Abstract
RNA helicases are a diverse group of RNA-dependent ATPases known to play a large number of biological roles inside the cell, such as RNA unwinding, remodeling, export and degradation. Understanding how helicases mediate changes in RNA structure is therefore of fundamental interest. The advent of single-molecule spectroscopic techniques has unveiled with unprecedented detail the interplay of RNA helicases with their substrates. In this review, we describe the characterization of helicase-RNA interactions by single-molecule approaches. State-of-the-art techniques are presented, followed by a discussion of recent advancements in this exciting field.
Keywords: single-molecule spectroscopy, FRET, PIFE, AFM, optical tweezers, RNA folding, RNA helicase, DEAD-box
Introduction
Originally thought of as simple intermediaries in protein synthesis from a DNA sequence, it has become increasingly clear that RNA molecules play essential roles in almost every aspect of cellular metabolism.1 RNA functions span diverse processes, such as protein synthesis (as messenger RNA, mRNA, ribosomal RNA, rRNA and transfer RNA, tRNA),2-6 post-transcriptional precursor mRNA processing (as small nuclear RNA, snRNA and small nucleolar RNA, snoRNA),7-12 maintenance of telomeres (as telomeric RNA)13 and control of gene expression (as microRNA, miRNA, small interfering RNA, siRNA, aRNA aRNA and riboswitches).14-18 Moreover, RNA molecules are crucial in the action of parasitic entities like retrotransposons, viruses and satellite RNA.19-21 Such functional diversity is brought about by the structural diversity of RNA, which can be somewhat surprising considering the limited number of building blocks: the four nucleobases adenine, guanine, cytidine and uracyl.22 In addition to canonical base pairing, RNA molecules are also capable of forming numerous interactions, such as base-stacking, non-canonical base-pairing, ribose 2'-hydroxyl bonding and metal ion binding.23,24 These interactions are essential to form the native, three-dimensional structures that make RNA molecules function inside living cells.23,25
Small functional motifs, such as G-quadruplexes in the untranslated regions of mRNA, are able to form readily.22,26 In contrast, long RNA molecules composed of several hundred nucleotides often form stable conformational intermediates that present local minima in the energy landscape. When separated by considerable energy barriers from the next minimum, these intermediates can result in folding kinetic traps,27-31 which prevent the RNA from readily folding into the more stable native structure.32 Accumulation of kinetic traps can result in reduced, altered or even loss of functionality, because the native conformation is reached slowly or not at all.33 As a consequence, kinetic traps must be resolved in vivo through the action of helper proteins, such as helicases. Helicases are ubiquitous enzymes that can catalyze different types of conformational changes and structural rearrangements, going far beyond the traditional view of double-strand separating motors.34 RNA helicases are involved in almost all aspects of cellular metabolism, including translation initiation,35,36 ribosome biogenesis,37,38 RNA splicing,39,40 miRNA function,41-43 RNA transport,34,44,45 replication by RNA viruses46 and unwinding of G-quadruplex structures.47 RNA helicases are RNA-dependent ATPases. Their mechanism can comprise sequences of complex steps involving as many as 75 rate constants in the case of DEAD-box RNA helicases (vide infra).48 Rapid RNA structural rearrangements occur upon ATP hydrolysis, followed by phosphate and ADP release, thereby turning over to another round of catalysis, in which each conformational intermediate displays altered affinity its cognate substrate.49,50 Interestingly, dsRNA unwinding is also observed in the absence of ATP, though with slower kinetics and to a lesser extent.49,50 Given the vast number of cellular processes RNA helicases are involved in, it is not surprising that individual helicases can strongly differ in translocation rate, directionality, processivity, active vs. passive mechanism and step size (i.e., number of base pairs translocated or unwound during each catalytic cycle).51 Through phylogenetic analysis, RNA helicases have been grouped and functionally classified into four superfamilies (SF). The most prevalent ones are the structurally similar SF1 and SF2, while a number of viral helicases belong to SF3 and SF4.52,53 The majority of RNA