Abstract
Effector functions of inflammatory IL-17–producing Th (Th17) cells have been linked to autoimmune diseases such as experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis (MS). However, what determines Th17 cell encephalitogenicity is still unresolved. Here, we show that after EAE induction, mice deficient for the NF-κB regulator MALT1 (Malt1–/– mice) exhibit strong lymphocytic infiltration in the CNS, but do not develop any clinical signs of EAE. Loss of Malt1 interfered with expression of the Th17 effector cytokines IL-17 and GM-CSF both in vitro and in vivo. In line with their impaired GM-CSF secretion, Malt1–/– Th cells failed to recruit myeloid cells to the CNS to sustain neuroinflammation, whereas autoreactive WT Th cells successfully induced EAE in Malt1–/– hosts. In contrast, Malt1 deficiency did not affect Th1 cells. Despite their significantly decreased secretion of Th17 effector cytokines, Malt1–/– Th17 cells showed normal expression of lineage-specific transcription factors. Malt1–/– Th cells failed to cleave RelB, a suppressor of canonical NF-κB, and exhibited altered cellular localization of this protein. Our results indicate that MALT1 is a central, cell-intrinsic factor that determines the encephalitogenic potential of inflammatory Th17 cells in vivo.
Introduction
CD4+ Th cells can be categorized into 3 major subsets that make complementary contributions to immunity. Th1 cells predominantly mediate cellular immunity and are characterized by their production of the signature cytokine IFN-γ. Th2 cells mainly support humoral immunity and secrete IL-4, IL-5, and IL-13 (1). Th17 cells are key inflammatory drivers and are characterized by their production of IL-17A, IL-17F, IL-21, IL-22, TNF, and GM-CSF (2–4). In particular, Th17 cells are regarded as the principal cell type responsible for the induction of EAE, an important mouse model of the human disease MS (5–7). The development of EAE is markedly impaired in mice that lack expression of IL-17, the IL-17 receptor (IL-17R), or GM-CSF, establishing these cytokines as the major encephalitogenic mediators in EAE (3–5, 8–10).
Differentiation of Th17 cells in vitro requires TGF-β in combination with IL-6 or IL-21 (5, 11, 12). IL-23 is thought to promote terminal differentiation of Th17 cells and triggers an encephalitogenic program that is closely associated with GM-CSF secretion (3, 4, 13). In contrast, IL-2 constrains Th17 cell differentiation (14). At the transcriptional level, Th17 cell differentiation requires the functions of a specific set of transcription factors that includes RORα (encoded by Rora), RORγt (encoded by Rorc), IRF4 (encoded by Irf4), STAT3 (encoded by Stat3), and Runx (encoded by Runx1) (15–19).
MALT1 plays an important role in peripheral T cell activation and proliferation (20, 21). TCR engagement leads to the formation of an oligomeric CBM complex, which contains the CARMA1 (CARD11), B cell lymphoma-10 (BCL10), and MALT1 proteins and is required for activation of downstream canonical NF-κB signaling (22). In addition to this scaffolding function, MALT1 has paracaspase activity that cleaves specific protein substrates, which then facilitate T cell activation (23–25). Although the CBM complex and its functions are dispensable for T cell development (26), their role in Th cell differentiation and specific Th cell subset functions is less clear. It is known that CARMA1 influences Th1 and Th2 responses and the development of natural Tregs (27–29). In the present study, we provided evidence that the CBM component MALT1 is essential for development of encephalitogenic Th17 cells during EAE induction.
Malt1–/– Th17 cells showed normal expression of all lineage-specific transcription factors and successfully infiltrated the CNS, but were nonpathogenic and produced low levels of IL-17 and GM-CSF. The noncanonical NF-κB subunit RelB was cleaved and thus inactivated in WT Th17 cells, but not in Malt1–/– Th17 cells, and was constitutively localized in the nucleus. Our findings indicate that MALT1 represents a central signal integrator for inflammatory responses mediated by Th17 cells.
Results
Malt1–/– mice are resistant to EAE, despite lymphocytic infiltration of the CNS.
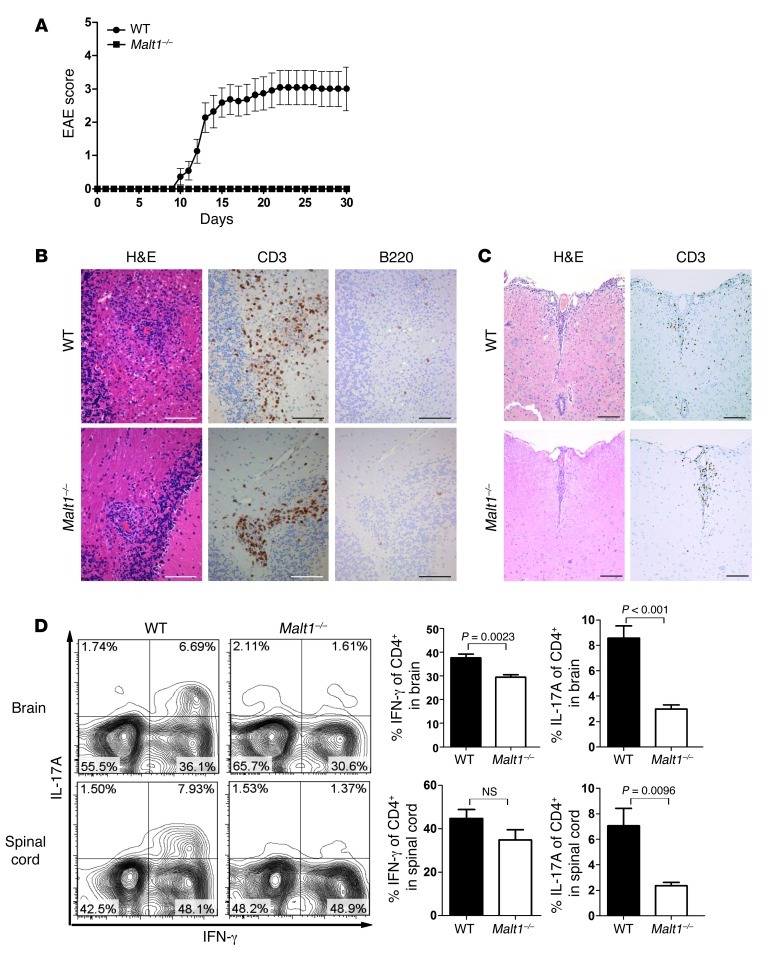
NF-κB is an important regulator of lymphocyte effector functions (22), but precisely how different Th cell subsets are controlled by this pathway is unclear. Among the Th subsets, inflammatory Th17 and Th1 cells have been reported to be important for EAE. To investigate the in vivo role of the NF-κB pathway in these subsets, we used immunization with myelin oligodendrocyte glycoprotein (MOG) peptide plus injection of pertussis toxin (PT) to induce EAE in WT and Malt1–/– mice (15). All WT mice showed signs of severe EAE by 30 days after induction, whereas no Malt1–/– mouse showed any signs of EAE (Figure 1A). Histopathological analyses at 30 days after MOG immunization showed dense immune cell infiltrates in the CNS tissue in both WT and Malt1–/– mice (Figure 1, B and C). There was no obvious disparity in T cell infiltrates observed in WT and Malt1–/– brains, but the distribution of these infiltrates exhibited striking differences. The white matter of WT brains contained a perivascularly centered, diffuse, widespread infiltrate consisting mainly of CD3+ T cells (Figure 1B). In contrast, in Malt1–/– brain tissue, most T cells were located in very close proximity to blood vessels. As expected, we did not detect any B cell infiltrates in WT or Malt1–/– brains (Figure 1B). Infiltrating cells were also clearly visible in the spinal cords of WT and Malt1–/– mice, although the differences between the genotypes were less pronounced (Figure 1C).
Figure 1. Malt1–/– mice are resistant to EAE induction.
