Abstract
The endoplasmic reticulum (ER) is a central hub where secreted or membrane-bound proteins are maturated and folded properly in eukaryotes. Maintenance of ER homeostasis is particularly important for human fungal pathogens, such as Cryptococcus neoformans, which encounter a plethora of host-mediated stresses during infection. Our previous study demonstrated that the unfolded protein response (UPR) pathway, composed of the evolutionarily conserved Ire1 kinase and the unique Hxl1 transcription factor, has pleiotropic roles in ER stress response, thermotolerance, antifungal drug resistance, and virulence in C. neoformans. Here, we functionally characterized an ER-resident molecular chaperone, Kar2/BiP, in C. neoformans. Conditional expression of KAR2 by the copper-regulated promoter revealed that Kar2 is essential for the viability of C. neoformans. Constitutive expression of KAR2 by the strong histone H3 promoter partially restores resistance to ER stress, cell wall stress, thermotolerance, and genotoxic stress in ire1Δ and hxl1Δ mutants, suggesting that Kar2 mainly functions downstream of the UPR pathway. Furthermore, Kar2 appears to control azole resistance in C. neoformans downstream of the UPR pathway without regulation of ERG11 or ERG3. Interestingly, we discovered that azole treatment is sensed as ER-stress and subsequently activates the Ire1-dependent Hxl1 splicing event and induction of KAR2 by the UPR pathway. In contrast, the constitutive expression of Kar2 is not sufficient to restore the Ire1-mediated regulation of capsule production in C. neoformans UPR mutants. In conclusion, this study demonstrates that Kar2 is not only essential for vegetative growth but also required for response and adaptation to the environmental stresses and antifungal drugs downstream of the UPR pathway in C. neoformans.
Introduction
Sensing, responding, and adapting to environmental cues, such as nutrient starvation, hypoxia, and temperature changes, are essential for all living organisms. To cope with such stresses, all organisms have evolutionarily conserved and unique signal transduction pathways depending on their biological niches. Particularly, those involved in homeostasis of the endoplasmic reticulum (ER) play a crucial role in protein quality control for eukaryotes because secreted or membrane proteins must be assembled or folded properly in the ER prior to their cellular localization. When misfolded or unfolded proteins accumulate beyond the protein folding capacity of the ER due to external environmental cues or internal physiological changes – a condition known as ER stress – the unfolded protein response (UPR) and ubiquitin dependent ER-associated degradation (ERAD) pathways are activated [1], [2].
The molecular mechanism of the UPR pathway is well characterized in budding yeast Saccharomyces cerevisiae [3]. Upon exposure to ER stress, the ER transmembrane-resident Ire1 kinase undergoes autophosphorylation and induces spliceosome-independent, unconventional splicing of HAC1 mRNA, which encodes the downstream transcription factor of Ire1 [4]. The activated Hac1 transcription factor induces diverse UPR target genes, including those involved in translocation, glycosylation/modification, and protein folding and degradation, to alleviate ER stress [5]. The UPR pathway is critical for virulence regulation in opportunistic human fungal pathogens [6]–[9]. In Aspergillus fumigatus, which is an ascomycete filamentous fungus causing invasive and systemic aspergillosis, deletion of the Hac1 transcription factor or the Ire1 kinase gene results in severe virulence attenuation [6], [9]. Hac1 regulates the morphology of Candida albicans, which is an ascomycete pleomorphic fungus causing superficial, vaginal, and systemic candidiasis, by modulating the expression of genes encoding cell surface proteins [8].
In the basidiomycete fungus Cryptococcus neoformans, the UPR pathway also has essential roles in controlling its virulence as well as the ER stress response [7]. Notably, the Cryptococcus UPR pathway comprises the evolutionarily conserved Ire1 kinase and the unique bZIP transcription factor, Hxl1, which is phylogenetically divergent from the conventional yeast Hac1 or human Xbp1 proteins. Interestingly, Ire1 appears to have both Hxl1-dependent and -independent functions. Ire1 modulates ER stress response, thermotolerance, maintenance of cell wall integrity, and azole drug resistance in an Hxl1-dependent manner. In contrast, Ire1 also has Hxl1-independent roles in controlling capsule production and certain stress responses. Interestingly, the hxl1Δ mutant shows a greater thermosensitivity than the ire1Δ mutant, suggesting that an Ire1-independent signaling circuit could partly contribute to Hxl1 regulation and activation [7].
Kar2, also known as BiP, is not only an ER-resident molecular chaperone but also a common negative regulator of the UPR pathway in yeast and animal cells. It is essential for viability and involved in diverse cellular processes, including protein translocation and ER-associated degradation [10]–[14]. As the ER luminal Hsp70 molecular chaperone, KAR2 is induced in response to heat shock and treatment with tunicamycin, which is an ER stress-inducer inhibiting N-linked glycosylation [15]. Kar2 induced by the UPR pathway alleviates ER stress to interact with misfolded or unfolded proteins as a molecular chaperone. Although KAR2 mRNA levels are modulated via the UPR pathway, Kar2 itself is an important regulator of the UPR pathway. In the absence of ER stress, Kar2 physically interacts with and inactivates the Ire1 sensor. In response to ER stress, Kar2 dissociates from Ire1, which subsequently dimerizes or oligomerizes for activation [16], [17]. Thereby, Kar2 limits unrestricted activation of the UPR pathway under unstressed conditions.
Our prior study identified a Cryptococcus gene (CNAG_06443.2) orthologous to yeast Kar2 and demonstrated that the expression of KAR2 is controlled by the UPR pathway in C. neoformans in response to ER stress and temperature upshifts [7]. Nevertheless, the cellular roles of Kar2 remain elusive in basidiomycete fungi, including C. neoformans. In this study, we functionally characterized the Cryptococcus Kar2 through the construction and phenotypic analysis of conditional and constitutive KAR2 overexpression strains. Here, we discovered that Kar2 is not only required for viability, but also as an important chaperone downstream of the Ire1/Hxl1-dependent UPR pathway in C. neoformans.
Results
Identification of the KAR2 gene in C. neoformans
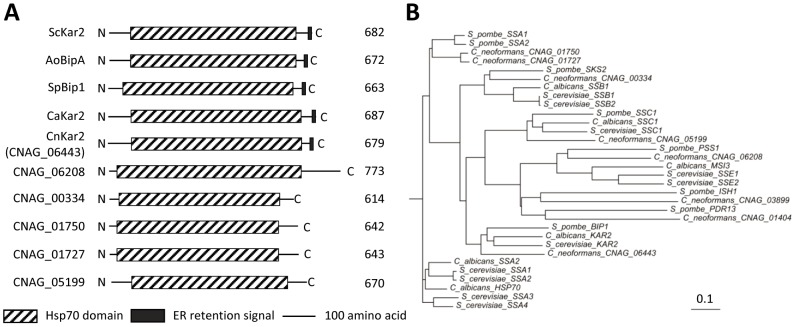
To identify a KAR2 ortholog in C. neoformans, we performed BLAST searches (blastp) with the S. cerevisiae Kar2 protein sequence as a query. In the genome from serotype A H99 strain, several Kar2 orthologous genes were discovered [CNAG_06443.2 (score: 786.563, e-value: 0), CNAG_01750.2 (score: 697.197, e-value: 0), CNAG_01727.2 (score: 693.345, e-value: 0), CNAG_00334.2 (score: 598.201, e-value: 0), CNAG_05199.2 (score: 515.383, e-value: 0), and CNAG_06208.2 (score: 258.84, e-value: 0)]. These may reflect the presence of a diverse Hsp70 class of proteins in C. neoformans. The protein domain analysis revealed that CNAG_06443.2 contains an ER retention signal at the C-terminus (HDEL), which is conserved among Kar2/BiP proteins in other fungi (Fig. 1A). The phylogenetic analysis showed that CNAG_06443.2 is evolutionarily more closely related to known Kar2/BiP proteins than to other Hsp70 family proteins (Fig. 1B). Therefore, the gene CNAG_06443.2 was named Kar2/BiP in C. neoformans.
Figure 1. Identification of Kar2/BiP in C. neoformans.
(A) Schematic outline of Kar2/BiP proteins in fungi and Hsp70 proteins in C. neoformans. The box with dashed line indicates an Hsp70 domain. The black box represents an ER retention signaling motif at the C-terminus [HDEL for S. cerevisiae Kar2 (ScKar2), Aspergillus oryzae BipA (AoBipA), C. albicans Kar2 (CaKar2) and C. neoformans Kar2 (CnKar2) or ADEL for Schizosaccharomyces pombe Bip1 (SpBip1)]. Each number represents the size of the protein in amino acid. (B) Phylogenetic tree of Kar2/BiP proteins and Hsp70 family proteins in C. neoformans and in other fungi. The phylogenetic tree was generated by the philodendron phylogenetic tree printer (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html). The scale bar represents the evolutionary distance of 0.1. Each protein sequence used for phylogenetic analysis was retrieved from the following genome database [Saccharomyces Genome Database (http://www.yeastgenome.org/), Candida Genome Database (http://www.candidagenome.org/), S. pombe GeneDB (http://old.genedb.org/genedb/pombe/), and C. neofomans var. grubii. H99 Database (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html)].
Kar2/BiP is essential for the viability of C. neoformans
Kar2 is known to be essential for the viability of S. cerevisiae and C. albicans, shown by the construction and analysis of the conditional null mutant [10]–[12]. To address whether Kar2 is also essential for the growth of C. neoformans, we constructed conditional null kar2 mutants by inserting a copper regulated promoter (CTR4 promoter; PCTR4) right upstream of the ATG start codon of the KAR2 gene (Fig. 2A). To verify the exact start codon of the KAR2 gene, we performed rapid amplification of cDNA ends (RACE) for the 5′ untranslated region (UTR) (GenBank accession number JX982102). After we confirmed the targeted insertion of the CTR4 promoter right upstream of the KAR2 gene by Southern blot analysis (Fig. 2B), we measured the growth defect of PCTR4:KAR2 strains in a YNB medium containing bathocuproine disulfonate (BCS, a copper chelator), which induces the CTR4 promoter, or copper sulfate (CuSO4), which represses the CTR4 promoter. The wild-type (WT) strain did not show any growth defects in both BCS and CuSO4-containing YNB media, whereas the PCTR4:YPD1 strain, which was spotted as a positive control, exhibited a severe growth defect in the YNB+CuSO4 medium as previously described [18] (Fig. 2C). The YPD1 gene encodes a histidine-containing phosphotransfer protein essential for the growth of C. neoformans [18]. Interestingly, the PCTR4:KAR2 strains displayed greater growth defects than those of the PCTR4:YPD1 strain in the YNB+CuSO4 medium, whereas their growth defects were suppressed in the YNB+BCS medium (Fig. 2C). To indirectly support these data, repeated trials to construct kar2Δ deletion mutants were not successful (data not shown). In conclusion, KAR2 is required for the viability of C. neoformans.
