 ml of TEN (20mM Tris, 50mM EDTA, 100mM NaCl) and digested them for 0.5 hours at 60°C in TEN plus 0.5% SDS and 0.1 mg/ml Proteinase K. The concentration of Proteinase K was increased to 0.2 mg/ml, and the digestion was extended for another hour. DNA was extracted from the digest via phenol/chloroform/isoamyl alcohol and precipitated with NaOAc/EtOH. The pellet was resuspended in TEN, RNase A was added to 40 mg/ml, and the digestion was incubated for an hour. The DNA was extracted and precipitated as above and resuspended in TE. DNA sequencing was conducted at the City of Hope and Beckman Research Institute’s DNA Sequencing/Solexa Core (Duarte, CA), where sequencing with the Illumina Genome Analyzer II platform was performed on two flow cell lanes with 80 bp paired-end reads from the ends of ∼350 bp fragments of the DNA, generating a total of 82,301,352 reads. Using the bioinformatics software Geneious Pro™ (version 5.6.2) 14.2% of the reads, or 11,687,711 reads, were mapped to Chromosome I (Accession no. NC_003279). This percentage corresponds closely to the percentage of the genome found on Chromosome I (15.8%) and provided an average depth of 62 reads per base pair.
Microinjection Transformation
Worms were transformed with PCR products from one of the two genes tested plus flanking sequence (W06D4.2 plus 1,545 bp upstream and 732 bp downstream; T24B1.1 plus 1,324 bp upstream and 677 bp downstream). The sequences were amplified from N2 DNA using the Expand High FidelityPLUS PCR System (Roche Diagnostics) following the manufacturer’s protocols. The amplicons were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega). An injection mix of the PCR product (15 ng/µl) along with a plasmid containing myo-3::mCherry (pCFJ104, 100 ng/µl) was microinjected into the gonads of young adult hermaphrodites. Transformed worms were identified by mCherry fluorescence in the body wall muscle. F2 transformants were scored for rescue, but transformed lines were not maintained past the rescue experiments because transgenes expressed in sperm are generally rendered ineffective by germline silencing after a small number of generations [46].
RT-PCR
RNA was extracted from mixed age populations of worms from the strains fem-1(hc17ts), fem-3(q23ts), spe-46(hc197), and N2. Large populations of each strain were collected and rinsed 4 times with M9 buffer. After freezing at -80°C, the worms were disrupted by sonication in TRIzol® Reagent, and the RNA was extracted following the manufacturer’s protocols (Life Technologies™). RNA samples were treated with RQ1 DNase (Promega) and subsequently purified via phenol-chloroform isoamyl alcohol extraction, and samples were diluted to 33 ng/µl. RT-PCR was performed with the MyTaq™ One-Step RT-PCR Kit (Bioline) according to manufacturer’s instructions. We multiplexed spe-46 specific primers (Exon2F: 5′-GGCAGAGAATAGTCCATTGG-3′; and Exon5R: 5′-TTGCTTGACTCTCTCCAACG-3′) with primers specific to act-2, the C. elegans homolog of β-Actin (actinF: 5′-GTATGGGACAGAAAGACTCG-3′; and actinR: 5′-CGTCGTATTCTTGCTTGGAG-3′).
Construction of a spe-46 RNAi Clone and Induction of RNA Interference
Using the MyTaq™ One-Step RT-PCR Kit a product was amplified from N2 RNA using primer pairs RNAiF and RNAiR (5′-gtgtGCGGCCGCgtgtcattgcatattacgatgg-3′ and 5′- gtgtACTAGTaagtggtaatgaagacgctg-3′, respectively). Primers were engineered with an added restriction site on the 5′ end for cloning: NotI on the forward primer and SpeI on the reverse primer (uppercase letters correspond to restriction enzyme sites; the extreme 5′-gtgt was added to each primer to maximize restriction enzyme activity). The amplicon was cloned into the L4440 RNAi vector [47], and transformed into E. coli HT115(DE3), a strain which lacks RNase III activity. Expression of dsRNA from the insert was achieved through isopropyl-β-D-thiogalactopyranoside (IPTG) induction of T7 promoter sites flanking the L4440 multiple cloning site.