(A) Malt1–/– (n = 9) and WT (n = 11) mice were immunized s.c. with MOG peptide in CFA, followed by i.p. injection of PT. Disease severity was scored daily. Data are the mean score ± SEM and are representative of 2 independent experiments. (B and C) Representative histopathological analyses of cross-sections of (B) brain and (C) spinal cord from mice in A at 30 days after MOG injection. Brain and spinal cord sections were stained with H&E (B and C) or immunostained to detect CD3+ T cells (B and C) or B220+ B cells (B). Scale bars: 100 μm. Data are from 1 mouse per group, representative of 2 independent experiments. (D) CNS-infiltrating CD4+ T cells from brains and spinal cords of Malt1–/– and WT mice were isolated by gradient centrifugation on day 14 after EAE induction. Expression levels of IL-17A and IFN-γ were analyzed by intracellular staining and flow cytometry. Data are gated on live CD4+ T cells. Also shown is quantitation of percent brain- and spinal cord–infiltrating CD4+ T cells producing IFN-γ and IL-17. Data are mean ± SEM (n = 4 per group).
We next assessed the functionality of the Th cells infiltrating the brains and spinal cords of MOG-immunized WT and Malt1–/– mice. At 14 days after EAE induction, we isolated CNS-infiltrating lymphocytes by density gradient centrifugation. Cells isolated from Malt1–/– brains and spinal cords showed a dramatic decrease in the percentage of infiltrating Th cells that secreted IL-17A (Figure 1D). In contrast, IFN-γ levels were comparable to those of controls in spinal cord Malt1–/– Th cells and were only marginally reduced in brain-infiltrating Malt1–/– Th cells. Interestingly, the majority of Th cells isolated from WT spinal cord or brain produced both IL-17A and IFN-γ, whereas double IL-17A/IFN-γ–producing Th cells were not detected in Malt1–/– mice (Figure 1D).
MALT1 is dispensable for Th1 cell priming, but required for Th17 cell priming.
Th17 cells are the major inflammatory cells driving EAE (6). The lack of IL-17 production by the CNS-infiltrating Th cells in our Malt1–/– mice subjected to EAE induction prompted us to investigate the influence of MALT1 on Th cell differentiation.
We isolated naive CD4+CD62L+ T cells from LNs and spleens of Malt1–/– and WT mice and primed them by stimulation for 72 hours in vitro with anti-CD3 plus anti-CD28 Abs. In addition, naive CD4+CD62L+ T cells were treated with specific cytokine cocktails to induce Th1 cell, Th2 cell, Th17 cell, or Treg differentiation. For Th1 cells, primed cultures received IL-12 with or without IL-2. As determined by IFN-γ production, Th1 cell generation was not altered in the absence of MALT1 (Figure 2A), consistent with the normal IFN-γ levels we found in vivo (Figure 1D). As expected, IL-17 was not present in cultures of either WT or Malt1–/– Th1 cells (Figure 2A). In addition, MALT1 deficiency did not impair secretion of IL-4 in Th2 cell cultures or expression of the Treg-specific transcription factor Foxp3 in Treg cultures (Supplemental Figure 1, A and B).
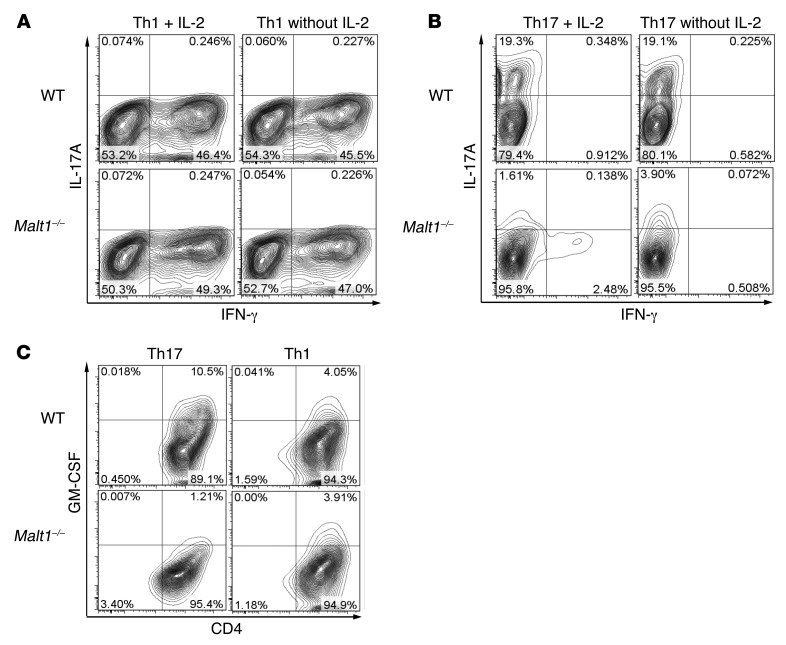
Figure 2. MALT1 is essential for effector cytokine production by Th17-polarized cells in vitro.
Th1 and Th17 cell subsets were generated from naive T cells of Malt1–/– and WT mice by priming with anti-CD3 and anti-CD28 and treating with subset-specific cytokines for 72 hours in the presence (A–C) or absence (A and B) of IL-2. Th cells were restimulated for 6 hours with PMA/Iono, followed by intracellular staining and flow cytometry. (A and B) Analysis of (A) Th1 and (B) Th17 subsets to detect cells producing IL-17A and IFN-γ. (C) Analysis of Th1 and Th17 subsets to detect cells producing GM-CSF. Data are gated on live cells and are representative of 3 (A and B) or 6 (C) independent experiments.
Th17 cell differentiation of naive Malt1–/– and WT CD4+CD62L+ T cells was induced by addition of TGF-β, IL-6, and anti–IFN-γ Abs, with or without IL-2. Production of cytokines was determined by intracellular staining and flow cytometry (Figure 2B), RT-PCR analysis of cytokine mRNA expression (Supplemental Figure 1C), and ELISA analysis of culture supernatants (Supplemental Figure 1D). As expected, WT Th17 cells produced IL-17, but no IFN-γ, and IL-17 levels were markedly reduced in Malt1–/– Th17 cell cultures (Figure 2B). Loss of MALT1 had no effect on IL-22 secretion (Supplemental Figure 1D).
Malt1 deficiency has previously been shown to reduce T cell proliferation (20, 21). However, although addition of IL-2 restored proliferation of Malt1–/– Th17 cells to near-WT levels (Supplemental Figure 2A), it further reduced IL-17 production (Figure 2B). These results indicate that the decreases in proliferation and IL-17 production exhibited by Malt1–/– Th17 cells are not associated with each other. Differentiation of Th17 cells induced by IL-21 plus TGF-β yielded similar results (data not shown). Thus, in vitro, Malt1 deficiency interfered with Th17 cell generation.
GM-CSF is the main effector cytokine in Th17 cell–driven EAE (3, 4). Interestingly, as assessed by intracellular flow cytometry, Malt1–/– Th17 cells generated in vitro showed a dramatic drop in GM-CSF expression, whereas GM-CSF expression in Malt1–/– Th1 cells was low, but comparable to that in WT Th1 cells (Figure 2C). Taken together, these data indicate that Th1 cell, Th2 cell, and Treg differentiation in vitro is normal in the absence of MALT1, but that the production of IL-17 and GM-CSF by Th17 cells requires MALT1.
Genome-wide expression for WT and Malt1–/– Th1 and Th17 cells at 48 and 72 hours after priming, performed on Illumina whole mouse genome microarrays, confirmed the above results. An unsupervised hierarchical clustering (Supplemental Figure 2B) of probes with the most variable expression across all arrays showed Th1 and Th17 cells separating into distinct groups. Following this hierarchy, Th1 cells next formed time-dependent subclusters. Interestingly, within these subclusters, Th1 cells from both genotypes clustered together, indicative of overall similarity of their expression profiles. This was in contrast to Th17 cells, for which WT and Malt1–/– cells formed the first branch of different subclusters, followed by separation into the different time points (Supplemental Figure 2B). This indicates that overall, MALT1 seems to play a greater role in differentiation of Th17 cells compared with Th1 cells.
CNS-infiltrating Malt1–/– Th cells cannot produce GM-CSF to sustain neuroinflammation in vivo.