Figure 2. Kar2/BiP is essential for the viability of C. neoformans.
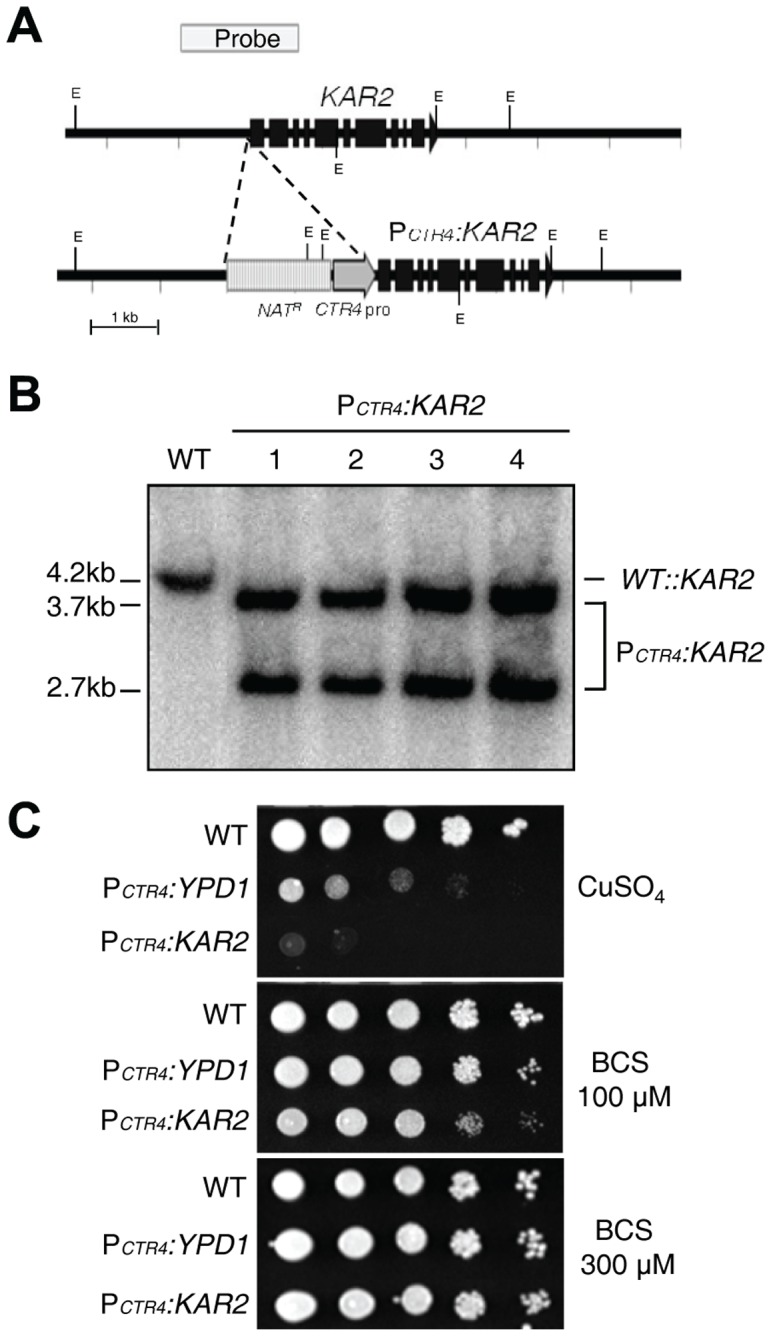
(A) The scheme for the construction of the PCTR4:KAR2 promoter insertion strain. A construct containing the CTR4 promoter and the NAT dominant selectable marker was inserted 8 bp upstream of the ATG start site of the KAR2 gene. Black boxes indicate exons of the KAR2 gene. (B) The correct genotypes of the PCTR4:KAR2 promoter strains [four independent strains (YSB1637, YSB1638, YSB1639, and YSB1640 as labeled 1 to 4, respectively)] compared to the WT H99 strain were confirmed by Southern blot analysis using genomic DNAs digested with the restriction enzyme EcoRΙ. The membrane was hybridized with a KAR2-specific probe, washed, and developed. (C) The WT H99, the PCTR4:KAR2 (YSB1637), and PCTR4:YPD1 (YSB859, a positive control) strains were cultured overnight at 30°C in a liquid YPD medium, 10-fold serially diluted, and spotted onto yeast nitrogen base agar (YNB) medium containing 12.5 µM CuSO4, 100 µM BCS, or 300 µM BCS. Cells were incubated at 30°C for 4 days and photographed.
Construction of KAR2 overexpression strains
Our previous study showed that expression of KAR2 is induced by tunicamycin (TM), which is an ER stress inducer, or by a temperature upshift (from 30°C to 37°C) in WT, but not in ire1Δ and hxl1Δ mutants, suggesting that Kar2 is one of the UPR downstream target genes in C. neoformans [7]. As described before, however, Ire1 and Hxl1 have both mutually dependent and exclusive roles in C. neoformans. Therefore, we next wished to determine which subsets of the Ire1- and Hxl1-dependent phenotypes are controlled by Kar2. For this purpose, we constructed constitutive KAR2 expression strains in a WT strain and ire1Δ and hxl1Δ mutant backgrounds by inserting the histone H3 gene promoter right upstream of the ATG start codon in the KAR2 gene (Fig. 3A). We verified the correct insertion of the PH3 :KAR2 alleles through Southern blot analysis (data not shown) and performed Northern blot analysis to measure the basal expression levels of KAR2 in the WT, ire1Δ, hxl1Δ, and PH3 :KAR2 strains (Fig. 3B). As expected, the expression levels of KAR2 were 3- to 5-fold higher in all PH3 :KAR2 strains in a WT, ire1Δ, or hxl1Δ strain background (Fig. 3B).
Figure 3. Construction of the constitutive KAR2 expression strain in C. neoformans.
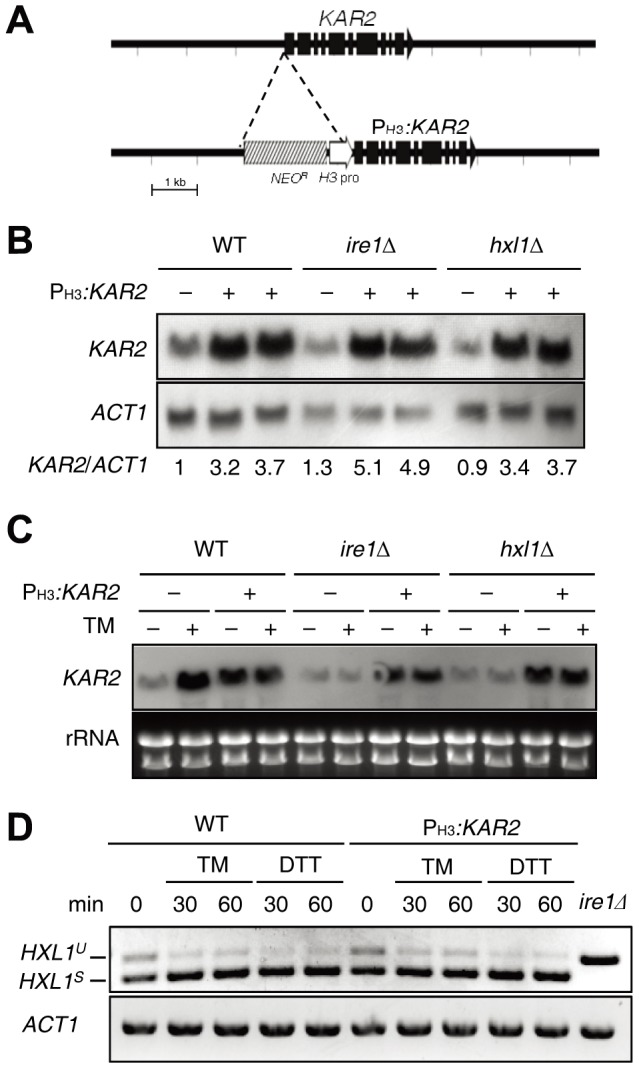
(A) The strategy for the construction of the PH3 :KAR2 strain containing the NEO resistance marker (NEO R) and the histone H3 gene promoter (H3 pro). (B) Northern blot analysis for measuring KAR2 expression in PH3 :KAR2 strains [YSB1751 (lane 2), YSB1752 (lane 3), YSB1741 (lane 5), YSB1744 (lane 6), YSB1745 (lane 8), and YSB1746 (lane 9)] and their parent strains [WT H99 strain (lane 1) and ire1Δ (lane 4) and hxl1Δ (lane 7) mutants]. KAR2 expression levels were quantitatively measured with a phosphorimager and normalized to ACT1 expression levels. Each KAR2/ACT1 is a value relative to that of the WT strain set to 1.0. (C) Northern blot analysis for measuring KAR2 expression in WT, ire1Δ, hxl1Δ mutants, and PH3 :KAR2 strains (YSB1751, YSB1741, and YSB1745) treated with or without TM (0.3 μg/ml) for 1 h. (D) RT-PCR analysis of UPR-induced HXL1 splicing with cDNA samples in WT H99 strain and PH3 :KAR2 strain (YSB1751) treated with or without TM (0.3 μg/ml) or DTT (20 mM). RT-PCR of HXL1 and ACT1 was performed with gene-specific primers listed in the Materials and Methods.