Worms were exposed to the RNAi feeding strain bacteria in petri dishes on agar prepared with 25 µg/ml carbenicillin and 1 mM IPTG. Plates were seeded with transformed HT115 bacteria that had been grown the previous night in a 2 ml liquid culture with LB media, 50 µg/ml carbenicillin, and 15 µg/ml tetracycline. In the final hour of liquid culture, IPTG was added to a final concentration of 1 mM. Large populations of worms were bleached [48] to obtain eggs, which were introduced onto plates. At the L4 larval stage, exposed hermaphrodites were transferred to their own RNAi plate, and their lifetime fecundity was measured. Identically handled control worms were exposed to HT115 bacteria containing the L4440 empty vector.
Construction of a spe-46::gfp Reporter
A transcriptional spe-46::gfp construct was integrated into Chromosome II following the Mos-SCI technique [49]. Initially, the spe-46 promoter was amplified from N2 DNA, as was the spe-46 3′ UTR. The promoter and 3′ UTR were stitched together with the GFP coding sequence following the PCR fusion technique described by Hobert [50]. The promoter-GFP-3′ UTR fusion was cloned into the multiple cloning site of the vector pCFJ151; the final construct contained 1,015 bp upstream of the spe-46 start codon (the next gene begins 846 bp upstream on the opposite strand), followed by the GFP coding sequence, and finally 1,020 bp downstream of the spe-46 stop codon. The pCFJ151 vector targets the ttTi5605 mos1 insertion on Chromosome II for homologous recombination, and we recovered a homozygous integrated copy of the construct [49].
Microscopy, in vitro Spermatid Activation, and Microinjection Transformation
Imaging was accomplished on a Nikon Eclipse Ti inverted microscope outfitted for Nomarski DIC and epifluorescence. Nuclear material was labeled with 30 ng/µl Hoechst 33342 (Life Technologies™) in SM1 buffer [51]. Fused MOs were visualized with SM1 containing 2µM FM® 1–43 (Life Technologies™), a vital fluorescent dye that labels the outer leaflet of plasma membrane [14]. On the other hand, all MOs were visualized with an Alexa Fluor® 594 conjugate of wheat germ agglutinin (WGA; Life Technologies™). Here, the sperm were fixed in 4% paraformaldehyde in SM1, rinsed 3 times in PBS, permeabilized with 0.5% Triton X-100 in PBS, rinsed another 3 times with PBS, and labeled with 5 µg/ml of WGA in PBS. In other experiments, sperm were activated in-vitro by exposure to SM1 containing either 200 µg/ml Pronase or 70 mM TEA (pH 7.8). Images were captured on a Nikon DS-Qi1 12 bit monochrome camera and analyzed with Nikon NIS Elements software. The same microscope system was used for microinjection of DNA into the gonads of recipient young adult hermaphrodites. The injection mix contained 10–15 ng/µl of the transgenes for rescue and 100 ng/µl of the transformation marker.
Acknowledgments
We are indebted to Paul Muhlrad, who originally isolated the hc197 mutation. We also thank Samuel Ward, who provided us with the large collection of spe-27 suppressor mutants. We are grateful to Steven L’Hernault, Diane Shakes, and Paul Muhlrad for helpful discussions, to Diane Shakes for improving an earlier draft of the manuscript, and to Aidyl Gonzalez-Serricchio and Gizelle Hacopian for technical assistance.