helicases studied to date belongs to the largest subfamily SF2, characterized by ATP-dependent translocation on the nucleic acid substrate and/or induction of RNA conformational changes and further sub-grouped as DEAD-, DEAH- and DExH-box helicases. The subgroup names derive from a highly conserved amino acid motif, in single letter code (e.g., D-E-A-D, Asp-Glu-Ala-Asp), in their helicase domain.51,52,54 Numerous biochemical studies, including gel mobility55,56 and unwinding assays,57,58 have yielded valuable insights into their function, substrate specificity and steady-state kinetics.59 However, due to ensemble-averaging, these experiments may fail to detect unwinding intermediates and/or conformational or functional heterogeneities within the sample.59 Through the advent of single-molecule methods, it has become possible to monitor individual macromolecules in real time, thus eliminating population averaging.60-62 Rare or transient intermediates can, therefore, be directly observed provided that (1) time resolution is sufficient, and (2) enough events can be captured to rule out experimental artifacts.61,63-66 Over the years, single molecule approaches have led to an increasing number of fascinating discoveries.67-72 Here, we provide an overview on single-molecule spectroscopy methods and how they have been successfully employed to characterize the interplay between helicases and their RNA substrates. Technical specifications, advantages and drawbacks associated with individual techniques are also briefly discussed. We would also like to direct the reader to other interesting reviews on related topics.59,73-77
Force-Based Approaches and their Application to Study Helicase-Mediated Duplex Unwinding
The application of force to single molecules allows for probing and manipulating the folding energy landscape, as well as measuring the force generated by single biomolecules.64,78 These approaches do not typically require labeled samples, as they extend the biopolymer between two handles, or attachment sites.64 Atomic force microscopy (AFM) and optical tweezers have been previously used to study the effect of helicases on RNA substrates (vide infra).79,80 Two other force-based techniques are magnetic tweezers and tethered-particle microscopy (TPM), which have both been successfully used to characterize DNA helicases, but they will not be presented here.81-84
Atomic force microscopy
Put forward by Binnig and coworkers,85 AFM has become a powerful tool for imaging and manipulating molecules at the nanoscale.86-89 At the heart of the mechanical microscope is a flexible cantilever, which acts as a linear spring according to Hooke's law and is used to detect force exerted onto its tip by means of a laser beam that is reflected from the cantilever onto a detector.90 Typically, the molecule under study is tethered to a surface at one end, and to the AFM tip at the other, such that force can be measured and/or applied (Fig. 1A),69,80 although other experimental approaches have also been used.79 AFM requires minimal sample preparation and experiments can be performed in many different environments to obtain data in nm3 spatial and 100 ms temporal resolution,90,91 offering the possibility of imaging entire proteins and their environment at once (Table 1).90 The advent of high-speed AFM using smaller cantilevers oscillating at higher frequencies has led to time resolutions over 1,000 frames/s.92 However, due to the requirement for physical contact with the sample, its application is currently limited to in vitro systems with isolated components. A more detailed description has been given elsewhere.69,78,90
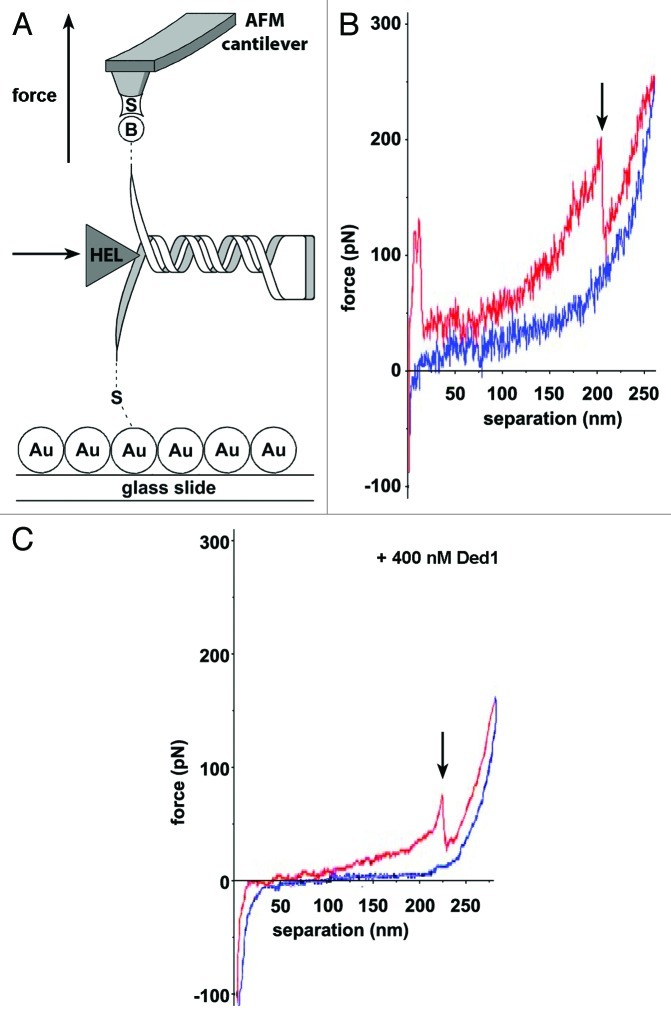
Figure 1. Helicase-induced RNA hairpin unfolding using AFM force spectroscopy. (A) RNA immobilization involves tethering the 5′-end to the gold-coated surface (“Au”) and attaching the biotinylated 3′-end (“B”) to a streptavidin-coated (“S”) silicon nitride (Si3Ni4) tip. Force-extension curves were recorded in the presence and absence of eIF4A or Ded1, represented as “HEL.” (B and C) Representative force-extension plots representing stretching of a single RNA molecule containing a GC-rich stem-loop. (C) Stretching of the same RNA in the presence of 400 nM Ded1. The pull curve (red) runs from left to right and the approach curve (blue) from right to left. The arrow indicates the force and extension when the hairpin unfolds. In the presence of Ded1, the hairpin unfolds at lower force. Adapted from reference 80.