To examine whether KAR2 in the PH3 :KAR2 strain is constitutively expressed under ER stress, we monitored the expression levels of KAR2 in response to TM. Supporting previous data, KAR2 expression was highly induced in response to TM in WT but not in ire1Δ and hxl1Δ mutants, whereas KAR2 overexpression levels in the PH3 :KAR2 strain were maintained stably under ER stress (Fig. 3C). Notably, TM-induced KAR2 expression levels in WT were higher than those in the PH3 :KAR2 strain (Fig. 3C), indicating that the native KAR2 promoter could be stronger than the H3 promoter under certain ER stressed conditions.
Finally, we examined whether increased basal expression of KAR2 in the PH3 :KAR2 strain might inhibit activation of the Ire1-mediated unconventional splicing of HXL1 mRNA, given the known fact that Kar2 serves as a negative regulator of Ire1 in yeast [16], [17]. As previously described [7], both spliced (HXL1S) and unspliced (HXL1U) versions of HXL1 mRNA are observed even under an unstressed condition in C. neoformans, but the spliced version is predominant upon treatment with ER stress inducers (Fig. 3D). The basal ratio of HXL1S and HXL1U under the unstressed condition and the Ire1-mediated unconventional splicing of HXL1 by ER stress occurred in a similar fashion in the PH3 :KAR2 strain, indicating that the increased basal expression of KAR2 by the H3 promoter does not affect basal activation and ER stress-mediated induction of the UPR pathway in C. neoformans.
Kar2 controls ER stress response, high temperature growth, and cell wall integrity downstream of the UPR pathway in C. neoformans
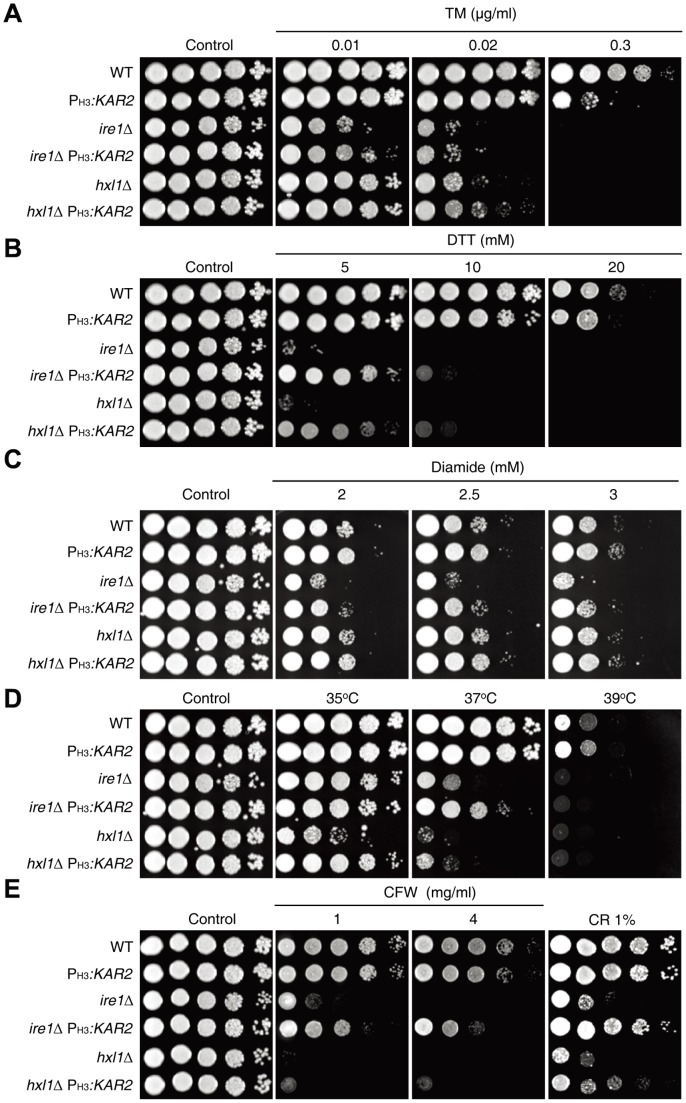
To elucidate the functions of Kar2 in ER stress response and adaptation, we examined whether KAR2 overexpression could suppress the ER stress sensitivity of UPR mutants. Previously, we have reported that the ire1Δ and hxl1Δ mutants exhibit equal levels of hypersensitivity to high concentrations of 2 ER stress-inducing agents, TM (0.075 µg/ml) and dithiothreitol (DTT; 10 mM). In this study, however, we have found that the ire1Δ mutant was more sensitive to TM than the hxl1Δ mutant at lower levels of TM (<0.02 µg/ml) (Fig. 4A), indicating that Ire1 may utilize other downstream effectors besides Hxl1 in counteracting ER stress. KAR2 overexpression partially suppressed the growth defects of ire1Δ and hxl1Δ mutants in response to low concentrations of TM (<0.02 µg/ml) and DTT (<5 mM) (Fig. 4A and 4B), indicating that Kar2 is one of the key effectors to counteract ER stress downstream of the Ire1/Hxl1-dependent UPR pathway in C. neoformans. At high concentrations of TM (>0.02 µg/ml) or DTT (>5 mM), however, the suppressive effect of KAR2 overexpression in the UPR mutants was not evident (Fig. 4A and 4B), probably because KAR2 expression levels by the H3 gene promoter might not be sufficient or other multiple UPR downstream effectors are required for a full-scale ER response and adaptation. In fact, KAR2 expression by the H3 gene promoter appeared to be less efficient in counteracting the ER stresses exerted by the high concentrations of TM (0.3 µg/ml) or DTT (20 mM) than by the native KAR2 promoter in WT strain (Fig. 4A and 4B).
Figure 4. Kar2 is involved in ER stress response, thermotolerance, and maintenance of cell wall integrity downstream of the UPR pathway.
The KAR2 overexpression strains [PH3 :KAR2 (YSB1751), ire1Δ PH3 :KAR2 (YSB1741), and hxl1Δ PH3 :KAR2 (YSB1745)] and their parent strains (WT H99 strain and ire1Δ and hxl1Δ mutants) were grown for 16 h at 30oC in a liquid YPD medium, 10-fold serially diluted, and spotted on a YPD agar medium containing the indicated concentrations of tunicamycin (TM; A), dithiothreitol (DTT; B), diamide (C), calcofluor white (CFW; E), and congo-red (CR; E). Strains were incubated at 30°C or at 35, 37, or 39°C for thermotolerance test (D) for 3–4 days and were photographed.
Notably, the suppressive effect of KAR2 overexpression in the UPR mutants was more evident under DTT than under TM (Fig. 4A and Fig. 4B). This may explain the molecular chaperonic role of Kar2 for resolving unfolded proteins given that DTT is a reducing agent, which disrupts protein disulfide bonds and normal protein structures to yield unfolded or misfolded proteins. With an action opposite to that of DTT, diamide is a thiol (SH)-specific oxidant, which induces abnormal disulfide bond formation and also perturbs natural protein structures. Our previous study revealed that the ire1Δ mutant, but not hxl1Δ mutant, exhibits increased diamide sensitivity [7], indicating that Ire1 has an Hxl1-independent manner in regulating diamide resistance. KAR2 overexpression also recovered diamide-resistance in the ire1Δ mutant (Fig. 4C).
Similarly, KAR2 overexpression was able to partly suppress the temperature-sensitive (TS) growth defect of the ire1Δ and hxl1Δ mutants. As previously reported [7], the hxl1Δ mutant exhibited even greater TS growth defects than the ire1Δ mutant compared with WT (Fig. 4D), which is in stark contrast to the case of TM-sensitivity. These data indicate that Hxl1 has an Ire1-independent role in controlling thermotolerance. At 37°C, KAR2 overexpression significantly restored the normal growth of the ire1Δ mutant but only slightly restored the growth of the hxl1Δ mutant (Fig. 4D). Growth recovery of the hxl1Δ mutant by KAR2 overexpression was evident at 35°C (Fig. 4D). At 39°C, however, KAR2 overexpression did not rescue the growth defects of ire1Δ and hxl1Δ mutants (Fig. 4D).
Related to the ER stress response and thermotolerance, the UPR pathway is required for maintenance of cell wall integrity in C. neoformans. Therefore, both ire1Δ and hxl1Δ mutants are highly sensitive to cell wall destabilizing agents, such as Congo red (CR) and calcofluor white (CFW). KAR2 overexpression partly restored CR- and CFW-resistance in the ire1Δ and hxl1Δ mutants, indicating that Kar2 is also involved in the maintenance of cell wall integrity. Interestingly, however, KAR2 overexpression recovered CR/CFW-resistance more efficiently in the ire1Δ mutant than in the hxl1Δ mutant (Fig. 4E). Taken together Kar2 has crucial roles in the ER stress response, thermotolerance, and maintenance of cell wall integrity downstream of the Ire1/Hxl1-mediated UPR pathway in C. neoformans.
Kar2 controls genotoxic stress response in an Ire1-dependent, but Hxl1-independent, manner
Genotoxic stress is likely to cause ER stress indirectly because DNA damage leads to production of truncated or mutated proteins, which could accumulate as misfolded or unfolded toxic proteins in the ER. Supporting this idea, the ire1Δ and hxl1Δ mutants were hypersensitive to methyl methanesulfonate (MMS), which is a DNA alkylating agent that causes DNA mutagenesis by base mispairing and replication blocking [19], compared to WT (Fig. 5A). Verifying the result, reintegration of IRE1 or HXL1 restored the WT levels of MMS- resistance (Fig. 5A). Overexpression of KAR2 partly restored MMS-resistance in the ire1Δ mutant, but not in the hxl1Δ mutant, suggesting that Kar2 is involved in the genotoxic stress response downstream of Ire1 (Fig. 5A). Similarly, the ire1Δ mutant exhibited a greater sensitivity to hydroxyurea (HU), which inhibits DNA replication by blocking ribonucleotide reductase [20], than WT whereas the hxl1Δ mutant showed only a slightly increased sensitivity to HU (Fig. 5B). KAR2 overexpression partly recovered the HU-resistance in the ire1Δ and hxl1Δ mutants (Fig. 5B).