Funding Statement
This research was funded by U.S. National Institutes of Health award 1 SC3 GM087212. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kaupp UB, Hildebrand E, Weyand I (2006) Sperm chemotaxis in marine invertebrates–molecules and mechanisms. J Cell Physiol 208: 487–494. [DOI] [PubMed] [Google Scholar]
- 2. Christen R, Schackmann RW, Shapiro BM (1982) Elevation of the intracellular pH activates respiration and motility of sperm of the sea urchin, Strongylocentrotus purpuratus. J Biol Chem 257: 14881–14890. [PubMed] [Google Scholar]
- 3. Neill AT, Vacquier VD (2004) Ligands and receptors mediating signal transduction in sea urchin spermatozoa. Reproduction 127: 141–149. [DOI] [PubMed] [Google Scholar]
- 4. Schackmann RW, Shapiro BM (1981) A partial sequence of ionic changes associated with the acrosome reaction of Strongylocentrotus purpuratus. Dev Biol 81: 145–154. [DOI] [PubMed] [Google Scholar]
- 5. Boatman DE, Robbins RS (1991) Bicarbonate: carbon-dioxide regulation of sperm capacitation, hyperactivated motility, and acrosome reactions. Biol Reprod 44: 806–813. [DOI] [PubMed] [Google Scholar]
- 6. Gadella BM, Harrison RA (2000) The capacitating agent bicarbonate induces protein kinase A-dependent changes in phospholipid transbilayer behavior in the sperm plasma membrane. Development 127: 2407–2420. [DOI] [PubMed] [Google Scholar]
- 7. Gadella BM, Harrison RA (2002) Capacitation induces cyclic adenosine 3′,5′-monophosphate-dependent, but apoptosis-unrelated, exposure of aminophospholipids at the apical head plasma membrane of boar sperm cells. Biol Reprod 67: 340–350. [DOI] [PubMed] [Google Scholar]
- 8. Wennemuth G, Carlson AE, Harper AJ, Babcock DF (2003) Bicarbonate actions on flagellar and Ca2+ -channel responses: initial events in sperm activation. Development 130: 1317–1326. [DOI] [PubMed] [Google Scholar]
- 9. Smith JR, Stanfield GM (2011) TRY-5 is a sperm-activating protease in Caenorhabditis elegans seminal fluid. PLoS Genet 7: e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nelson GA, Ward S (1980) Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell 19: 457–464. [DOI] [PubMed] [Google Scholar]
- 11. Ward S, Hogan E, Nelson GA (1983) The initiation of spermiogenesis in the nematode Caenorhabditis elegans. Dev Biol 98: 70–79. [DOI] [PubMed] [Google Scholar]
- 12.L’Hernault SW (1997) Spermatogenesis. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C elegans II. Cold Spring Harbor, NY USA: Cold Spring Harbor Laboratory Press. 271–294. [PubMed] [Google Scholar]
- 13. Bandyopadhyay J, Lee J, Lee J, Lee JI, Yu JR, et al. (2002) Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement, fertility, egg laying, and growth in Caenorhabditis elegans. Mol Biol Cell 13: 3281–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Washington NL, Ward S (2006) FER-1 regulates Ca2+ -mediated membrane fusion during C. elegans spermatogenesis. J Cell Sci 119: 2552–2562. [DOI] [PubMed] [Google Scholar]
- 15. Muhlrad PJ, Ward S (2002) Spermiogenesis Initiation in Caenorhabditis elegans Involves a Casein Kinase 1 Encoded by the spe-6 Gene. Genetics 161: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gosney R, Liau WS, Lamunyon CW (2008) A novel function for the presenilin family member spe-4: inhibition of spermatid activation in Caenorhabditis elegans. BMC Dev Biol 8: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhlrad PJ (2001) A Genetic and Molecular Analysis of Spermiogenesis Initiation in Caenorhabditis elegans PhD thesis. Tucson, AZ: University of Arizona. 192 p. [Google Scholar]
- 18. Shakes DC, Ward S (1989) Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev Biol 134: 189–200. [DOI] [PubMed] [Google Scholar]
- 19. Nance J, Minniti AN, Sadler C, Ward S (1999) spe-12 encodes a sperm cell surface protein that promotes spermiogenesis in Caenorhabditis elegans. Genetics 152: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geldziler B, Chatterjee I, Singson A (2005) The genetic and molecular analysis of spe-19, a gene required for sperm activation in Caenorhabditis elegans. Dev Biol 283: 424–436. [DOI] [PubMed] [Google Scholar]
- 21. Minniti AN, Sadler C, Ward S (1996) Genetic and molecular analysis of spe-27, a gene required for spermiogenesis in Caenorhabditis elegans hermaphrodites. Genetics 143: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nance J, Davis EB, Ward S (2000) spe-29 encodes a small predicted membrane protein required for the initiation of sperm activation in Caenorhabditis elegans. Genetics 156: 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burke DJ, Ward S (1983) Identification of a large multigene family encoding the major sperm protein of Caenorhabditis elegans. J Mol Biol 171: 1–29. [DOI] [PubMed] [Google Scholar]