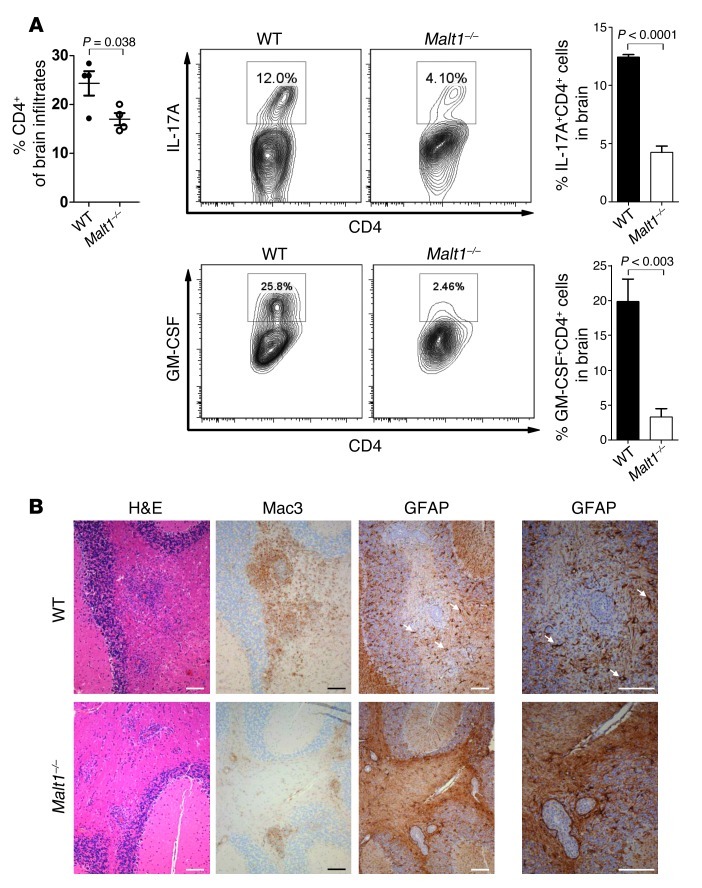
The decreased IL-17 production by Malt1–/– Th17 cells suggested that the cells’ effector functions are impaired. We therefore assessed the functionality of the Th cell subsets infiltrating the brains of MOG-immunized WT and Malt1–/– mice. At the WT disease peak, 20 days after EAE induction, we sacrificed WT and Malt1–/– mice and isolated CNS-infiltrating lymphocytes by density gradient centrifugation. At this time point, WT and Malt1–/– brains showed no appreciable differences in total infiltrating CD4+ T cells (Figure 3A). However, compared with WT brain, there were dramatic decreases in Malt1–/– brain in the percentages of infiltrating Th cells that secreted IL-17A (WT, 12.0%; Malt1–/–, 4.10%) or GM-CSF (WT, 25.8%; Malt1–/–, 2.46%; Figure 3A).
Figure 3. Malt1–/– Th cells do not produce GM-CSF in vitro or recruit macrophages.
(A) Left: CNS-infiltrating CD4+ T cells from brains of Malt1–/– and WT mice were isolated by gradient centrifugation on day 20 after EAE induction and quantitated. Middle: Expression levels of IL-17A and GM-CSF were analyzed by intracellular staining and flow cytometry. Data shown are gated on live CD4+ T cells. Right: Quantitation of percent of CNS-infiltrating CD4+ T cells producing IL-17A or GM-CSF. Data are mean ± SEM (n = 4). (B) Representative histopathological analyses of brain cross-sections from mice in Figure 1A at 30 days after MOG injection. Brain sections were stained with H&E or to detect Mac3 (macrophages) or GFAP (reactive astrocytes; white arrows). Scale bars: 100 μm. Data are from 1 mouse per group and are representative of 2 independent experiments.
The recruitment of bone marrow–derived myeloid cells has been associated with encephalitogenic Th17 cell responses and GM-CSF secretion (3, 5, 10, 12, 30, 31). Consistent with our observation of impaired IL-17A and GM-CSF secretion by CNS-infiltrating Malt1–/– Th17 cells, we detected a striking reduction in the number of activated macrophages/microglia in Malt1–/– brains. In contrast to the diffuse distribution of these Mac3+ cell infiltrates in WT brains, they were found only in close proximity to blood vessels in Malt1–/– brains (Figure 3B). Moreover, whereas WT brains showed large numbers of reactive GFAP+ astrocytes within the infiltrated areas (a nonspecific sign of severe CNS damage), hardly any GFAP+ astrocytes were detectable in Malt1–/– brains (Figure 3B). Thus, loss of MALT1 did not affect the capacity of Th cells to infiltrate brain tissue, but did compromise their effector functions and impair their ability to recruit other inflammatory cells. This lack of inflammatory cell infiltration could explain the resistance of Malt1–/– mice to EAE.
To consolidate these findings, we isolated brain-infiltrating cells on day 14 after EAE induction, a time point prior to the WT disease peak (day 20). In contrast to our day 20 results, Th cell infiltrates on day 14 were reduced in Malt1–/– versus WT brains (Supplemental Figure 3A), which suggests that Malt1–/– Th cells exhibit delayed kinetics. Consistent with our histological analysis, recruitment of inflammatory macrophages (F4/80hiMHCIIhi) to the CNS was significantly impaired in Malt1–/– mice (Figure 3B and Supplemental Figure 3, A and B). However, MALT1 deficiency did not alter the recruitment of granulocytes (Supplemental Figure 3A), which are important inflammatory mediators in EAE to the CNS (32).
Malt1–/– Th17 cells are not pathogenic in vivo.
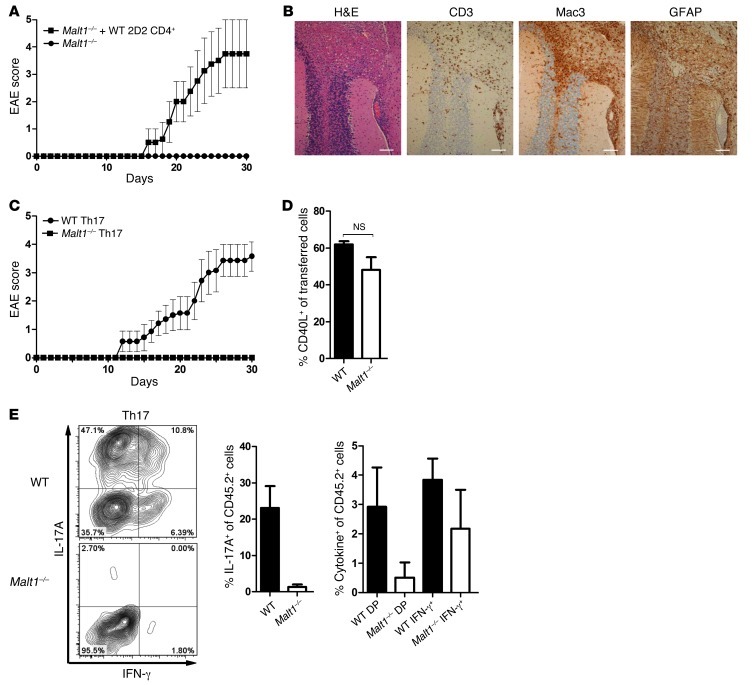
Our observations suggested that T cells are the main determinants of EAE resistance in Malt1–/– mice. We reinforced this contention by demonstrating that Malt1–/– bone marrow–derived DCs elicited normal T cell proliferation and cytokine secretion and showed no defects in the expression of molecules related to antigen presentation, including CD86 and MHCII (Supplemental Figure 4). To confirm our theory experimentally, we injected WT 2D2 CD4+ T cells into Malt1–/– mice and induced EAE. Consistent with our previous data, transfer of WT 2D2 T cells enabled EAE induction in the formerly resistant Malt1–/– mice (Figure 4A). Histopathological analyses of brain tissue from Malt1–/– mice injected with WT 2D2 CD4+ T cells revealed strong infiltration of immune cells (Figure 4B), with cellular infiltrate distribution similar to that in WT mice (Figure 1B). T cell infiltrates were diffuse and widespread, and Malt1–/– macrophage/microglia infiltration and/or activation was prominent (Figure 4B). Punctate GFAP staining of reactive astrocytes was also observed, indicative of extensive neuroinflammation. Thus, although other mechanisms could theoretically contribute, we believe the EAE resistance of Malt1–/– mice is caused primarily by impaired T cell responses.
Figure 4. Malt1–/– Th17 cells are not encephalitogenic.