Figure 5. Kar2 mediates genotoxic stress responses downstream of the UPR pathway.
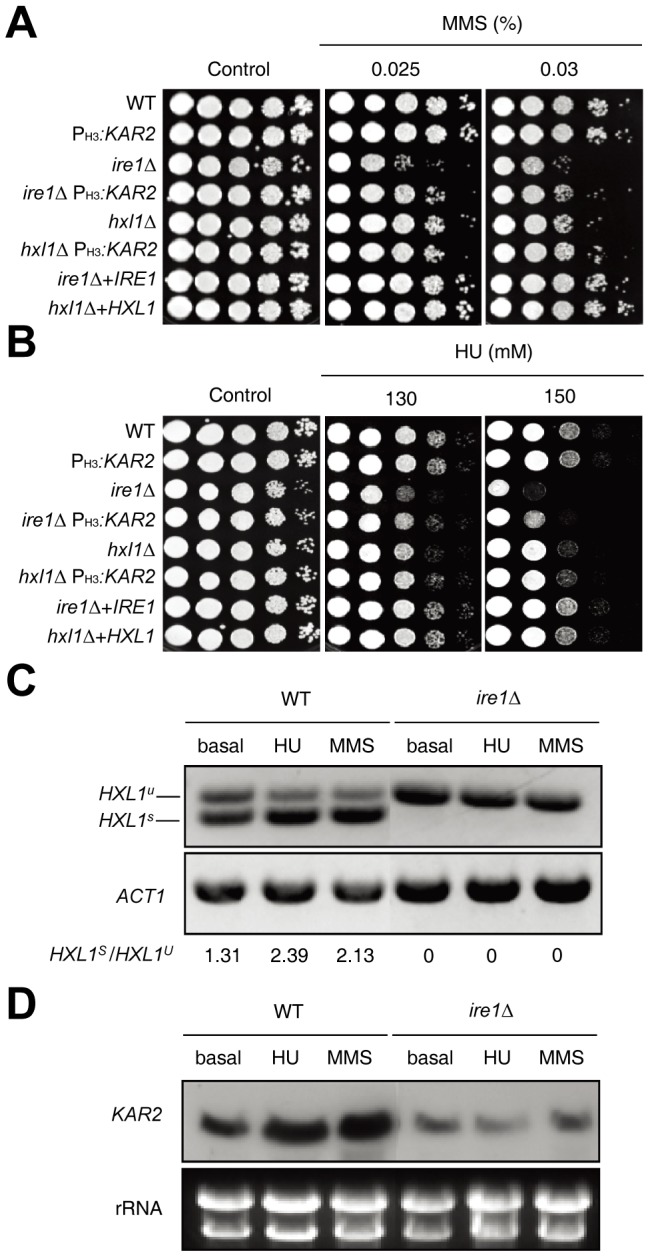
(A and B) Cells [the WT H99 strain, ire1Δ and hxl1Δ mutants, ire1Δ+IRE1 and hxl1Δ+HXL1 complemented strains, and PH3 :KAR2 strains (YSB1751, YSB1741, and YSB1745)] were spotted on a YPD agar medium containing the indicated concentrations of DNA damage inducers, including methyl methanesulfonate (MMS; A) or hydroxyl urea (HU; B), incubated at 30oC for 2–4 days, and photographed. (C) The RT-PCR analysis of UPR-induced HXL1 splicing was performed with cDNA samples prepared from total RNA samples of the WT H99 strain and ire1Δ mutants treated with HU (90 mM) or MMS (0.03%) for 1 h. (D) Using the total RNA set from (C), Northern blot assay was performed to measure KAR2 induction levels in the WT H99 strain and ire1Δ mutants. The Northern blot membrane was hybridized with the KAR2 specific probe, washed, and developed.
To test the hypothesis that genotoxic stress causes ER stress, which could activate UPR pathway and induces KAR2 expression, we performed RT-analysis of HXL1 splicing and Northern blot assay to monitor KAR2 induction treated with the two DNA damaging agents, MMS and HU. Interestingly, the unconventional splicing event in the HXL1 mRNA was enhanced in WT when treated with HU or MMS (Fig. 5C). Furthermore, the treatment of HU or MMS induced KAR2 expression in WT strain, but not in the ire1Δ (Fig. 5D), indicating that the HU or MMS treatment activates the KAR2 induction in the Ire1/Hxl1-dependent manner.
Therefore, genotoxic stress causes ER stress and activates the UPR pathway, partly through the Kar2-dependent manner.
Kar2 controls azole drug susceptibility downstream of the UPR pathway in Erg11- and Erg3-independent manners in C. neoformans
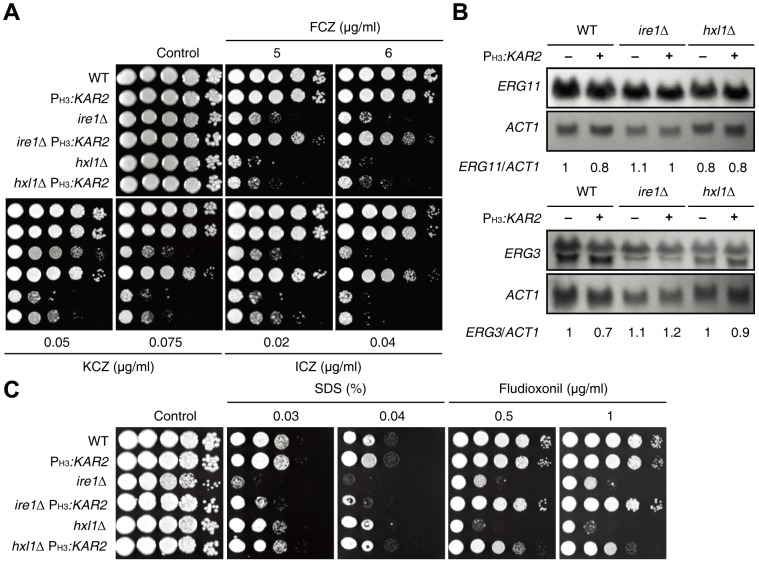
Ergosterol is the major sterol found in the membrane of fungi and has critical roles in controlling membrane stability and fluidity [21]. Since ergosterol is replaced by cholesterol in the human cell membrane, ergosterol or its synthesis pathway has been exploited as the major target for modulating fungal infection. For example, a polyene class of amphotericin B binds to ergosterol and produces pores in the fungal membrane, which lead to a lack of ions such as potassium. The azole class of drugs directly inhibits ergosterol synthesis from lanosterol at different steps [22]–[24]. Since the ER is the place where sterol biosynthesis occurs, the UPR pathway could be related to ergosterol biosynthesis and antifungal drug resistance. In fact, UPR signaling mutants are significantly susceptible to azole drugs [6], [7], [9]. In C. neoformans, both ire1Δ and hxl1Δ mutants are hypersensitive to azole drugs, such as fluconazole, ketoconazole, and itraconazole [7] (Fig. 6A), indicating that Ire1 and Hxl1 may have redundant and discrete roles in controlling azole resistance. Here, we determined whether Kar2 is involved in azole resistance downstream of the UPR pathway. Surprisingly, the overexpression of KAR2 partly but significantly restored the azole resistance of the ire1Δ mutant, but only slightly restored the azole resistance of the hxl1Δ mutant (Fig. 6A). Similar to the cell wall stress response, KAR2 overexpression recovered the azole drug resistance more efficiently in the ire1Δ mutant than in the hxl1Δ mutant at low levels of the azole drug. These data further support the hypothesis that Ire1 and Hxl1 control azole resistance in different ways and that the role of Ire1 in azole resistance mainly depends on Kar2.
Figure 6. Kar2 has a role in azole drug resistance downstream of the UPR pathway without affecting ERG11 and ERG3 expression.
(A) WT H99 strain, ire1Δ and hxl1Δ mutants, and PH3 :KAR2 strains (YSB1751, YSB1741, and YSB1745) were grown for 16 hr at 30oC in a liquid YPD medium, 10-fold serially diluted and spotted on a YPD agar medium containing the indicated concentrations of azole drugs and photographed. (B) The expression levels of ERG11 and ERG3 in strains described in (A). Each membrane was hybridized with an ERG11 or ERG3-specific probe, washed, and developed. Subsequently, the same membrane was stripped, subjected to re-hybridization with the ACT1-specific probe, washed, and developed. Expression levels of ERG11 or ERG3 were quantitatively measured with a phosphorimager and normalized with those of ACT1. Each KAR2/ACT1 is a value relative to that of the WT strain set to 1.0. (C) Strains described in (A) were grown for 16 hr at 30°C in a liquid YPD medium, 10-fold serially diluted, spotted on a YPD agar medium containing the indicated concentrations of SDS and fludioxonil, and photographed after incubation for 3 days.
Transcriptome analysis in A. fumigatus revealed that the expression levels of ergosterol biosynthesis genes, such as ERG11 and ERG3, in ire1Δ and hac1Δ mutants are lower than those in WT [14]. This finding led us to examine the expression patterns of the ergosterol synthesis genes in the WT strain and ire1Δ and hxl1Δ mutants with or without KAR2 overexpression by Northern blot analysis in C. neoformans. In stark contrast to the results in A. fumigatus, the expression levels of ERG11 and ERG3 were not significantly affected by the ire1Δ and hxl1Δ mutations in C. neoformans (Fig. 6B). Furthermore, KAR2 overexpression did not significantly change the expression levels of ERG11 and ERG3 (Fig. 6B). All of these data suggest that the role of the UPR pathway in azole resistance is mainly independent of the regulation of ERG11 or ERG3 expression in C. neoformans.