- 24. Roberts TM, Pavalko FM, Ward S (1986) Membrane and cytoplasmic proteins are transported in the same organelle complex during nematode spermatogenesis. J Cell Biol 102: 1787–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sepsenwol S, Ris H, Roberts TM (1989) A unique cytoskeleton associated with crawling in the amoeboid sperm of the nematode, Ascaris suum . J Cell Biol 108: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts TM, Ward S (1982) Membrane flow during nematode spermiogenesis. J Cell Biol 92: 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arduengo PM, Appleberry OK, Chuang P, L’Hernault SW (1998) The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci 111: 3645–3654. [DOI] [PubMed] [Google Scholar]
- 28. L’Hernault SW, Arduengo PM (1992) Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J Cell Biol 119: 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Varkey JP, Jansma PL, Minniti AN, Ward S (1993) The Caenorhabditis elegans spe-6 gene is required for major sperm protein assembly and shows second site non-complementation with an unlinked deficiency. Genetics 133: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reinke V, Smith HE, Nance J, Wang J, Van Doren C, et al. (2000) A global profile of germline gene expression in C. elegans. Mol Cell 6: 605–616. [DOI] [PubMed] [Google Scholar]
- 31. Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet 28: 160–164. [DOI] [PubMed] [Google Scholar]
- 32. Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, et al. (2005) Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. [DOI] [PubMed] [Google Scholar]
- 33. del Castillo-Olivares A, Kulkarni M, Smith HE (2009) Regulation of sperm gene expression by the GATA factor ELT-1. Dev Biol 333: 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Veljkovic E, Bacconi A, Stetak A, Hajnal A, Stasiuk S, et al. (2004) Aromatic amino acid transporter AAT-9 of Caenorhabditis elegans localizes to neurons and muscle cells. J Biol Chem 279: 49268–49273. [DOI] [PubMed] [Google Scholar]
- 35. Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, et al. (2006) CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell 17: 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gould CM, Diella F, Via A, Puntervoll P, Gemund C, et al. (2010) ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res 38: D167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dell’Angelica EC, Payne GS (2001) Intracellular cycling of lysosomal enzyme receptors: cytoplasmic tails’ tales. Cell 106: 395–398. [DOI] [PubMed] [Google Scholar]
- 38. Blom N, Gammeltoft S, Brunak S (1999) Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294: 1351–1362. [DOI] [PubMed] [Google Scholar]
- 39. Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S (2009) Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183: 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stanfield GM, Villeneuve AM (2006) Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Curr Biol 16: 252–263. [DOI] [PubMed] [Google Scholar]
- 41. Sadler PL, Shakes DC (2000) Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development 127: 355–366. [DOI] [PubMed] [Google Scholar]
- 42.Kulkarni M, Shakes DC, Guevel K, Smith HE (2012) SPE-44 implements sperm cell fate. PLoS Genet 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. L’Hernault SW, Shakes DC, Ward S (1988) Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics 120: 435–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reinke V, Gil IS, Ward S, Kazmer K (2004) Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- 45. Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kelly WG, Fire A (1998) Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125: 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Timmons L, Court DL, Fire A (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- 48.Stiernagle T (1999) Maintenance of C. elegans In: Hope I, editor. C elegans: A Practical Approach: Oxford University Press. 51–67. [Google Scholar]
- 49. Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, et al. (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nature Genetics 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hobert O (2002) PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C-elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- 51. Machaca K, DeFelice LJ, L’Hernault SW (1996) A novel chloride channel localizes to Caenorhabditis elegans spermatids and chloride channel blockers induce spermatid differentiation. Dev Biol 176: 1–16. [DOI] [PubMed] [Google Scholar]
- 52. Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, et al. (2010) WormBase: a comprehensive resource for nematode research. Nucleic Acids Res 38: D463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Han X, Gomes JE, Birmingham CL, Pintard L, Sugimoto A, et al. (2009) The role of protein phosphatase 4 in regulating microtubule severing in the Caenorhabditis elegans embryo. Genetics 181: 933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]