(A) Malt1–/– mice were injected with 1 × 105 WT 2D2 CD4+ cells or PBS as control. EAE was initiated, and animals were monitored as in Figure 1A. Data (mean ± SEM) are representative of 2 independent experiments. (B) Histopathological analyses of CNS cross-sections from mice in A at 30 days after MOG injection, showing cerebellar white matter, cerebellar cortex, and leptomeningeal blood vessels. Brain sections were stained with H&E or immunostained to detect CD3 (T cells), Mac3 (macrophages), or GFAP (reactive astrocytes). Scale bars: 100 μm. Data are representative of 2 independent experiments. (C) EAE was initiated in Malt1–/– and WT mice by MOG injection. At 10 days after injection, CD4+ T cells from spleens and LNs were isolated and cultured for 5 days with MOG peptide, IL-23, and anti–IFN-γ before injection (2 × 107 cells/mouse) into WT recipients (n = 7 per group). Recipients were injected with PT on the day of T cell transfer and 2 days thereafter. Mice were monitored daily as in Figure 1A. (D and E) Cells generated as in C were injected into transgenic CD45.1+ WT recipients. At day 18 after injection, mice were sacrificed, and CD4+CD45.2+ T cells from spleens and LNs were analyzed for CD40L surface expression (D) and IFN-γ and IL-17A production (E) by intracellular flow cytometry. DP, double IFN-γ/IL-17A producers. Data are mean ± SEM (n = 6).
Based on the above-described dissection of Th cell populations, we reasoned that the EAE resistance of Malt1–/– mice might be due to their nonfunctional Th17 cells. To test this hypothesis in vivo, WT and Malt1–/– mice were injected with MOG plus PT to induce EAE and sacrificed on day 10 after immunization. Cells were isolated from spleens and draining LNs and expanded under Th1- or Th17-polarizing conditions. These MOG-specific Th1 and Th17 cells of Malt1–/– and WT origin were injected into WT recipients, and animals were monitored for signs of EAE. Injection of MOG-specific WT Th1 or WT Th17 cells resulted in severe EAE, as expected (Figure 4C and Supplemental Figure 5A). However, in accordance with our previous results, MOG-specific Malt1–/– Th17 cells failed to cause any disease in their WT hosts (Figure 4C), which indicates that Malt1–/– Th17 cells are not pathogenic. Surprisingly, transfer of MOG-specific Malt1–/– Th1 cells also did not elicit any signs of EAE (Supplemental Figure 5A). One potential interpretation of these results is that Malt1 deficiency causes a general decrease in antigen-specific Th cell generation. To explore this issue, we took advantage of the fact that CD40L upregulation can be used to identify antigen-specific T cells in EAE (33). However, we detected no differences in the frequencies of CD40L+ MOG-specific WT and Malt1–/– Th cells at day 18 after transfer (Figure 4D and Supplemental Figure 5B), which indicates that antigen-specific Th cell generation is not altered by Malt1 deficiency.
Plasticity between the Th1 and Th17 cell subsets in terms of overlapping cytokine production profiles is crucial in several diseases, including EAE (34, 35). To test whether Th1/Th17 plasticity is affected by Malt1 deficiency, we injected CD45.1 transgenic WT mice with MOG-specific CD45.2+ Th cells that were derived from WT or Malt1–/– mice and polarized in vitro toward the Th1 or Th17 subset. Mice were sacrificed at day 18 after injection and analyzed for frequencies of Th1 and Th17 cells. In mice receiving WT Th1 or Th17 cells, a small population producing both IFN-γ and IL-17A was detected (Figure 4E and Supplemental Figure 5C), indicative of Th1/Th17 cell plasticity. In contrast, in mice receiving Malt1–/– Th1 or Th17 cells, there were no Th cells with the capacity to produce both IFN-γ and IL-17A. This lack of double IFN-γ/IL-17A producers in the absence of MALT1 suggests a defect in Th1/Th17 cell plasticity that could explain the inability of MOG-specific Malt1–/– Th1 and Th17 cells to cause EAE in WT mice. In any case, these in vivo data clearly showed that Malt1–/– Th cells were not pathogenic and were the main determinants of EAE resistance in Malt1–/– mice. We therefore conclude that MALT1 is critical for the proinflammatory and encephalitogenic functions of WT Th cells in vivo.
MALT1 deficiency does not alter expression of transcription factors crucial for Th17 cell differentiation.
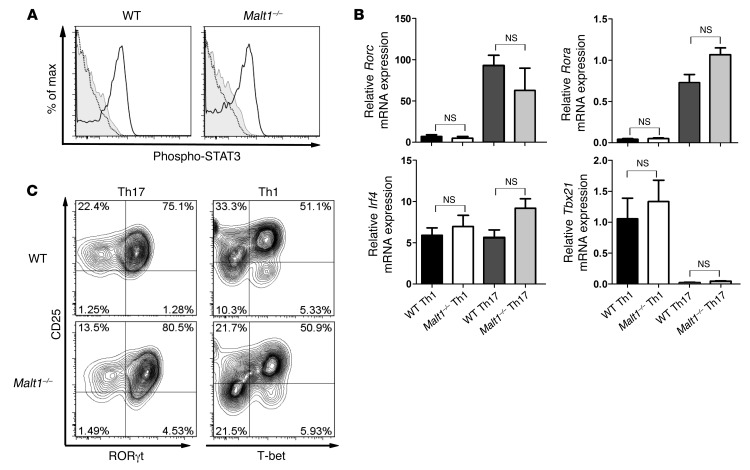
TGF-β and IL-6 are essential for Th17 cell development in vitro (11, 12). As shown in Supplemental Figure 1B, TGF-β–driven Treg induction was normal in the absence of MALT1, which suggests that TGF-β signaling is intact in these cells. IL-6 signaling triggers phosphorylation and activation of the transcription factor STAT3 (36). To determine whether defective IL-6 signaling caused a defect in Malt1–/– Th17 cell differentiation, we stimulated Malt1–/– and WT naive CD4+ T cells with IL-6 and monitored STAT3 phosphorylation by intracellular staining and flow cytometry. Phospho-STAT3 levels were comparable between Malt1–/– and WT CD4+ T cells (Figure 5A), which indicates that the dysfunctional Th17 cell differentiation of Malt1–/– naive T cells is not due to altered IL-6 signaling.
Figure 5. MALT1 does not interfere with expression of Th17-associated transcription factors.
(A) Malt1–/– and WT naive CD4+CD62L+ T cells were treated with IL-6 for 30 minutes (black line), and phospho-STAT3 levels were assessed by intracellular staining and flow cytometry. Gray line, isotype control; dotted line, without IL-6. Data are gated on live CD4+ T cells and representative of 3 independent experiments. (B) Levels of Th17 cell lineage–specific transcription factors were determined in Malt1–/– and WT Th1 and Th17 cells. Th cell differentiation was induced as described in Methods. After 16 hours of priming, mRNA levels of the indicated transcription factors were determined by RT-PCR. Results are expressed as the mRNA level normalized to Hprt and relative to that in naive WT Th cells. Data are mean ± SEM of 3 independent experiments. (C) In vitro differentiation of WT and Malt1–/– Th1 and Th17 cells was induced as in Figure 2. Expression of the indicated transcription factors was measured by intracellular flow cytometry. Data are gated on live CD4+ T cells and are representative of 4 independent experiments.
Th17 cells are characterized not only by their distinct cytokine secretion profile, but also by their expression of lineage-specific transcription factors. In particular, RORγt, RORα, and IRF4 have been implicated in Th17 cell differentiation (18). We used RT-PCR and intracellular flow cytometry to determine whether decreased expression of these transcription factors correlates with the inability of Malt1–/– CD4+ T cells to differentiate into Th17 cells. We primed Malt1–/– and WT naive CD4+CD62L+ Th cells in the presence of IL-6 plus TGF-β to induce Th17 cell differentiation. As controls, parallel cultures of Malt1–/– and WT CD4+ Th cells were treated with IL-12 and IL-2 to generate Th1 cells. As expected, Th1 cells of either Malt1–/– or WT origin expressed the Th1 lineage–specific transcription factor T-bet (encoded by Tbx21), but not RORγt or RORα (Figure 5, B and C). However, to our surprise, there were no significant alterations to the expression of Rorc, Rora, or Irf4 in WT or Malt1–/– CD4+ Th cells treated with Th17-polarizing cytokines (Figure 5, B and C). These results imply that Malt1 deficiency does not abrogate the initiation of Th17 cell differentiation per se, but might interfere with the development of Th17 cell effector functions.