Given the above, it remains to be answered how the UPR pathway is involved in azole resistance, partly through the Kar2 molecular chaperone, in C. neoformans. Accumulation of toxic sterol intermediates by treatment with azole disrupts the membrane integrity [25], [26]. Therefore, there is a possibility that mutation of the UPR pathway may disrupt membrane stability, which may synergize with azole treatment for antifungal activity. Supporting this idea, there is a report that Hsp90, a molecular chaperone, suppresses azole susceptibility of cells by stabilizing a catalytic subunit of calcineurin that is required for azole tolerance [25]. To address this possibility, we tested the membrane stability of the UPR mutants by using SDS (sodium dodecyl sulfate), which is an ionic detergent, disrupting cell membrane stability. The ire1Δ mutant exhibited increased sensitivity to SDS and overexpression of KAR2 restored the SDS-resistance in the ire1Δ mutant (Fig. 6C). Interestingly, however, the hxl1Δ mutant was almost as susceptible or only slightly more susceptible to SDS as that of WT. We tested another membrane destabilizer, fludioxonil. Fludioxonil is a phenylpyrrole antifungal drug and hyperactivates Hog1 MAP kinase (MAPK) to induce over-accumulation of intracellular glycerol, which increases intracellular turgor pressure and indirectly affects cell membrane stability, resulting in defective cytokinesis and cell swelling [27]. KAR2 overexpression suppressed the increased fludioxonil susceptibility in both the ire1Δ and hxl1Δ mutants. Interestingly, similar to the azole drug test, KAR2 overexpression rescued fludioxonil-resistance more efficiently in the ire1Δ mutant than in the hxl1Δ mutant (Fig. 6C).
Taken together, the UPR pathway appears to be involved in azole resistance by controlling membrane stability, partly through the Kar2 molecular chaperone, but not by affecting the expression of ergosterol biosynthesis genes, such as ERG11 and ERG3, in C. neoformans.
Azole treatment is sensed as ER stress and activates the UPR pathway and induction of KAR2 in C. neoformans
As an additional explanation for the role of the UPR pathway in azole susceptibility, it is possible that azole drugs may cause ER stress, which could activate the UPR pathway and induction of KAR2 to counteract their effects. To test this hypothesis, we examined whether azole treatment could activate the UPR pathway. As a first hallmark for the UPR activation, the Ire1-mediated Hxl1 splicing event was monitored in cells treated with fluconazole (Fig. 7A). Surprisingly, the unconventional splicing event in the HXL1 mRNA significantly increased upon fluconazole treatment (Fig. 7A). Because such an HXL1 splicing event was not present in the ire1Δ mutant, the azole-mediated HXL1 mRNA splicing clearly depends on the Ire1 kinase (Fig. 7A).
Figure 7. Azole treatment induces the HXL1 unconventional splicing event in the UPR pathway and upregulation of KAR2 in C. neoformans.
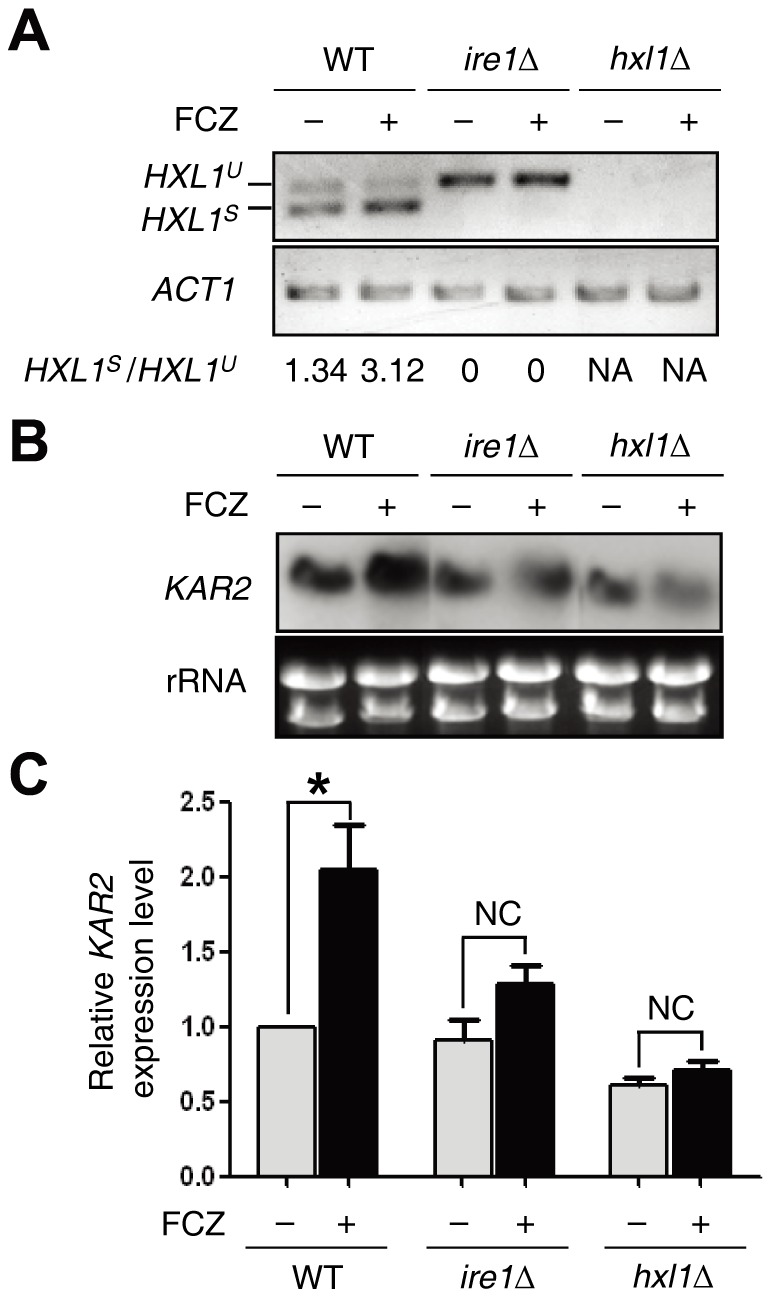
(A) The RT-PCR analysis of UPR-induced HXL1 splicing was performed with cDNA samples prepared from total RNA samples of the WT H99 strain and ire1Δ and hxl1Δ mutants treated with or without fluconazole (FCZ, 10 μg/ml) for 1h. NA, not available. (B and C) Using the same total RNA set of (A), Northern blot assay and qRT-PCR analysis were performed to monitor KAR2 induction levels. For quantitative qRT-PCR analysis, KAR2 expression levels were normalized with ACT1 as a control. Relative KAR2 expression levels indicate the ratio of the normalized KAR2 expression level of each strain with or without FCZ (10 µg/ml) to that of WT H99 strain at zero time point without FCZ. RT-PCR of HXL1 and ACT1, qRT-PCR analysis, and Northern blot analysis were performed with gene-specific primers or probes as described in the Materials and Methods.
As a second hallmark for the UPR activation, we examined whether the azole treatment could induce the expression of KAR2. Northern blot analysis revealed that the treatment of fluconazole slightly induced KAR2 expression in WT strain, but not in the ire1Δ or hxl1Δ mutant (Fig. 7B). This result was further confirmed by quantitative reverse transcription-PCR (qRT-PCR) analysis (Fig. 7C), indicating that the azole treatment activates the KAR2 induction in the Ire1/Hxl1-dependent manner. Taken together, these data suggest that the activation of the UPR pathway is required for counteracting ER stress caused by azole treatment in C. neoformans.
KAR2 is not involved in Ire1-mediated capsule regulation in C. neoformans
The capsule is one of the key virulence factors in C. neoformans because it prevents cells from being phagocytized by macrophages. Capsule production is regulated by the iron concentration and physiological CO2 levels [28]. A polysaccharide capsule is secreted by Sec4 or Sec6-mediated exocytosis into the extracellular space [29], [30]. Our previous study reported that the ire1Δ mutant, but not the hxl1Δ mutant, is highly defective in capsule production [7]. To address whether Kar2 is involved in Ire1-mediated capsule biosynthesis, we compared the capsule production levels between the ire1Δ mutant and the ire1Δ PH3:KAR2 strain. Overexpression of KAR2 did not rescue capsule defects in the ire1Δ mutant in both qualitative (Fig. 8A) and quantitative measurement (Fig. 8B). This indicates that Ire1 in the UPR pathway is involved in capsule production in an Hxl1- and Kar2-independnet manner or KAR2 overexpression is not sufficient to restore the capsule production in the ire1Δ mutant without concomitant expression of other factors.
Figure 8. Kar2 is not sufficient for Ire1-mediated regulation of capsule production in C. neoformans.
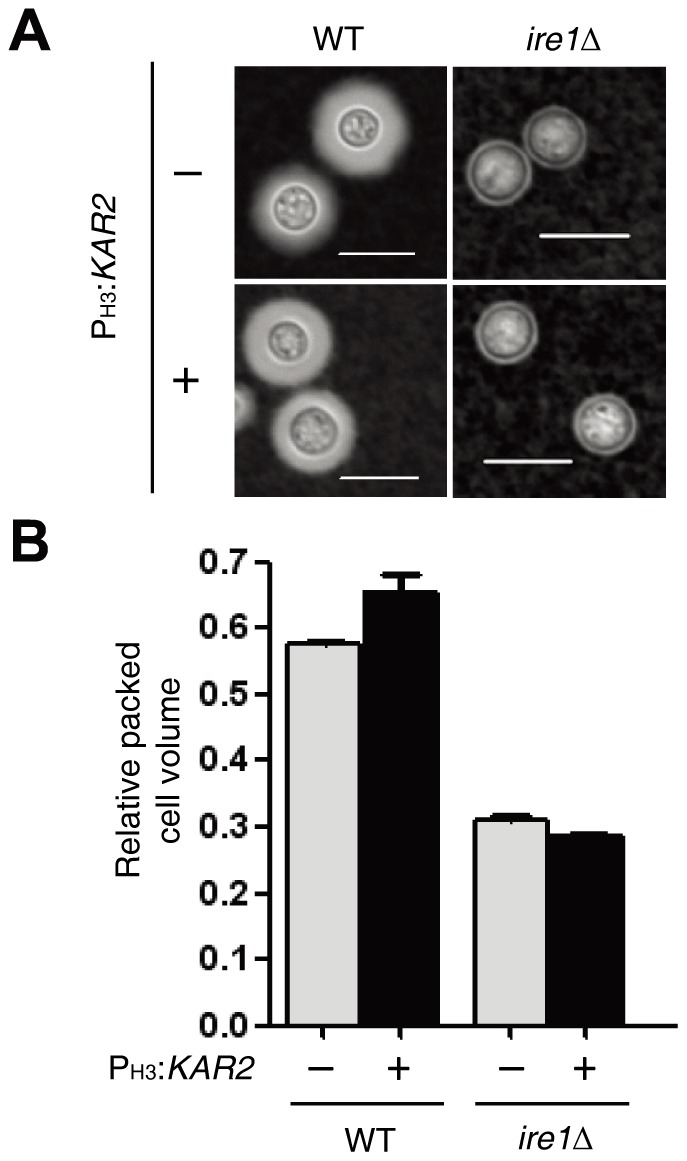
To measure the capsule production, each strain [WT H99 strain, the ire1Δ mutant (YSB552), and PH3 :KAR2 strains (YSB1741 and YSB1751)] was spotted and cultured on a DME agar medium at 30oC for 2 days. Capsules were visualized by India ink staining (A), and the relative capsule volume was measured by calculating the ratio of the length of the packed cell volume phase per length of the total volume phase (B). Three independent experiments with technical triplicates were performed. The scale bar represents 10 µm. Error bars represent the standard deviation.