Table 1. Single-molecule techniques applied in studying helicase-mediated RNA folding and their technical specifications.
| Optical tweezers69,94 | AFM69,90 | smFRET70,175 | PIFE131,136 | |
|---|---|---|---|---|
| Spatial resolution |
0.1–2 nm |
0.1 nm in z plane 5 nm in x-y plane |
3–8 nm, depending on R0 |
below 4 nm |
| Temporal resolution |
10−4 s |
10−5 s |
Confocal: ≥ 5 × 10−5 s TIRFM: ≥ 10−3 s |
TIRFM: ≥ 10−3 s |
| Force range |
0.1–100 pN |
10–104 pN |
- |
- |
| Typical applications |
3D manipulation tethered assays Interaction assays |
high-force pulling and interaction assays |
protein and nucleic acid folding interaction assays, enzymology |
docking/undocking dynamics, protein displacement along RNA |
| Features |
low-noise and low-drift dumbbell geometry no labeling required, in vivo measurements (in principle) possible |
high-resolution imaging, no labeling, measurements under near-physiological conditions |
confocal, observe one molecule at a time; TIRFM, image several hundreds of single molecules simultaneously |
Image > 100 molecules at a time, rather robust against photo-physics |
| Limitations | photodamage, sample heating, nonspecific | large high-stiffness probe, large minimal force, nonspecific, surface technique | labeling required, photobleaching, dye-sample interaction | labeling, photo-bleaching, dye-sample interaction |
Optical tweezers
In the same year that AFM was invented, Chu and coworkers demonstrated particle trapping in a laser beam and suggested its application to study “biological particles.”93 The technique is based on light-matter interaction; the optical trap is generated by focusing laser light to a diffraction-limited spot using a high numerical aperture objective.69 In the vicinity of the laser focus, the dielectric object to be trapped experiences a three-dimensional force directed toward the beam waist.69 Therefore, the laser can be regarded as a microscopic lens that focuses the electro-magnetic field, which causes the particle to move toward the highest intensity point.94 Particle displacements from the trap center are used to follow and quantify the forces exerted onto a biomolecule, such as an RNA molecule, tethered to the bead. Optical tweezers readily enable measurements of forces as small as 25 fN and sub-nanometer resolution (Table 1).78,94-98 Since their discovery, experiments with optical tweezers have enabled ground-breaking studies of actin-myosin interactions, kinesin motion along microtubules, viral packaging, RNA polymerase transcription and DNA and RNA folding among others.94,99-102 While the use of optical tweezers has become popular due their purely optical nature, they can also present some experimental hindrances such as challenging calibrations,69 local heating resulting from high-intensity and focused laser beams. These issues can complicate the implementation of optical tweezers measurements in live cells, even though the technique has been successfully adapted for in vivo studies on other systems.103 For a more detailed description on optical tweezers, we direct the reader to the following reviews.69,94,104
RNA duplex unwinding in the presence of the DEAD-box helicases eIF4A and Ded180
Secondary structure motifs along mRNA molecules can impede the translational machinery search for the start codon required for protein synthesis initiation.105-108 Two eukaryotic initiation factors, eIF4A and Ded1 (Table 2),73 belonging to the DEAD-box family (SF2) of RNA helicases, have been shown to resolve RNA secondary structures and unfold RNA hairpins in vitro.80,109,110 Using AFM, McCarthy and coworkers studied the mechanism of these two helicases at the single molecule level.80 The authors used RNA sequences derived from the naturally occurring GCN4 5′-UTR, which were surface-tethered through 5′-end thiolation and attached to the cantilever tip using biotinylated nucleotides in the poly-A tail (Fig. 1A). The RNA was then stretched by pulling the cantilever away from the surface. The force exerted on the RNA was determined by measuring the bending of the cantilever with the reflected laser beam (Fig. 1A). The resulting force-extension curves (Fig. 1B and C) revealed the force necessary to stretch (red) and refold (blue) the RNA at a certain distance. Each molecule could be pulled ~20–30 stretching/refolding cycles before dissociation of the RNA, backbone cleavage or disruption of the biotin/streptavidin bond. Incorporation of a stable 25 GC basepair stem-loop has been shown to inhibit translational initiation in S. cerevisiae.107 In the AFM experiments, incorporation of the same hairpin resulted in an abrupt drop in force in the stretching curve (arrow, Fig. 1B), which was absent in the refolding curve. This drop was assigned to the opening of the hairpin.107 In the presence of saturating Ded1, the force required to open the hairpins decreases from ~150 pN to ~90 pN (Fig. 1C). Comparison between the two helicases showed that Ded1 is more effective in reducing the force necessary to unfold the hairpin, even in the presence of elF4B, an elF4A cofactor that enhances its unwinding rate. In addition, the unwinding activity of eIF4A/B saturates well before the stem loop is fully unwound. Based on these results, the authors concluded that Ded1 is a more efficient facilitator of stem-loop unwinding than elF4A/B, and likely the major unwinding factor on natural mRNA substrates. Based on earlier bulk studies,110 the authors proposed that eIF4A acts via an ATP-dependent steric mechanism, whereby the helicase binds to single-stranded RNA adjacent to the stem loop structure, thus causing its partial unwinding, whereas Ded1 dynamically unwinds the stem loop through low processive, ATP-dependent translocation.80
Table 2. Some RNA helicases studied by single-molecule approaches, in order of appearance in text.