MALT1 is highly expressed in Th17 cells and cleaves the NF-κB subunit RelB.
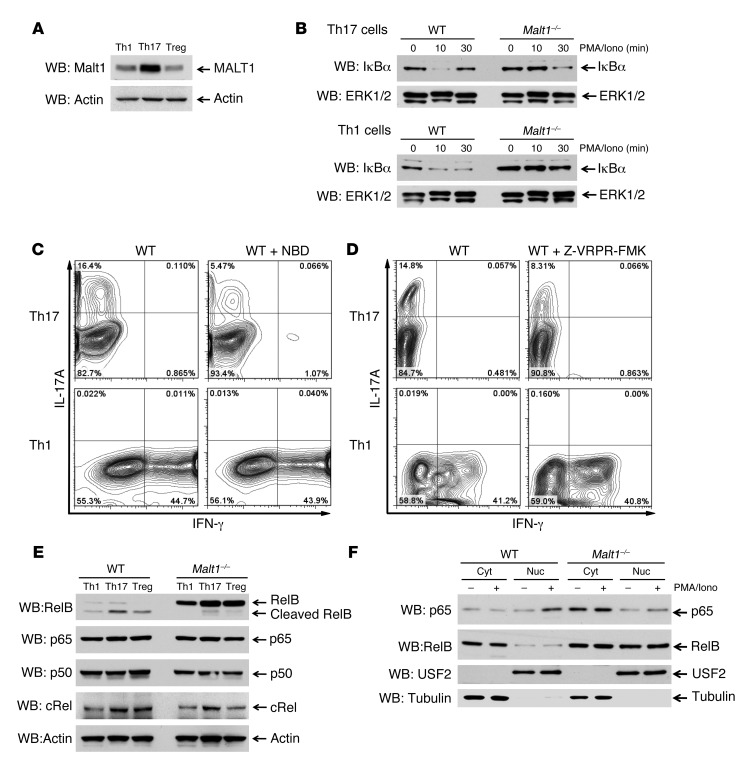
To gain more mechanistic insight into MALT1’s role in Th17 cell differentiation, we first measured MALT1 protein levels in various WT Th cell subsets. Surprisingly, MALT1 expression was substantially higher in Th17 cells than in Th1 cells or Tregs, as determined by immunoblotting (Figure 6A). Because we found that MALT1 influenced the differentiation of Th17 cells, but not Th1 cells, we speculated that canonical NF-κB signaling might not be of equal importance in these subsets. Although it is believed that MALT1’s structural components are generally important for IκBα degradation (21, 37), Th subset–specific differences in this involvement have yet to be investigated. We found that IκBα degradation in Malt1–/– Th17 cells was delayed but still clearly obvious at 30 minutes after restimulation with PMA plus ionomycin (PMA/Iono), whereas restimulated Malt1–/– Th1 cells showed almost no IκBα degradation (Figure 6B). In addition, restimulated WT Th17 cells showed almost complete IκBα degradation and faster resynthesis of the bona fide NF-κB target IκBα compared with restimulated WT Th1 cells. These results suggest that IKK activation is normally stronger in Th17 cells than in Th1 cells. Next, we induced the differentiation of naive WT T cells into Th1 or Th17 cells in the presence or absence of the pharmacological inhibitor Nemo-binding domain (NBD), which prevents IKK assembly and therefore blocks canonical NF-κB activation (38). Th17-polarized WT Th cells differentiating in the presence of NBD showed a drastic reduction in IL-17A secretion, whereas secretion of IFN-γ by Th1-polarized cells differentiating in the presence of NBD was not affected (Figure 6C). These data support our hypothesis that IKK signaling and canonical NF-κB activation is more important for Th17 cells than for Th1 cells.
Figure 6. MALT1 is highly expressed in Th17 cells and inactivates RelB.
In vitro differentiation of WT and Malt1–/– Th1 cells, Th17 cells, and Tregs was induced as described in Methods; cells were stimulated for 5 hours (unless otherwise indicated) with PMA/Iono. (A and B) Expression of the indicated proteins in lysates was analyzed by immunoblotting. Actin and ERK1/2 were loading controls. Data are representative of 5 (A) and 3 (B) independent experiments. (C and D) WT Th cells were induced to differentiate into Th1 or Th17 cells with or without (C) 10 μM NBD or (D) 50 μM Z-VRPR-FMK. After 6 hours of PMA/Iono restimulation, production of IL-17A and IFN-γ was measured by intracellular flow cytometry. Data are representative of 3 independent experiments. (E and F) WT and Malt1–/– cells induced to differentiate into Th1 cells, Th17 cells, or Tregs were restimulated for 1 hour with PMA/Iono. (E) Whole cell lysates were immunoblotted to detect the indicated proteins. (F) Lysates were subjected to subcellular fractionation, and cytosolic (cyt) and nuclear (nuc) fractions were analyzed by immunoblotting to detect the indicated proteins. USF2 served as a marker for nuclear localization; tubulin served as a marker for cytosolic localization. Data are representative of 3 independent experiments.
The paracaspase activity of MALT1 is important for NF-κB activation in antigen receptor–stimulated T cells (24, 25) and for the survival of an aggressive subset of diffuse large B cell lymphomas (39, 40). In contrast to MALT1’s scaffolding function, its protease activity is not crucial for IκBα degradation and acts downstream of IKK (41). We found that inhibition of MALT1’s paracaspase activity by the peptide caspase inhibitor Z-VRPR-FMK reduced IL-17A production by Th17-polarized WT Th cells, but had no effect on Th1 cell differentiation (Figure 6D). Thus, IKK-driven canonical NF-κB activation (Figure 6, B and C) and IKK-independent MALT1 protease activity (Figure 6D) were crucial for the differentiation of Th17 cells, but not Th1 cells. These data further suggest that Th17 cell differentiation is regulated by 2 independent pathways that both involve MALT1. A logical scenario might be that MALT1 paracaspase activity is important for cleaving negative regulators of Th17 cell differentiation
It was recently shown in a tumor cell line setting that MALT1 cleaves the noncanonical NF-κB subunit RelB (23), but the physiological role of this cleavage is not known. To analyze the RelB status during Th cell differentiation, we treated differentiating Th1 cells, Th17 cells, and Tregs with the protease inhibitor MG132 for 3 hours and analyzed RelB protein levels by immunoblotting. Interestingly, RelB levels were dramatically reduced in WT Th1 cells, Th17 cells, and Tregs (Figure 6E). In addition, the small amount of RelB present in these cells was already cleaved, and therefore primed for degradation. In contrast, RelB was highly expressed in Malt1–/– Th1 cells, Th17 cells, and Tregs and was not cleaved at all (Figure 6E), which indicates that RelB remains functional in these cells. Importantly, expression levels of the NF-κB subunits p65, p50, and cRel were not altered by Malt1 deficiency (Figure 6E).
Several studies have demonstrated that RelB inhibits the canonical NF-κB pathway by directly interfering with p65 (23, 42, 43). In cellular fractionation experiments, we found that p65 and RelB were reciprocally regulated in Malt1–/– Th17 cells (Figure 6F). In the absence of MALT1, only a minor fraction of p65 translocated to the nucleus upon stimulation. In sharp contrast, RelB was constitutively detected in cytosolic and nuclear fractions of Malt1–/– Th17 cells, whereas in WT Th17 cells, RelB expression was far lower and exclusively found in the cytosol (Figure 6F), which indicates that MALT1 also regulates RelB translocation. Taken together, these results show that IKK-induced canonical NF-κB activation and the paracaspase activity of MALT1 are important for Th17 cell differentiation. We conclude that RelB is inactivated during Th cell differentiation in WT cells, but not in Malt1–/– cells, and serves as a transcriptional repressor of canonical p65, which is of particular importance for the generation of Th17 cells.