Discussion
Cellular functions of the ER-resident molecular chaperone BiP/Kar2, and its connection to the UPR signaling pathway, have been characterized in models including budding yeast and animal cells, but not in basidiomycete fungi. In this study, we for the first time functionally characterized a Kar2/BiP protein in the UPR pathway in a basidiomycete fungus, C. neoformans. Our previous study revealed that expression of KAR2 is induced by the evolutionarily conserved Ire1 kinase and a unique Hxl1 transcription factor under ER stress induced by TM or a temperature upshift in C. neoformans [7]. During preparation of this manuscript, the function of Kar2 in sexual differentiation of C. neoformans was reported [31]. However, the roles of Kar2 in the UPR pathway as a molecular chaperone remain unexplored in C. neoformans.
The requirement of Kar2 for cell survival has been investigated in opportunistic fungal pathogens as well as budding yeast [10], [11], [31]. Our study also demonstrated that Kar2 is essential for the viability of C. neoformans, as it is for other fungi. Conditional null kar2 mutant strains exhibited severe growth defects under repressed conditions (Fig. 2C). This result is in agreement with a previous study proposing that Kar2 is essential for the viability of C. neoformans [31]. Recent studies have reported that Kar2 has a wide range of roles including translocation, protein folding, and nuclear fusion [10]–[14]. Protein translocation, especially, is essential for localization of secreted or folded proteins to their correct sites during the secretory process. Impairment of protein folding and secretion leads to an accumulation of toxic proteins, which results in cell death. Therefore, Kar2 is likely to be involved in key cellular processes for the survival of eukaryotes.
In this study, several lines of evidence demonstrated that Kar2 acts as one of the downstream effectors of the UPR signaling pathway to counteract ER stress, high temperature stress, and cell wall destabilizing stress. First, the overexpression of KAR2 by the H3 promoter partly restored resistance to ER stress, high temperature, and cell wall destabilizing stress in the ire1Δ and hxl1Δ mutants (Fig. 4). Interestingly, however, overexpression of KAR2 recovered cell wall stress resistance in the ire1Δ mutant more efficiently than in the hxl1Δ mutant, further supporting the idea that Ire1 and Hxl1 do not strictly have a linear relationship in the UPR pathway. Second, the abundance of KAR2 mRNA is controlled by Ire1 kinase and Hxl1 transcription factor [7]. KAR2 expression is induced by ER stress or a temperature upshift in both Ire1- and Hxl1-dependent manners [7] (Fig. 3C). In conclusion, Kar2 operates downstream of the Ire1 and Hxl1 in the UPR pathway to control ER stress, high temperature growth, and maintenance of cell wall integrity.
The finding that KAR2 overexpression restores DTT resistance more efficiently than TM resistance in the ire1Δ and hxl1Δ mutants implies that Kar2 is better suited as a molecular chaperone for resolving unfolded proteins generated by the perturbed redox state of the ER rather than by a lack of N-glycosylation-dependent folding capacity. In the ER, non-glycoproteins undergo proper folding through protein disulfide isomerase (PDI) and Kar2/BiP, whereas glycoproteins undergo N-glycan-dependent protein folding mediated by PDI and the Calnexin cycle [32]. Given that the ER has an oxidized environment that assists in efficient disulfide bond formation for protein folding [33], treatment with a reducing agent, such as DTT, significantly affects normal protein folding in the ER. Therefore, it is conceivable that accumulated misfolded non-glycoproteins by DTT treatment could be efficiently resolved by KAR2 overexpression. In contrast, TM inhibits the first step of N-linked glycosylation by blocking the transfer of the N-acetylglucosamine-1-phosphate (GlcNAc-1-P) group of UDP-GlcNAc to dolichol-p, which subsequently blocks PDI/Calnexin-mediated N-glycan-dependent protein folding and causes the accumulation of unfolded proteins [34]. Therefore, KAR2 overexpression may have only a limited role in resolving the TM-mediated protein misfolding unless proper N-linked glycosylation is provided. Similar to DTT, treatment with diamide, a diazine compound inducing the formation of disulfide bonds, may also generate unnaturally folded proteins in the ER, which could be efficiently resolved by KAR2 overexpression. In summary, our data indicate that the Kar2 molecular chaperone is more suitable for dealing with misfolded or unfolded proteins resulting from a change in the redox state of the ER rather than for the quality control of glycoproteins in the ER.
One of the notable findings in this study is that the UPR pathway controls the genotoxic stress response partly through Kar2. It is not surprising that the UPR pathway is involved in defense against genotoxic stress because DNA damage leads to the production of truncated or mutated proteins, which could accumulate as misfolded or unfolded toxic proteins in the ER. In fact, the involvement of the UPR pathway in the genotoxic stress response has been reported in other organisms [35]. It is possible that Kar2 proteins prevent misfolded proteins caused by genotoxic stress from aggregating by acting as a molecular chaperone. In S. cerevisiae, strains having defects in KAR2 induction become greatly sensitive to increased expression of mutated carboxypeptidase Y (CPY*), which is widely used as a model misfolded protein [36]. It was not unexpected to find that Kar2 overexpression only partly suppressed genotoxic sensitivity in the ire1Δ mutant because Ire1 must have other downstream effector(s), other than Kar2, to counteract genotoxic stresses. Furthermore the fact that the ire1Δ mutant is much more susceptible to genotoxic agents than the hxl1Δ mutant indicates that Ire1 may control genotoxic stress response in both Hxl1-dependent and independent manners. These data provide further support for the idea that Ire1 has both Hxl1-independent and dependent roles in C. neoformans.
The striking role of the UPR pathway in azole susceptibility is of clinical importance, but its mode of action remains puzzling. Previously, we have shown that the inhibition of the UPR pathway greatly increases azole susceptibility in C. neoformans, suggesting that the signaling components of the UPR pathway, such as Ire1 and Hxl1, could be excellent antifungal drug targets for combination therapy with azole drugs [7]. Particularly because Hxl1 is a unique transcription factor, which is phylogenetically distinct from the Xbp1 transcription factor in humans, it is an attractive antifungal drug target. Askew and colleagues reported a similar finding in A. fumigatus. The A. fumigatus strain with a hacA gene deletion, which encodes a yeast Hac1 ortholog, is more susceptible to azoles (e.g. ICZ and FCZ) and polyene (e.g. amphotericin B, AMB) than the WT strain [9]. More recently, they proposed a potential mechanism explaining how the UPR pathway mutants are involved in azole susceptibility [6]. Feng et al. performed DNA microarray analysis and found that the expression of ERG11, encoding a lanosterol 14α-demethylase that is a target of most azole drugs, decreases in the hacA and ireA mutants. Supporting this finding, cellular ergosterol levels were also shown to decrease in the UPR mutants [6]. The explanation remains elusive, however, for why the hacA mutant is highly susceptible to AMB in A. fumigatus [9] given that the decreased ergosterol content could confer resistance to AMB, which binds ergosterol. Probably multiple reasons exist for the increased susceptibility of the UPR mutants to both azole and polyene drugs in A. fumigatus. Interestingly, the hac1Δ mutant in S. cerevisiae is as resistant to azole drugs as the wild-type. Therefore, the role and regulatory mechanisms of the UPR pathway in azole resistance appears to be highly divergent between fungal species.
This study provides a potential mechanism for the role of the UPR pathway in azole susceptibility in C. neoformans. Under unstressed condition, the UPR pathway is not involved in ergosterol biosynthesis, given the fact that the expression levels of ERG11 and ERG3 are not significantly affected by mutations in IRE1 or HXL1 (Fig. 6B). Supporting this, the UPR mutants of C. neoformans that are highly susceptible to most azole drugs are also susceptible to AMB [7]. Treatment with azole drugs, however, may confer an ER stress to cells, which the UPR pathway must be activated to counteract. Supporting this, it was discovered that the Ire1-mediated Hxl1 splicing increases in response to FCZ treatment (Fig. 7). Furthermore, KAR2 expression was found to be induced by FCZ treatment in the Ire1/Hxl1-dependent manner. These findings may explain why KAR2 overexpression considerably recovers normal azole resistance in the ire1Δ mutant without affecting the ERG11 and ERG3 expression levels. Nevertheless, Ire1 and Hxl1 appear to differentially control azole resistance in C. neoformans because restoration of azole resistance by KAR2 overexpression is not as efficient in the hxl1Δ mutant as in the ire1Δ mutant. In addition to this defensive role of the UPR pathway in azole treatment, the requirement of the UPR pathway for maintaining membrane stability may also contribute to the synergism with azole treatment for antifungal activity.