| Helicase | Full name (www.uniprot.org) | Subgroup | Biological role | Substrate | Method | Ref. |
|---|---|---|---|---|---|---|
| eIF4A |
eukaryotic initiation factor 4A |
DEAD-box |
translation initiation |
dsRNA |
AFM, smFRET |
80
,
111
|
| Ded1 |
ATP-dependent RNA helicase DED1 |
DEAD-box |
translation initiation |
ssRNA, dsRNA |
AFM, smFRET |
80
,
140
|
| HCV NS3 |
Hepatitis C virus non-structural protein NS3 |
DEAH-box |
viral replication |
dsRNA/DNA with 3′ ss overhang55,114 |
Optical tweezers, smFRET* |
112
,
115
,
118
,
119
|
| NPH-II |
Nucleoside triphosphate phosphohydrolase II |
DEAD-box |
transcription termination, RNA export146 |
viral dsRNA |
smFRET |
121
|
| Mss116 |
ATP-dependent RNA helicase MSS116, mitochondrial |
DEAD-box |
splicing |
group I and II introns |
smFRET |
124
|
| RIG-I |
retinoic acid inducible gene I |
DExD/H-box |
RNA interference,176 immune response168 |
viral dsRNA |
PIFE |
135
|
| YxiN | ATP-dependent RNA helicase YxiN | DEAD-box | ribosome biogenesis177 | 23S rRNA178 | smFRET | 126 - 129 |
Study performed using dsDNA.
In this study, the relative efficiency of two eukaryotic helicases to separate was assessed from the decrease in unwinding force. Future studies may involve eIF4G, which has been shown to stimulate eIF4A via a conformational guidance mechanism.111 A valuable extension of analogous studies will involve time-dependent force-extension plots (vide infra). This will allow finding key mechanistic features of processive helicases, such as the number of base pairs unwound/ATP hydrolyzed, the physical step size (mean distance/translocation step), as well as the kinetic step size (mean number of base pairs unwound/rate-limiting step).59
RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP112
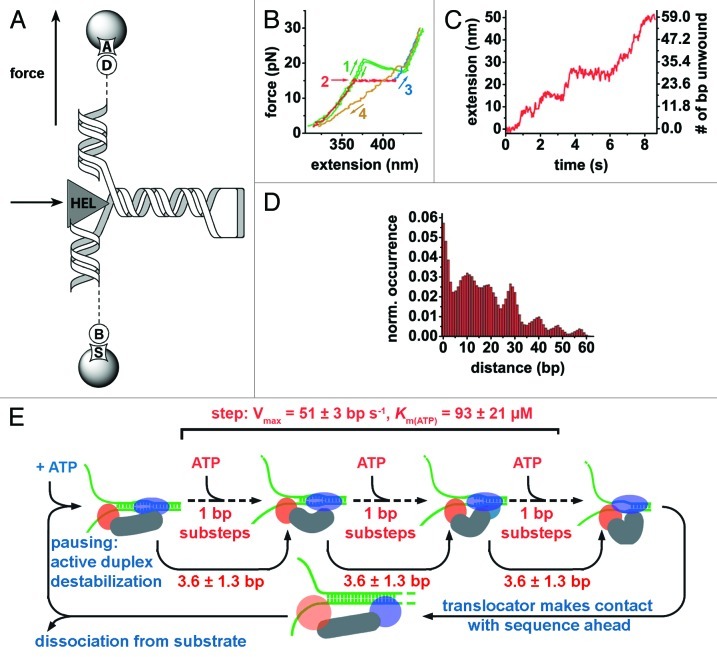
The hepatitis C virus (HCV) non-structural protein 3 (NS3) is an SF2 DEAH-box helicase that plays a key role in viral RNA replication and an important drug target against HCV infection (Table 2).113,114 In a study by Dumont and coworkers, NS3-catalyzed unwinding of dsRNA and translocation along the substrate was characterized using optical tweezers.112 A 60 bp RNA hairpin with handles was attached to two beads and held under constant strain (Fig. 2A). The end-to-end distance of the RNA was measured in the presence and absence of NS3 to monitor helicase-induced RNA unwinding. Force-extension curves show that the presence of NS3 decreased the force required for unfolding from 20.4 pN (Fig. 2B, green) to 5 pN, though this depends on the GC-content of the sequence.115 To monitor NS3 unwinding activity, the RNA substrate was held at constant force in the 5–17 pN range in the presence of the helicase (Fig. 2B, red). The resulting time trajectories revealed stepwise increases of inter-bead distance, which was subsequently converted into basepairs (Fig. 2C),116 and interpreted as unwinding steps and pauses. A histogram of pairwise distances (Fig. 2D) revealed a periodic pattern of steps, suggesting that the cyclic behavior of NS3 is coordinated by ATP in discrete steps of 11 ± 3 bp that include rapid smaller substeps of 3.6 ± 1.3 bp, which reflect actual unwinding. However, in a follow-up study, step size was shown to be decreased to 9 ± 2 bp in the presence of long GC-rich stretches that were absent in the substrate used in the initial study.115 Interestingly, the authors also observed apparent backward steps of 12.0 ± 3.6 bp corresponding to the stepwise refolding of the substrate after unwinding. This pronounced processivity is somewhat surprising, given that NS3 differs in only one of six conserved motifs from its phylogenetically close relatives from the DEAD-box family of helicases, which are almost non-processive.34
Figure 2. Revealing NS3 translocation and unwinding mechanism by optical tweezers. (A) A 60 bp RNA hairpin is flanked by two RNA/DNA duplexes. The bottom duplex (599 bp) is attached to a bead via biotin-streptavidin linkage (“B,” “S”). The top duplex (535 bp) is attached to an anti-digoxigenin-coated bead via digoxigenin (“A,” “D”). NS3-mediated (“HEL”) duplex unwinding is followed upon application of external force. (B) Unwinding experimental steps: mechanical folding and unfolding of substrate in the absence of NS3 shows a transition at 20.4 pN, the force required for hairpin unwinding (green), the presence of NS3 decreases the force required for duplex unwinding (red), force is