Discussion
How Th17 cells are generated in vivo is the subject of much ongoing debate, particularly as to the identity of the cytokines involved. In vitro, IL-6, IL-21, and IL-23 — with or without TGF-β — have been reported to induce Th17 cell differentiation or stabilization (5, 11, 12, 44, 45). However, the roles of TCR stimulation and costimulatory signaling in the differentiation of specific Th subsets have yet to be clearly delineated (46–49). Engagement of both the TCR and CD28 results in activation of the IKK complex, which relieves NF-κB from its suppression by the IκB proteins. MALT1 is known to be an important modifier in the signaling pathway linking the TCR to NF-κB activation (22). In the present study, we revealed what we believe to be previously unknown functions for MALT1 and canonical NF-κB in the development of Th17 cells and their effector functions.
Our in vitro and in vivo studies showed that Malt1–/– mice exhibited normal differentiation of Th1 cells capable of normal IFN-γ production, but a profound reduction in the frequency of IL-17–producing Th cells. Malt1–/– mice were also completely resistant to the development of EAE, a disease linked to the effector functions of Th17 cells. However, previous research has established that IL-17 production alone is not sufficient for EAE induction (50). Several reports have shown that Th17 cell functionality is needed to recruit myeloid cells into the CNS (5, 10, 12, 30, 31) and that the GM-CSF produced by Th17 cells is the key driver of this recruitment and subsequent EAE onset (3, 4, 51). Indeed, in addition to their IL-17 deficit, we found that Malt1–/– Th17 cells produced much less GM-CSF than did WT Th17 cells, consistent with the substantially fewer inflammatory macrophages observed in histological sections of brains of Malt1–/– mice subjected to EAE induction. All these data point to a defective inflammatory Th17 cell response in the absence of MALT1, with the observed defects in IL-17 and GM-CSF production accounting for the EAE resistance of Malt1–/– mice.
The proliferation of peripheral T cells has been previously reported to depend on MALT1 (20, 21). In line with these studies, we found that Malt1–/– cells cultured under Th17-polarizing conditions proliferated only slightly less vigorously than did WT cells. In vivo, Malt1–/– Th cells infiltrated the CNS with delayed kinetics. At day 14 after EAE induction, T cell infiltration in Malt1–/– CNS was significantly reduced compared with WT animals, whereas at the disease peak (day 20), Malt1–/– CNS showed only a modest reduction in T cell infiltration. However, since these Malt1–/– mice never developed clinical signs of EAE, we can conclude that these Malt1–/– Th cells are no longer pathogenic, despite their near-normal expansion and CNS infiltration.
The differentiation of self-reactive T cells is a prerequisite for establishing autoimmunity. Our data suggest that the EAE resistance of Malt1–/– mice is mainly a T cell–intrinsic effect. Transfer of transgenic WT 2D2 CD4+ T cells into Malt1–/– mice allowed for EAE induction in the formerly resistant animals. Histological analysis indicated that WT 2D2 Th cells were able to recruit inflammatory Malt1–/– macrophages to the CNS, resulting in severe neuroinflammation. This result suggests that Malt1–/– mice do not have other defects that prevent EAE development and that MALT1 regulates Th17 cell functionality in a cell-autonomous fashion. We confirmed this hypothesis by transferring Malt1–/– Th17-polarized cells into WT recipients: the mutant Th cells could not induce EAE in their WT hosts. Moreover, Malt1–/– mice showed a normal frequency of antigen-specific T cells, ruling out a deficit in this capacity as the cause of the observed EAE resistance. Thus, we conclude that MALT1 is a central regulator of Th17 cell differentiation and determines the pathogenic potential of these cells. However, EAE resistance was also observed in mice receiving MOG-specific Malt1–/– Th1 cells. Our analyses established that a substantial proportion of WT Th17 cells and a small fraction of WT Th1 cells acquired the capacity to produce both IFN-γ and IL-17. However, this plasticity was completely lost in the absence of MALT1. Th1/Th17 plasticity is reportedly important in EAE (34, 35), which might explain the inability of Malt1–/– Th1 cells to induce the disease. This theory is further supported by the work of several others showing that mouse strains with impairments of Th1 or Th17 cells are resistant to EAE independent of the functionality of the other Th subset (9, 13, 16, 52), indicative of a tight connection within the different Th subsets.
The differentiation of naive Th cells into Th17 cells is governed by several lineage-specific transcription factors, including RORα, RORγt, and IRF4 (18). To our surprise, the expression of these factors was not influenced by Malt1 deficiency. Recent studies have suggested that c-Rel, which may act downstream of MALT1, controls Th17 cell differentiation by inducing RORγ and RORγt expression (53, 54). However, in our experimental system, MALT1 appeared to act independently of c-Rel, as we did not detect any changes in c-Rel protein levels or in RORγt expression in Malt1–/– Th cells. Importantly, we demonstrated that MALT1-driven NF-κB activity was essential for Th17 cell effector function. Although it is theoretically possible that NF-κB is a direct transcriptional activator of the Il17a gene, a previous computer-based analysis to detect putative NF-κB elements in the mouse Il17a locus identified only 1 nonfunctional sequence (53), making it unlikely that NF-κB is a direct activator of this gene.
Although IKK activity is crucial for IκBα degradation, the paracaspase activity of MALT1 is important for NF-κB activation, but not for IκBα degradation (22, 41). However, in our study, application of either an IKK inhibitor or a specific caspase inhibitor led to the same result: a block in Th17 cell differentiation. We propose that MALT1 independently regulates 2 pathways that influence NF-κB activation and are of equal importance for Th17 cell differentiation. Previous work in T cells has shown that the scaffolding function of MALT1 is important for IKK activation and IκBα degradation (21, 37). In contrast, in a tumor cell line–based model, the catalytic activity of MALT1 targeted the noncanonical NF-κB subunit RelB (23), which was identified as a transcriptional repressor of the canonical NF-κB proteins p65 and cRel (42, 43). We showed under physiological conditions that MALT1 negatively regulated RelB protein stability during Th cell differentiation. Malt1–/– Th cells showed dramatically higher RelB levels than did WT Th cells. More importantly, RelB was constitutively localized in the nucleus of Malt1–/– Th17 cells, where it could potentially block p65 or c-Rel. Thus, we propose that MALT1 regulates RelB’s localization in addition to its protein stability. It should be noted that both inhibitory and stimulatory functions have been described for RelB, depending on the cell type and context (55). In that respect, it is interesting that RelB is essential for IL-17 production by γδ T cells (56).
In conclusion, our present findings demonstrated an essential role for MALT1 in the differentiation of functional Th17 cells and the development of EAE. Importantly, we showed that MALT1 determines the encephalitogenicity of the Th17 cells driving EAE, which may position MALT1 as an attractive drug target for the treatment of human MS.
Methods
Mice.
Malt1–/– mice were originally generated in our laboratory (20, 37) and backcrossed for more than 10 generations into the C57BL/6 background. Rag1–/–, 2D2 transgenic, CD45.1 congenic, and C57BL/6 mice were from Jackson Laboratories.
CD4+ Th cell purification and differentiation conditions.
For in vitro Th cell differentiation assays, naive CD4+CD62L+ Th cells were isolated from spleens and LNs and sorted using a magnetic bead cell purification kit according to the manufacturer’s instructions (Miltenyi). For priming, enriched Th cells were stimulated for 72 hours with 1 μg/ml plate bound anti-CD3 Ab plus 1 μg/ml anti-CD28 Ab (both from BD Biosciences). Naive cells were induced to differentiate into Tregs by addition of 3 ng/ml recombinant human TGF-β (rhTGF-β; R&D), 50 U rhIL-2 (Peprotech), and 5 μg/ml anti–IFN-γ (BD Biosciences); into Th1 cells by addition of 4 ng/ml IL-12 (Peprotech) plus 50 U rhIL-2; or into Th2 cells by addition of 10 ng/ml IL-4 (Peprotech), 5 μg/ml anti–IFN-γ, and 50 U rhIL-2. After 72 hours of induction, Th2 cells were transferred from anti-CD3–coated plates to noncoated plates. The Th2 cultures were treated with an additional 50 U rhIL-2, and cells were rested for 2 days prior to use in experiments. For differentiation of Th17 cells, naive Th cells received 5 μg/ml anti–IFN-γ, 30 ng/ml rmIL-6 (Peprotech), 2 ng/ml rhTGF-β, and 50 U rhIL-2.
Isolation of lymphocytes.