Currently the nature of the ER stress by azole treatment still remains elusive. In eukaryotes, sterols are synthesized in the ER and then transported to the plasma membrane (PM) mainly independent of a classical secretory pathway. Sterols are highly enriched in the PM but their concentration is low in the ER membrane [37]. Although it is not clear how the inhibition of sterol biosynthesis by azole drugs activates the UPR pathway and Kar2/BiP induction, accumulation of ER membrane cholesterol induces ER stress and apoptosis in mammals and the UPR pathway regulates expression of genes involved in lipid metabolism in S. cerevisiae [5], [38], [39]. Furthermore, it has been reported that disruption of ER function induces lipid dysregulation [40] and vice versa, lipotoxicity triggered by lipid imbalance leads ER stress, which activates the UPR pathway [41]. Therefore, it is conceivable that perturbation of sterol/lipid metabolism by azole drug treatment may cause ER stress and activate the UPR pathway in C. neoformans. Functional correlation between the UPR and sterol/lipid metabolic pathways needs to be further investigated in future studies.
Thus far, most of the Ire1-dependent phenotypes, including the ER stress response, thermotolerance, maintenance of cell wall/membrane integrity, genotoxic stress response, and antifungal drug resistance, have been found at least partly to depend on the functions of the Kar2 molecular chaperone, except in the case of capsule production. KAR2 overexpression did not restore capsule production defects in the ire1Δ mutants at all. Therefore, the role of Ire1 in capsule synthesis appears to be independent of Hxl1 and Kar2. In fact, Ire1 may not directly affect capsule biosynthesis per se, but control the secretion of polysaccharide capsular precursors onto the cell surface. In yeast, it is known that the UPR pathway is involved in the secretory pathway. Our previous study also showed that some of the secretion-related genes, including SEC61, are regulated by the UPR pathway [7]. However, the exact regulatory mechanism of Ire1 in capsule production remains to be elucidated further in future studies.
In conclusion, the molecular chaperone Kar2/BiP has pleiotropic roles in cell viability, ER stress response, thermotolerance, maintenance of cell wall/membrane integrity, genotoxic stress response, and azole drug resistance, but not in capsule production, downstream of the Ire1/Hxl1-dependent UPR signaling pathway in C. neoformans.
Materials and Methods
Strains and growth conditions
C. neoformans strains used in the study are listed in Table 1 and were cultured on a yeast extract-peptone-dextrose (YPD) medium. For capsule production assay, the agar-based Dulbecco Modified Eagle (DME, Invitrogen, Carlsbad, CA) medium was used [42], [43].
Table 1. Strains used in this study.
| Strain | Genotype | Parent | Reference |
| C. neoformans | |||
| H99 | MATα | [49] | |
| YSB552 | MATα ire1Δ::NAT-STM#224 | H99 | [7] |
| YSB723 | MATα hxl1Δ::NAT-STM#295 | H99 | [7] |
| YSB1000 | MATα ire1Δ::NAT-STM#224 IRE1-NEO | YSB552 | [7] |
| YSB762 | MATα hxl1Δ::NAT-STM#295 HXL1-NEO | YSB723 | [7] |
| YSB1637 | MATα PCTR4:KAR2 NAT | H99 | This study |
| YSB1638 | MATα PCTR4:KAR2 NAT | H99 | This study |
| YSB1639 | MATα PCTR4:KAR2 NAT | H99 | This study |
| YSB1640 | MATα PCTR4:KAR2 NAT | H99 | This study |
| YSB1741 | MATα ire1Δ::NAT-STM#224 P H3 :KAR2 NEO | YSB552 | This study |
| YSB1744 | MATα ire1Δ::NAT-STM#224 P H3 :KAR2 NEO | YSB552 | This study |
| YSB1745 | MATα hxl1Δ::NAT-STM#295 P H3 :KAR2 NEO | YSB723 | This study |
| YSB1746 | MATα hxl1Δ::NAT-STM#295 P H3 :KAR2 NEO | YSB723 | This study |
| YSB1751 | MATα P H3 :KAR2 NEO | H99 | This study |
| YSB1752 | MATα P H3 :KAR2 NEO | H99 | This study |
Each NAT-STM# indicates the Natr marker with a unique signature tag.
Rapid amplification of cDNA ends (RACE) analysis of 5′ and 3′ untranslated regions (UTRs) of the KAR2 gene
To characterize 5′ and 3′ untranslated regions (UTRs) and coding sequence of the KAR2 genes, we performed rapid amplification of cDNA ends (RACE) analysis with GeneRacer Kit (Invitrogen, Carlsbad, CA). The total RNA of the WT H99 strain incubated overnight at 30°C was isolated with RiboEX (GeneAll, Korea) according to the manufacturer's instructions. Each 5′ and 3′ RACE products of KAR2 were cloned into the pTOP-V2 (Enzynomics) and sequenced. The 5′ and 3′ UTR and coding sequences of KAR2 have been deposited in GenBank (accession number JX982102).
Construction of the PCTR4:KAR2 and PH3:KAR2 strains
To replace the native KAR2 promoter with the copper-regulated CTR4 promoter, we generated a KAR2 promoter replacement cassette as follows. The 5′-flanking region of KAR2 and the 3′-flanking region of KAR2 were amplified for homologous recombination by PCR with primer pairs B3551 (5′-CGTAGGGTATGTCTCTGATGAG-3′)/B3650 (5′-CACTCGAATCCTGCATGCAAAGTCTTGAGGAATAGACAA-3′) and B3651 (5′-CGACAACGACTTCACCAATCTGCCACCATGGCATACCCT-3′)/B3652 (5′-ACTCCTGTTTGCCACTTCG-3′), respectively. B354 (5′-GCATGCAGGATTCGAGTG-3′) and B355 (5′-GATTGGTGAAGTCGTTGTCG-3′) primers were used for PCR-amplification of the NAT-CTR4 promoter using pNAT-CTR4-2 as a template [18]. The KAR2 promoter replacement cassette was produced by double joint PCR with primer pairs, B3551/B1455 (5′-AACTCCGTCGCGAGCCCCATCAAC-3′) for the 5′-franking region and B3652/B1454 (5′-AAGGTGTTCCCCGACGACGAATCG-3′) for the 3′-franking region, and the WT H99 strain was biolistically transformed, as previously described [44], [45]. The correct insertion was confirmed with Southern blot analysis, as previously described [46].
To replace the native KAR2 promoter with the histone H3 promoter, the PH3:KAR2 cassette was constructed as follows. Primers B3551 and B4270 (5′-CACTCGAATCCTGCATGCGGTGGCAAAAGTCTTGAGGA-3′) for the 5′-flanking region of the KAR2 gene and primers B4264 (5′-CAAGACCTCAAAGACACCG-3′) and B4271 (5′-ACCACAACACATCTATCACATGGCATACCCTTCAAGAAT-3′) for the exon of the KAR2 gene were used in the first round PCR. The 5′- and 3′-regions of the dominant selectable NEO marker (neomycin/G418-resistant marker) were amplified with primer pairs B4017 (5′-GCATGCAGGATTCGAGTG-3′)/B1887 (5′-ATTGTCTGTTGTGCCCAG-3′) and B4018 (5′-GTGATAGATGTGTTGTGGTG-3′)/B1886 (5′-TGGAAGAGATGGATGTGC-3′), respectively. Next, the 5′ and 3′ regions of the NEO-marked histone H3 promoter cassette were amplified by double joint PCR with primer pairs B3551/B1887 and B4271/B1886, respectively. The PH3:KAR2 strains were generated by introducing the NEO-marked H3 promoter cassette into C. neoformans serotype A H99 strain and ire1Δ and hxl1Δ mutants by biolistic transformation, as previously described [44], [45]. Stable transformants selected on the YPD medium containing G418 were screened by diagnostic PCR with primers B3550 (5′-TCCCAATCTACTGACCTATCG-3′) and B79 (5′-TGTGGATGCTGGCGGAGGATA-3′). Next, the correct genotypes were verified by Southern blot analysis, as previously described [46]. A probe for the KAR2 gene was amplified with primers B3551 and B3555 (5′- CAAGCAGGGACAGTAACAAC-3′).
Total RNA isolation, Northern blot assay, and qRT-PCR analysis
To evaluate KAR2 expression levels and patterns, total RNA isolation and Northern blot or qRT-PCR were performed as follows. Strains were cultured in a 30 ml YPD liquid medium for 16 hr at 30°C. Then 5 ml of overnight culture was inoculated into 50 ml of fresh YPD medium, further incubated at 30°C until the optical density at 600 nm (OD600) of the culture medium reached approximately 1.0, and then pelleted by centrifugation for total RNA isolation. To monitor expression levels of KAR2 in WT and PH3:KAR2 strains in response to TM, the overnight culture was inoculated into 150 ml of fresh YPD medium and incubated at 30°C up to an OD600 of 1.0. For the zero time sample, 50 ml out of the 150 ml culture was sampled and pelleted by centrifugation. The remaining 100 ml culture was treated with the indicated concentrations of TM. During incubation, a 50 ml culture was sampled at 30 and 60 min. Total RNA was isolated by the TRIzol reagent (RiboEx) as previously described [47]. Northern blot analysis was performed with 10 μg of total RNA from each strain. Electrophoresis, membrane transfer, hybridization, and washing were performed by following the protocol previously described [46]. A probe for the KAR2 gene was amplified with primers B4978 (5′- AGGCAGTCTGGAGTGTCATC-3′) and B3652. The qRT-PCR for quantitatively measuring relative expression level of KAR2 was performed with gene specific primers B679 (5′-CGCCCTTGCTCCTTCTTCTATG-3′)/B680 (5′-GACTCGTCGTATTCGCTCTTCG-3′) for ACT1, as a reference, and B5253 (5′-CTCTGAGGACGACAAGGACA-3′)/B5254 (5′-AGCTCAGAAAGCTGCTCCTC-3′) for KAR2.