brought to 30 pN to fully extend the nucleic acid strand (blue). The RNA hairpin is allowed to refold at 2 pN force (yellow). (C) Extension of bead separation during RNA unwinding by NS3 reveals discrete steps and pausing. (D) A pairwise distance histogram of the unwinding trace shown in (C) is later subjected to Fourrier analysis to determine the apparent unwinding step size. (E) Unified model for NS3 helicase activity. Nucleic acid substrate binding is followed by ATP-dependent destabilization duplex by the translocator domain (blue ellipse).115 The RNA hairpin is unwound by helix-opening domain (orange circle) in fast ATP-dependent 1 bp-substeps adding up to steps of 3.6 ± 1.3 bp, unwinding 7–13 basepairs per apparent step depending on the GC content.112,115,118,119 Subsequently, the translocator moves forward to start a new catalytic cycle.112 At this stage, GC-rich sequences increase the probability of helicase dissociation.115 Adapted from references 112 and 115.
Mean pause duration of the NS3 helicase was found to be inversely proportional to ATP concentration, indicating that exiting from pauses requires ATP hydrolysis. Analysis of pause distributions below and above the Michaelis-Menten constant pointed the authors to propose a kinetic mechanism by which the helicase exits from pauses in two steps, only one of which involves ATP hydrolysis. The occurrence and duration of stalling was shown to depend upon the presence of GC islands of six or more basepairs but not three.115 Furthermore, the stepping velocity of NS3 between pauses increases with ATP concentration, indicating that each step consists of ATP-dependent substeps. Based on the kinetic data, the authors concluded that NS3 must bind ATP in each substep during unwinding.112 The processivity of NS3 did not significantly depend upon either ATP or NS3 concentration above KD, but was strongly dependent on the force applied and also found to be affected by the base composition of the duplex.115 In contrast, pausing and stepping velocity were independent of force indicating that NS3-mediated RNA unwinding activity is not limited by strand separation but by translocation.
Although the stepping velocity and Km were in good agreement with prior bulk experiments, the single molecule assays were seemingly in conflict with previous ensemble experiments reporting a step size of 18 ± 2 bp as compared with 11 ± 3 bp.117 The authors proposed that NS3 dimerization in bulk experiments may account for this difference, while the 3′-overhang used in the single-molecule study did not allow for dimer formation.112 This hypothesis was further confirmed in subsequent bulk experiments.115 In the proposed model, the NS3 monomer has two RNA binding sites; one site acts as a translocator and the other as a helix opener.112 Both translocation and unwinding take place as ATP-dependent substeps, resulting in 11-basepair unwinding bursts. Following results further indicated that these substeps are preceded by active destabilization of the double-stranded region to be unwound, as opposed to opportunistic forward motion of the enzyme upon thermal fraying of the duplex.115 In an independent fluorescence-based study performed with double-stranded DNA, Myong et al. proposed an alternative model analogous to a loaded spring involving three successive ATP hydrolysis events driving the two domains forward in one-base-pair steps, followed by an abrupt 3 bp-separation using DNA.118 To reconcile these findings, Cheng and coworkers have recently used a homopolymeric GC sequence to avoid sequence-dependent variation in step size and to slow down the helicase.119 Hence, their experiments at very low ATP concentrations (to further slow down the helicase) achieve 1 bp resolution.119 They observed both large 11-bp unwinding steps and 1-bp unwinding events, thus reconciling fluorescence experiments with force spectroscopy studies. However, they also observed statistically significant higher-order 1.5-, 2- and 2.5-bp unwinding events.119 As the rate constants for unwinding were independent of substepping size, the authors proposed that higher-order steps are due to release of nucleotides held in previous rounds of unwinding, in agreement with recent crystallographic data (Fig. 2E).119,120 This example illustrates how different single molecule approaches can be used alongside to improve our understanding of helicase-mediated double-stranded RNA unwinding.