On day 14 or 20 after EAE induction, mice were sacrificed, and their cardiovascular system was flushed with 20 ml PBS applied to the left ventricle. Brain and spinal cord were isolated and digested separately for 1 hour at 37°C in RPMI containing 50 μg/ml collagenase D (Roche) and 10 μg/ml DNAseI (Roche). Lymphocytes were enriched by centrifugation through a 2-layer Percoll gradient (40% and 70%) and washed twice in PBS before further analysis.
Cytokine production.
For analysis of cytokines by intracellular staining, T cells directly isolated from brain, spinal cord, or spleen or harvested after culture in vitro were washed twice with HBSS before stimulation for 6 hours with 50 nM PMA (Sigma-Aldrich) plus 750 nM Iono (Sigma-Aldrich) in the presence of Golgi Plug (1:1,000; BD Biosciences). Stimulated T cells were washed once in PBS containing 2% FBS, incubated with Fc Block (BD Biosciences) for 15 minutes, and surface stained for an additional 30 minutes with anti-CD4, anti-Gr1, anti-F4/80, anti-MHCII, anti-CD40L, anti-CD45.1, or anti-CD45.2 Abs (BD Biosciences). Permeabilization and intracellular cytokine staining to detect IL-17A, IFN-γ, IL-4, and GM-CSF were performed using Cytofix/Cytoperm kits (BD Biosciences) and to detect Foxp3, RORγt, and T-bet using specific staining kits (eBioscience) according to the manufacturers’ instructions. Flow cytometry was performed on a Canto II instrument (BD Biosciences) and analyzed using FlowJo 7.5 software (Tree Star).
For analysis of cytokines by quantitative real-time RT-PCR (qRT-PCR), RNA was isolated from cell pellets via TRIzol extraction. The iScript cDNA synthesis kit (Bio-Rad) was used for cDNA synthesis according to the manufacturer’s instructions. Sybrgreen master mix (ABI) and primers (Supplemental Table 1) were used for qRT-PCR, and products were run on an ABI 7500HT fast qRT-PCR instrument. Data were analyzed using the ΔΔCt method; accordingly, all data were normalized to Hprt transcription, and mRNA expression relative to the mean ΔCt value of 4 naive WT samples was calculated.
For quantitation of cytokines in culture supernatants, ELISAs were performed using the following kits, according to the manufacturers’ instructions: IL-17A (R&D); IL-22, IFN-γ (eBioscience).
Gene expression analysis.
RNA was extracted from Th1 and Th17 subpopulations of Malt1–/– and WT mice 48 and 72 hours after priming initiation using RNA purification kit and DNAse digestion (Qiagen) according to the manufacturer’s instructions. Processing and hybridization of RNA to Illumina Mouse Whole Genome (version 2, release 3) arrays was performed at the University Health Network Microarray Facility according to standard protocols (57). Data from the arrays were quantified in Illumina’s GenomeStudio software (version 2010.2) and passed quality metrics using the Bioconductor package Lumi (CITE ref 1) in R (version 2.14.1). Statistical analysis and visualization of results were conducted in GeneSpring (version 12.0; Agilent). All data were normalized with a quantile and median centering algorithm followed by logging (base 2). Samples were grouped based on cell type, time point, and presence of MALT1. Data were prefiltered (80% of samples in any 1 of the 8 groups having expression between the 20th and 100th percentile of measurements) in order to remove transcripts with essentially no signal across the set of arrays. 41,306 of 45,281 probes were included for final analysis. For unsupervised analysis, probes across all samples with a standard deviation greater than 0.5 were chosen (1,344 probes) and clustered using 2-way hierarchical clustering (Pearson centered distance measure under average linkage rules; ref. 58). All gene expression array data used are available under GEO (accession no. GSE40931).
STAT3 phosphorylation.
Malt1–/– and WT naive T cells were stimulated for 30 minutes with 50 ng/ml IL-6 (Peprotech) and simultaneously surface stained with anti-CD4 and anti-CD62L Abs (BD Biosciences). Intracellular staining was performed as above using phospho-STAT3 (pY705) Ab (BD Biosciences) followed by flow cytometry.
Immunoblotting.
Proteins were resolved by SDS-PAGE and transferred to a Hybond nitrocellulose membrane (Amersham Pharmacia Biotech). Membranes were incubated with 5% nonfat dry milk in TBS/T (TBS with 0.05% Tween-20) for 1 hour, washed with TBS/T, and incubated for 18 hours at 4°C with primary Ab diluted in TBS/T. Blots were developed using chemoluminescence following the manufacturer’s protocol (Millipore). Primary Abs were as follows: anti-MALT1 (gift of V. Dixit, Genentech, South San Francisco, California, USA); anti-actin and anti-tubulin (Sigma-Aldrich); anti-IκBα, anti-p65, anti-p50, and anti-USF2 (Santa Cruz); and anti-RelB and anti–c-Rel (Cell Signaling).
EAE induction.
Mice were s.c. immunized with 115 μg MOG35–55 peptide (Washington Biotech) emulsified in CFA (Difco) supplemented with 400 μg/ml Mycobacterium tuberculosis (Difco). On days 0 and 2 after immunization, mice were i.p. injected with 300 ng PT (List Biological). Clinical signs of EAE were monitored daily, according to the following criteria: 0, no disease; 1, decreased tail tone; 2, hind limb weakness or partial paralysis; 3, complete hind limb paralysis; 4, front and hind limb paralysis; 5, moribund state. Mice were sacrificed when they reached an EAE disease score of 4; these animals were assigned a score of 5 for the rest of observation period for the purpose of calculating mean EAE disease score. At the end of the 30-day observation period, any surviving mice were sacrificed, and their brains and spinal cords were subjected to histological analyses.
Histology and immunohistochemistry.
Brains and spinal cords were obtained from mice at 30 days after MOG injection and fixed in 10% buffered formalin for several days. Fixed tissues were cut into approximately 1.5-mm-thick coronal sections and paraffin embedded according to standard laboratory procedures. For histological and immunohistochemical assessments, 5-μm-thick paraffin sections were cut and stained with H&E or analyzed by immunohistochemistry according to standard laboratory procedures. For immunohistochemical staining, the following Abs and dilutions were used: anti-CD3 (1:200; Dako), anti-B220 (1:3,000; BD Biosciences — Pharmingen), anti-Mac3 (1:100; BD Biosciences — Pharmingen), and anti-GFAP (1:2,000; Dako).
Transfer of transgenic or Th1/Th17-polarized T cells.
EAE was initiated in Malt1–/– and WT mice by MOG injection as described above. At 10 days after injection, mice were sacrificed, and Malt1–/– and WT CD4+ T cells from spleens and LNs were cultured for 5 days in the presence of 25 μg/ml MOG peptide and 10 ng/ml IL-23 plus 5 μg/ml anti–IFN-γ to generate Th17 cells, or with MOG peptide and 10 ng/ml IL-12 plus 5 μg/ml anti–IL-23 to generate Th1 cells. WT CD4+ 2D2 T cells were isolated from spleens and LNs and sorted using a magnetic bead cell purification kit according to the manufacturer’s instructions (Miltenyi). WT CD4+ 2D2 T cells (1 × 105 cells/mouse) or polarized Th cells (2 × 107 cells/mouse) were transferred into recipient mice that had been sublethally irradiated 1 day before transfer. Transplanted mice were injected with PT on the day of T cell transfer and again 2 days later. EAE progression was monitored as described above.
DC differentiation and antigen presentation assay.
DCs were induced to differentiate from bone marrow stem cells by culture for 10 days in RPMI containing 40 ng/ml GM-CSF (Peprotech), as previously described (59). On day 10, cells were harvested, washed twice in PBS, and stimulated with 10 ng/ml LPS (Sigma-Aldrich) for 16 hours. DCs were then pulsed for 3 hours with 1 μM MOG peptide, washed twice with PBS, and cocultured with WT CD4+ 2D2 T cells labeled with 2.5 μM CellTrace Violet (Invitrogen) according to the manufacturer’s instructions. Proliferation was measured 72 hours later using a Canto II plate-reader (BD Biosciences) and analyzed using FlowJo 7.5 software.
Statistics.
Where appropriate, all differences were evaluated using 2-tailed Student’s t test, as calculated using Prism 5.0 software (GraphPad). Data are presented as mean ± SEM. A P value of 0.05 or less was considered significant.
Study approval.