Stress sensitivity test
Each strain was incubated overnight (about 16 hr) at 30°C in a liquid YPD medium, washed, serially diluted (1 to 104 dilutions) with dH2O, and spotted (3 µl) onto a solid YPD medium containing the indicated concentration of stress inducers. To test ER stress and cell wall stress, cells were spotted onto a solid YPD medium containing the indicated concentration of ER stress inducers, such as tunicamycin (TM, Sigma) or dithiothreitol (DTT, Sigma), cell wall stress inducers, such as calcofluor white (CFW, Sigma) or congo red (CR, Sigma), or cell membrane destabilizers, such as sodium dodecyl sulfate (SDS, Sigma) and fludioxonil (Sigma). To examine antifungal drug resistance, azole drugs including fluconazole, ketoconazole, and itraconazole were used. For genotoxic DNA damage stress, cells were spotted onto a solid YPD medium containing 130 and 150 mM hydroxyurea (HU, Sigma) and 0.02, 0.025, and 0.03% methylmethan sulfonate (MMS, Sigma). To test thermosensitivity, cells were incubated at 30, 35, 37, and 39°C and photographed after 2 to 3 days.
Monitoring HXL1 splicing event
To monitor splicing levels of HXL1 in WT and PH3:KAR2 strains, samples were prepared as follows. Strains were cultured in a 50 ml YPD liquid medium for 16 hr at 30°C. Then the overnight culture was inoculated at 1:20 dilution into a fresh 150 ml YPD medium and incubated at 30°C until the OD600 of the culture medium reached approximately 1.0. For the zero time sample, 50 ml out of the 150 ml culture was sampled. The remaining 100 ml culture was treated with indicated concentrations of TM, DTT, fluconazole, MMS, and HU. During incubation, a 50 ml culture was sampled at 30 and/or 60 min. Total RNAs were isolated with the TRIzol reagent (RiboEx) as previously described. Single strand cDNA was synthesized using a reverse transcriptase (Fermentas). RT-PCR of HXL1 and ACT1 was performed with gene specific primers C19 (5′-CACTCCATTCCTTTCTGC-3′)/C20 (5′- CGTAACTCCACTGTGTCC-3′) and B3294 (5′-GCACCATACCTTCTACAATGAG-3′)/B3295 (5′-ACTTTCGGTGGACGATTG-3′), respectively.
Capsule test
For the capsule assay, cells were incubated overnight at 30°C in a liquid YPD medium, spotted onto agar-based DME medium, and further incubated for 2 days at 30°C. For quantitative measurement of capsule production, the relative packed cell volume was measured with hematocrit capillary tubes, as previously described [46], [48]. For visualization of capsule production, each cell scraped from the DME medium was resuspended in a phosphate buffer saline (PBS) buffer, and stained with India ink, and visualized by Nikon eclipse Ti microscope.
Acknowledgments
We would like to thank Seon-Ah Cheon for critically reading this manuscript.
Funding Statement
This work was supported by the National Research Foundation of Korea grants (no. 2010-0029117 and no. 2008-0061963) from MEST. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529. [DOI] [PubMed] [Google Scholar]
- 2. Romisch K (2005) Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol 21: 435–456. [DOI] [PubMed] [Google Scholar]
- 3. Cox JS, Walter P (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404. [DOI] [PubMed] [Google Scholar]
- 4. Sidrauski C, Walter P (1997) The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039. [DOI] [PubMed] [Google Scholar]
- 5. Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, et al. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258. [DOI] [PubMed] [Google Scholar]
- 6. Feng X, Krishnan K, Richie DL, Aimanianda V, Hartl L, et al. (2011) HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus . PLoS Pathog 7: e1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheon SA, Jung KW, Chen YL, Heitman J, Bahn YS, et al. (2011) Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans . PLoS Pathog 7: e1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, et al. (2008) Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans . Fungal Genet Biol 45: 1235–1247. [DOI] [PubMed] [Google Scholar]
- 9. Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, et al. (2009) A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus . PLoS Pathog 5: e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morrow MW, Janke MR, Lund K, Morrison EP, Paulson BA (2011) The Candida albicans Kar2 protein is essential and functions during the translocation of proteins into the endoplasmic reticulum. Curr Genet 57: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J (1989) S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell 57: 1223–1236. [DOI] [PubMed] [Google Scholar]
- 12. Rose MD, Misra LM, Vogel JP (1989) KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57: 1211–1221. [DOI] [PubMed] [Google Scholar]
- 13. Vogel JP, Misra LM, Rose MD (1990) Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol 110: 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T (2001) Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol 153: 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Gemeren IA, Punt PJ, Drint-Kuyvenhoven A, Broekhuijsen MP, van't Hoog A, et al. (1997) The ER chaperone encoding bipA gene of black Aspergilli is induced by heat shock and unfolded proteins. Gene 198: 43–52. [DOI] [PubMed] [Google Scholar]
- 16. Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K (2000) Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 279: 445–450. [DOI] [PubMed] [Google Scholar]
- 17. Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, et al. (2010) BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8: e1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JW, Ko YJ, Kim SY, Bahn YS (2011) Multiple roles of Ypd1 phosphotransfer protein in viability, stress response, and virulence factor regulation in Cryptococcus neoformans . Eukaryot Cell 10: 998–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beranek DT (1990) Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res 231: 11–30. [DOI] [PubMed] [Google Scholar]
- 20. Rittberg DA, Wright JA (1989) Relationships between sensitivity to hydroxyurea and 4-methyl-5-amino-1-formylisoquinoline thiosemicarbazone (MAIO) and ribonucleotide reductase RNR2 mRNA levels in strains of Saccharomyces cerevisiae . Biochem Cell Biol 67: 352–357. [DOI] [PubMed] [Google Scholar]
- 21. Sturley SL (2000) Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim Biophys Acta 1529: 155–163. [DOI] [PubMed] [Google Scholar]
- 22. Ghannoum MA, Rice LB (1999) Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev 12: 501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Akins RA (2005) An update on antifungal targets and mechanisms of resistance in Candida albicans . Med Mycol 43: 285–318. [DOI] [PubMed] [Google Scholar]
- 24. Odds FC, Brown AJ, Gow NA (2003) Antifungal agents: mechanisms of action. Trends Microbiol 11: 272–279. [DOI] [PubMed] [Google Scholar]
- 25. Cowen LE (2008) The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol 6: 187–198. [DOI] [PubMed] [Google Scholar]
- 26. Cowen LE, Steinbach WJ (2008) Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot Cell 7: 747–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kojima K, Bahn YS, Heitman J (2006) Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans . Microbiology 152: 591–604. [DOI] [PubMed] [Google Scholar]
- 28. Aksenov SI, Babyeva IP, Golubev VI (1973) On the mechanism of adaptation of micro-organisms to conditions of extreme low humidity. Life Sci Space Res 11: 55–61. [PubMed] [Google Scholar]
- 29. Rodrigues ML, Djordjevic JT (2012) Unravelling secretion in Cryptococcus neoformans: more than one way to skin a cat. Mycopathologia 173: 407–418. [DOI] [PubMed] [Google Scholar]
- 30. Zaragoza O, Fries BC, Casadevall A (2003) Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2 . Infect Immun 71: 6155–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee SC, Heitman J (2012) Function of Cryptococcus neoformans KAR7 (SEC66) in Karyogamy during Unisexual and Opposite-Sex Mating. Eukaryot Cell 11: 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, et al. (2007) The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci U S A 104: 11676–11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Csala M, Margittai E, Banhegyi G (2010) Redox control of endoplasmic reticulum function. Antioxid Redox Signal 13: 77–108. [DOI] [PubMed] [Google Scholar]
- 34. Elbein AD (1987) Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem 56: 497–534. [DOI] [PubMed] [Google Scholar]
- 35. Henry KA, Blank HM, Hoose SA, Polymenis M (2010) The unfolded protein response is not necessary for the G1/S transition, but it is required for chromosome maintenance in Saccharomyces cerevisiae . PLoS ONE 5: e12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hsu CL, Prasad R, Blackman C, Ng DT (2012) Endoplasmic reticulum stress regulation of the Kar2p/BiP chaperone alleviates proteotoxicity via dual degradation pathways. Mol Biol Cell 23: 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sullivan DP, Ohvo-Rekila H, Baumann NA, Beh CT, Menon AK (2006) Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast. Biochem Soc Trans 34: 356–358. [DOI] [PubMed] [Google Scholar]
- 38. Feng B, Yao PM, Li Y, Devlin CM, Zhang D, et al. (2003) The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol 5: 781–792. [DOI] [PubMed] [Google Scholar]
- 39. Maxfield FR, Tabas I (2005) Role of cholesterol and lipid organization in disease. Nature 438: 612–621. [DOI] [PubMed] [Google Scholar]
- 40. Colgan SM, Hashimi AA, Austin RC (2011) Endoplasmic reticulum stress and lipid dysregulation. Expert Rev Mol Med 13: e4. [DOI] [PubMed] [Google Scholar]
- 41. Brookheart RT, Michel CI, Schaffer JE (2009) As a matter of fat. Cell Metab 10: 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bahn YS, Hicks JK, Giles SS, Cox GM, Heitman J (2004) Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot Cell 3: 1476–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hicks JK, D'Souza CA, Cox GM, Heitman J (2004) Cyclic AMP-dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans . Eukaryot Cell 3: 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, et al. (2002) A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148: 2607–2615. [DOI] [PubMed] [Google Scholar]
- 45. Kim MS, Kim SY, Yoon JK, Lee YW, Bahn YS (2009) An efficient gene-disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochem Biophys Res Commun 390: 983–988. [DOI] [PubMed] [Google Scholar]
- 46. Jung KW, Kim SY, Okagaki LH, Nielsen K, Bahn YS (2011) Ste50 adaptor protein governs sexual differentiation of Cryptococcus neoformans via the pheromone-response MAPK signaling pathway. Fungal Genet Biol 48: 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ko YJ, Yu YM, Kim GB, Lee GW, Maeng PJ, et al. (2009) Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell 8: 1197–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, et al. (2002) Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans . Eukaryot Cell 1: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perfect JR, Ketabchi N, Cox GM, Ingram CW, Beiser CL (1993) Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol 31: 3305–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]