Fluorescence-Based Methods and their Applications in Elucidating RNA-Helicase Interactions
Single-molecule Förster resonance energy transfer (smFRET)
FRET is a process by which energy is non-radiatively transferred between two dipoles, typically the transition dipole moments of two fluorophores that are referred to as donor and acceptor. Efficient energy transfer requires the emission spectrum of the donor to overlap with the excitation spectrum of the acceptor, which satisfies the resonant (equal energy) condition. Energy transfer efficiency depends very strongly on the distance between the two dipoles (R), and is calculated as E = (1 + (R/R0)6)−1, where R0 is the Förster radius at which half of the energy is transferred. The Förster radius is typically in the 30–80 Å range, but depends on the spectral properties of the fluorophores, their relative orientation and their local environment.70 Owing to its distance-dependence, FRET is regularly referred to as a “molecular ruler” to probe inter- and intramolecular distances in biological systems.54,121 Experimentally, the lack of information on relative fluorophore orientation makes it challenging to determine the exact energy transfer efficiency.122 Therefore, the apparent energy transfer efficiency is often simply calculated as FRET = IA/(IA+ID), where ID and IA are the fluorescent intensities of the donor and acceptor, respectively.
smFRET is a popular and adaptable approach to study biomolecular mechanisms of isolated molecules because FRET provides the sensitivity and selectivity necessary to detect single molecules dynamics in real time (Table 1).70,123 It has, therefore, been applied to study both the interaction of surface-immobilized RNAs with a helicase,121,124,125 and to probe helicase conformational dynamics in solution.126-129 By monitoring individual molecules, smFRET can help reveal sample heterogeneity and the presence of transient intermediates.130
Protein-induced fluorescence enhancement (PIFE)
An important limitation in the interpretation of smFRET data arises from fluorophore blinking (intersystem crossing to long-lived, non-fluorescent triplet states) and photobleaching (light-induced chemically modified non-fluorescent adducts).131 Despite recent advancements in the suppression of such photophysics through addition of reducing (or oxidizing) agents,132-134 dark states, blinking and unstable emission still present a challenge in single-molecule fluorescence microscopy.130 An interesting solution to this problem is the use of protein-induced fluorescence enhancement (PIFE), which requires only one label.135,136
Cyanine dyes, which consist of two aromatic moieties linked by a polymethylene chain, can adopt cis and trans conformations, but the latter conformer exhibits considerably higher fluorescence quantum yield.137 Therefore, cis-trans isomerization decreases the apparent quantum yield of cyanine dyes and their fluorescence lifetime.137 The principle behind PIFE relies on a local increase of the viscosity of the fluorophore environment due to the proximity of a protein, resulting in hindrance of the cis-trans isomerization and hence, altered fluorophore quantum yield. This allows to directly monitor protein binding and dissociation dynamics.135,136,138 However, in contrast to the ratiometric smFRET method that minimizes the effect of inherent emission fluctuations of single fluorophores,139 PIFE data are prone to produce noisy data. Because specific environmental changes in the vicinity of fluorophores are challenging to predict, interpretation of experimental data requires careful calibration and well-designed control experiments.136
Helicase-assisted secondary structure conversion toward and against thermodynamic equilibrium140
Helicases are multifunctional enzymes that, not only separate double-stranded nucleic acids, but they can also drive structural rearrangements required for function.141,142 Interestingly, stable secondary RNA structures have been observed to be converted into thermodynamically less stable ones.141 Shortly after the single molecule characterization of Ded1-catalyzed unwinding of RNA duplexes,80 Jankowsky and coworkers used smFRET in conjunction with bulk biophysical methods to characterize Ded1-catalyzed RNA structure remodeling (Table 2).140 The authors studied whether Ded1 catalyzes conversion of a stable RNA duplex into a thermodynamically less stable one by first disassembling existing duplex structures, or through recruitment of multiple strands to form a tripartite intermediate (Fig. 3A). To address this question, they designed a three-strand RNA substrate such that two mutually exclusive duplex structures of distinct stabilities could be formed (Fig. 3A, left and right). One of the strands contained a biotin label for surface-tethering, the second strand contained the donor fluorophore (Cy3) and the third strand contained the acceptor fluorophore Cy5 (Fig. 3A). Therefore, the more stable duplex is characterized by Cy3 emission alone (i.e., zero FRET), the least stable duplex is characterized by the loss of the Cy3 strand (and, thus, Cy3 fluorescence) and a possible tripartite complex by the presence of both Cy3 and Cy5 in close proximity (i.e., emission of both fluorophores via FRET). Having determined the equilibrium ratio of the two duplex structures under different conditions, the authors observed structural interconversion against thermodynamic equilibrium upon addition of Ded1 and ATP using non-denaturing gel electrophoresis. A gel mobility shift was accompanied by the generation of single-stranded RNA. In the absence of ATP, strand exchange was also observed, but to a lesser extent, and single-stranded RNA was not detected.