All mouse procedures were approved by the University Health Network Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank all Mak laboratory members for their support; the Histological Services of the Toronto General Hospital for expert technical assistance; M. Lohoff and M. Huber for enlightening discussions; M. Saunders for insightful scientific editing; and A. Pfefferle, F. Wu, and S.O. Gilani for technical assistance. This work was supported by grants to T.W. Mak by the Canadian Institutes of Health Research and the Terry Fox Foundation. A. Brüstle was, and D. Brenner currently is, supported by a postdoctoral fellowship from the German Research Foundation (DFG), and D. Brenner and C.B. Knobbe were supported by Feodor-Lynen fellowships from the Alexander von Humboldt Foundation. P.A. Lang is a Sofja Kovalevskaja fellow of the Alexander von Humboldt Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2012;122(12):4698–4709. doi:10.1172/JCI63528.
References
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136(7):2348–2357. [PubMed] [Google Scholar]
- 2.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140(6):845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 3.Codarri L, et al. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12(6):560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 4.El-Behi M, et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12(6):568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6(11):1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langrish CL, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201(2):233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croxford AL, Kurschus FC, Waisman A. Mouse models for multiple sclerosis: historical facts and future implications. Biochim Biophys Acta. 2011;1812(2):177–183. doi: 10.1016/j.bbadis.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Garcia I, et al. IL-17 signaling-independent central nervous system autoimmunity is negatively regulated by TGF-beta. J Immunol. 2009;182(5):2665–2671. doi: 10.4049/jimmunol.0802221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, et al. IL-17RC is required for IL-17A- and IL-17F-dependent signaling and the pathogenesis of experimental autoimmune encephalomyelitis. J Immunol. 2010;184(8):4307–4316. doi: 10.4049/jimmunol.0903614. [DOI] [PubMed] [Google Scholar]
- 10.Komiyama Y, et al. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J Immunol. 2006;177(1):566–573. doi: 10.4049/jimmunol.177.1.566. [DOI] [PubMed] [Google Scholar]
- 11.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 12.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441(7090):231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 13.McGeachy MJ, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10(3):314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26(3):371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Brustle A, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8(9):958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126(6):1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 17.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28(1):29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Littman DR. Transcriptional regulatory networks in Th17 cell differentiation. Curr Opin Immunol. 2009;21(2):146–152. doi: 10.1016/j.coi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9(11):1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19(5):749–758. doi: 10.1016/S1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- 21.Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302(5650):1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- 22.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2(9):a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hailfinger S, et al. Malt1-dependent RelB cleavage promotes canonical NF-kappaB activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci U S A. 2011;108(35):14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rebeaud F, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008;9(3):272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 25.Coornaert B, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol. 2008;9(3):263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 26.Jost PJ, et al. Bcl10/Malt1 signaling is essential for TCR-induced NF-kappaB activation in thymocytes but dispensable for positive or negative selection. J Immunol. 2007;178(2):953–960. doi: 10.4049/jimmunol.178.2.953. [DOI] [PubMed] [Google Scholar]
- 27.Jun JE, et al. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003;18(6):751–762. doi: 10.1016/S1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 28.Medoff BD, et al. CARMA1 is critical for the development of allergic airway inflammation in a murine model of asthma. J Immunol. 2006;176(12):7272–7277. doi: 10.4049/jimmunol.176.12.7272. [DOI] [PubMed] [Google Scholar]
- 29.Molinero LL, Yang J, Gajewski T, Abraham C, Farrar MA, Alegre ML. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J Immunol. 2009;182(11):6736–6743. doi: 10.4049/jimmunol.0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6(11):1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 31.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24(2):179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 32.McColl SR, Staykova MA, Wozniak A, Fordham S, Bruce J, Willenborg DO. Treatment with anti-granulocyte antibodies inhibits the effector phase of experimental autoimmune encephalomyelitis. J Immunol. 1998;161(11):6421–6426. [PubMed] [Google Scholar]
- 33.Rothhammer V, et al. Th17 lymphocytes traffic to the central nervous system independently of alpha4 integrin expression during EAE. J Exp Med. 2011;208(12):2465–2476. doi: 10.1084/jem.20110434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurschus FC, Croxford AL, Heinen AP, Wortge S, Ielo D, Waisman A. Genetic proof for the transient nature of the Th17 phenotype. Eur J Immunol. 2010;40(12):3336–3346. doi: 10.1002/eji.201040755. [DOI] [PubMed] [Google Scholar]
- 35.Hirota K, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12(3):255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28(4):477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruland J, et al. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell. 2001;104(1):33–42. doi: 10.1016/S0092-8674(01)00189-1. [DOI] [PubMed] [Google Scholar]
- 38.May MJ, D’Acquisto F, Madge LA, Glockner J, Pober JS, Ghosh S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000;289(5484):1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 39.Ferch U, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2009;206(11):2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hailfinger S, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2009;106(47):19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duwel M, et al. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182(12):7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 42.Marienfeld R, May MJ, Berberich I, Serfling E, Ghosh S, Neumann M. RelB forms transcriptionally inactive complexes with RelA/p65. J Biol Chem. 2003;278(22):19852–19860. doi: 10.1074/jbc.M301945200. [DOI] [PubMed] [Google Scholar]
- 43.Marienfeld R, Berberich-Siebelt F, Berberich I, Denk A, Serfling E, Neumann M. Signal-specific and phosphorylation-dependent RelB degradation: a potential mechanism of NF-kappaB control. Oncogene. 2001;20(56):8142–8147. doi: 10.1038/sj.onc.1204884. [DOI] [PubMed] [Google Scholar]
- 44.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467(7318):967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korn T, et al. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2008;105(47):18460–18465. doi: 10.1073/pnas.0809850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purvis HA, et al. Low-strength T-cell activation promotes Th17 responses. Blood. 2010;116(23):4829–4837. doi: 10.1182/blood-2010-03-272153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouguermouh S, Fortin G, Baba N, Rubio M, Sarfati M. CD28 co-stimulation down regulates Th17 development. PLoS One. 2009;4(3):e5087. doi: 10.1371/journal.pone.0005087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ying H, et al. Cutting edge: CTLA-4--B7 interaction suppresses Th17 cell differentiation. J Immunol. 2010;185(3):1375–1378. doi: 10.4049/jimmunol.0903369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paulos CM, et al. The inducible costimulator (ICOS) is critical for the development of human T(H)17 cells. Sci Transl Med. 2010;2(55):55ra78. doi: 10.1126/scitranslmed.3000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haak S, et al. IL-17A and IL-17F do not contribute vitally to autoimmune neuro-inflammation in mice. J Clin Invest. 2009;119(1):61–69. doi: 10.1172/JCI35997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponomarev ED, Shriver LP, Maresz K, Pedras-Vasconcelos J, Verthelyi D, Dittel BN. GM-CSF production by autoreactive T cells is required for the activation of microglial cells and the onset of experimental autoimmune encephalomyelitis. J Immunol. 2007;178(1):39–48. doi: 10.4049/jimmunol.178.1.39. [DOI] [PubMed] [Google Scholar]
- 52.Bettelli E, Sullivan B, Szabo SJ, Sobel RA, Glimcher LH, Kuchroo VK. Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J Exp Med. 2004;200(1):79–87. doi: 10.1084/jem.20031819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruan Q, et al. The Th17 immune response is controlled by the Rel-ROR{gamma}-ROR{gamma}T transcriptional axis. J Exp Med. 2011;208(11):2321–2333. doi: 10.1084/jem.20110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen G, et al. The NF-{kappa}B transcription factor c-Rel is required for Th17 effector cell development in experimental autoimmune encephalomyelitis. J Immunol. 2011;187(9):4483–4491. doi: 10.4049/jimmunol.1101757. [DOI] [PubMed] [Google Scholar]
- 55.Saccani S, Pantano S, Natoli G. Modulation of NF-kappaB activity by exchange of dimers. Mol Cell. 2003;11(6):1563–1574. doi: 10.1016/S1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 56.Powolny-Budnicka I, Riemann M, Tanzer S, Schmid RM, Hehlgans T, Weih F. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in gammadelta T cells. Immunity. 2011;34(3):364–374. doi: 10.1016/j.immuni.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 57.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 59.Lutz MB, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223(1):77–92. doi: 10.1016/S0022-1759(98)00204-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.