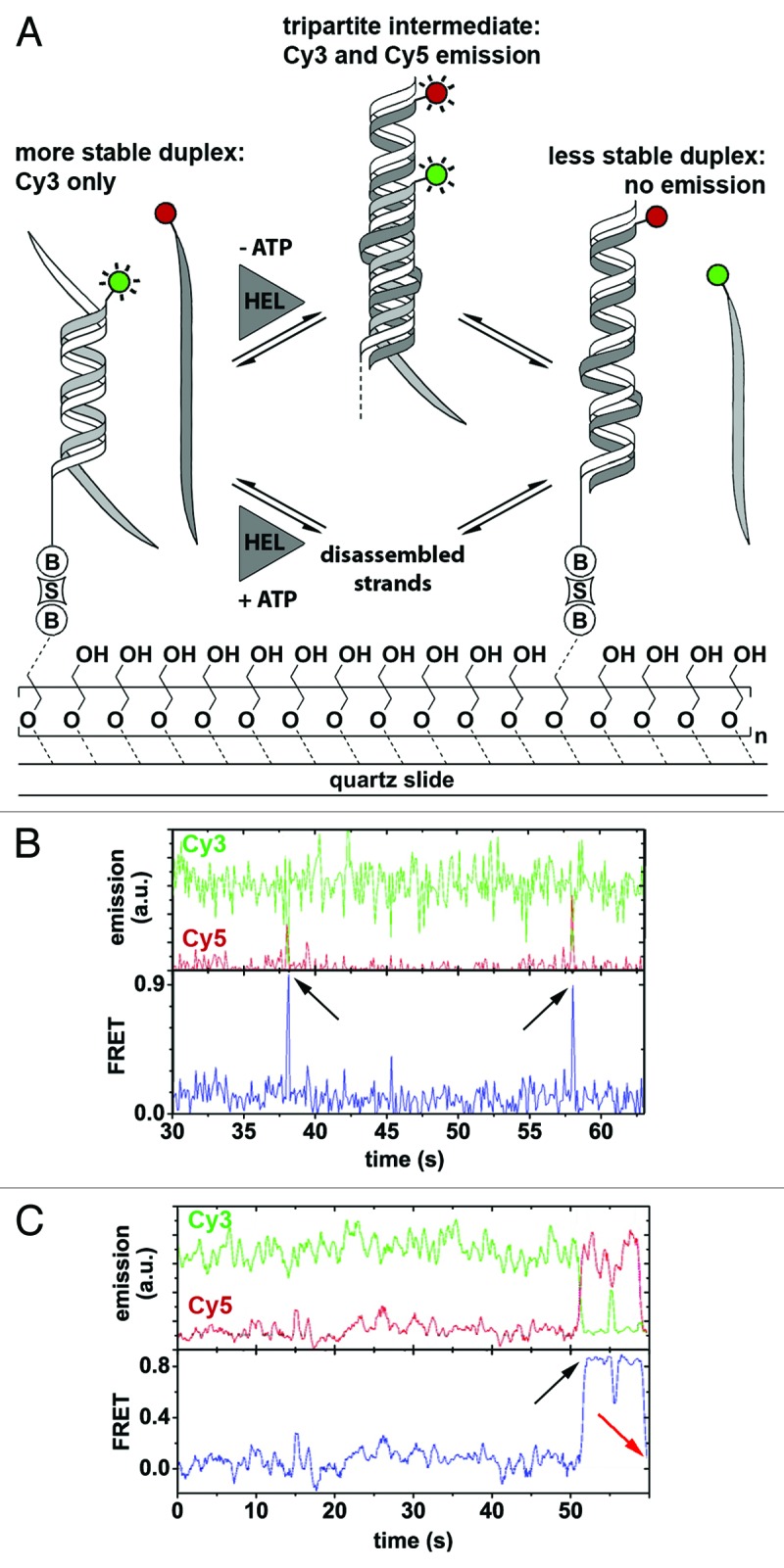
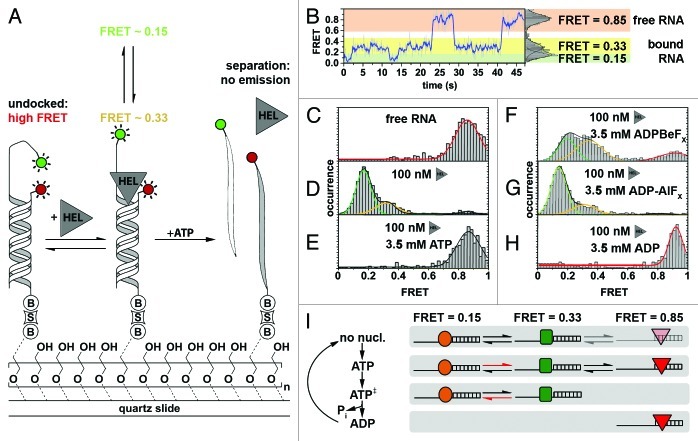
Figure 3. Ded1-mediated RNA remodeling monitored by smFRET. (A) Experimental design and results. The Cy3-labeled RNA strand (light gray; Cy3, green circle) forms a thermodynamically stable duplex with the immobilization strand (white) that is surface-tethered via a biotin-streptavidin linkage (“B,” “S”), while the Cy5-labeled strand (dark gray; Cy5, red circle) is in solution. Strand exchange in the presence of Ded1 (“HEL”) and in the absence of ATP proceeds via a tripartite intermediate characterized by a sudden increase in Cy5 emission through FRET, followed by the formation of the thermodynamically less stable duplex accompanied by a complete loss of fluorophore emission (upper pathway). In the presence of both Ded1 and ATP, strand exchange involves complete disassembly of the more stable duplex and subsequent formation of the less stable one (lower pathway). Passivation of the quartz slide is achieved through coating with polyethylene glycol to prevent non-specific binding of the protein to the slide.161 (B and C) Representative fluorophore emission and FRET time traces. Cy3 and Cy5 emission over time (green and red traces) displays FRET-typical anti-correlated behavior (upper plots). FRET over time reveals the formation of the tripartite intermediate, as indicated by a sudden burst of FRET (lower plots, black arrows), followed either by dissociation of the Cy5-labeled strand (B) or strand exchange and complete absence of fluorophore emission (C, red arrow). Figure modified from reference 140.
In single-molecule FRET experiments, the resulting time trajectories clearly showed the reversible formation of a transient tripartite intermediate complex (Fig. 3B and C, black arrows). In rare instances, the authors also observed the whole remodeling pathway with initial formation of the tripartite intermediate followed by dissociation of the Cy3-labeled strand (Fig. 3C, red arrow). The authors concluded that Ded1-assisted structure conversion can proceed via two pathways: (1) ATP-dependent strand exchange involving complete dissociation of the RNA strands, followed by protein-facilitated annealing; (2) An ATP-independent pathway promoting the formation of a tripartite intermediate complex (Fig. 3A).140
These results confirmed previous studies in which Ded1 was shown to be a low processive, ATP-dependent helicase.80,125 The characterization and identification of the transient, tripartite intermediate and the remodeling pathway would not have been possible using solely ensemble-averaged techniques, underscoring the strength of single-molecule techniques to resolve transient intermediates in the reaction pathway. The authors thereby revealed an RNA remodeling activity of the Ded1 helicase in addition to its previously known duplex unwinding activity. However, a limitation in this assay is that formation of the less stable duplex, which is characterized by the loss of the Cy3 strand and, thus, loss of both Cy3 and Cy5 fluorescence, could not be distinguished from Cy3 photobleaching. One possible alternative approach is to invert the labeling scheme, because formation of the tripartite complex would result in a high FRET signal followed by loss of Cy5 fluorescence, indicating strand exchange. An elegant way to rule out photophysical artifacts is by using alternating laser excitation (ALEX), which consists of exciting both fluorophores alternatively, thereby determined both FRET and fluorophore stoichiometry which unambiguously reports on photobleaching and blinking.143,144
Unwinding initiation by the viral RNA helicase NPH-II121
NPH-II is a viral SF2 helicase with NTP-dependent unwinding activity, which plays an important role in transcription termination and viral RNA export.145-147 Experiments in vitro have shown that NPH-II unwinding initiation can be two orders of magnitude slower than strand separation, thus making initiation the rate-limiting step.147 Interestingly, the enzyme can even hydrolyze hundreds of ATP molecules before unwinding.147 This makes NPH-II an interesting model system to study unwinding initiation. After their initial smFRET study with Ded1, Jankowsky and coworkers characterized NPH-II-mediated dsRNA unwinding initiation using single-molecule FRET. The authors surface-immobilized a Cy3-labeled dsRNA with a Cy5-labeled 3′-single-stranded overhang (Fig. 4A). The overhang is required in vitro for NPH-II recruitment.55,145,147,148 Single-molecule time trajectories recorded under various experimental conditions (Fig. 4B) were used to build time-binned FRET histograms that report on the conformational distribution of the 3′-end overhang (Fig. 4C–H).71
Figure 4. Characterizing NPH-II-catalyzed RNA duplex unwinding. (A) Experimental design and results. A fluorophore-labeled 19-bp RNA duplex (Cy3, green circle; Cy5, red circle) with a 24-nt 3′ extension was immobilized on the PEG-passivated quartz slide via a biotin-streptavidin linkage (“B,” “S”).161 The initial high FRET value of 0.85 is diminished in response to NPH-II binding (“HEL”) and fluctuates between two discrete low FRET values, indicating an increased inter-dye distance. Addition of ATP triggers duplex unwinding ultimately leading to complete loss of emission upon strand separation. (B) Representative smFRET trajectory (1 nM NPH-II, no ATP) showing transition between the helicase-unbound state (FRET 0.85, highlighted in orange), and the helicase-bound states (FRET 0.15 and 0.33, highlighted in green and yellow, respectively). (C–H) Averaged FRET histograms, each built from over 100 individual FRET time traces. Imaging conditions: (C) only RNA, (D) RNA and 100 nM NPH-II, (E) 100 nM NPH-II, 3.5 mM ATP, (F) 100 nM NPH-II, 3.5 mM 'ADP-BeFx (a ground-state analog), (G) 100 nM NPH-II, 3.5 mM ADP-AlFx (a transition state analog), (H) 100 nM NPH-II, 3.5 mM ADP. (I) Basic model for unwinding initiation by NPH-II relying on altered substrate affinities along the ATP hydrolysis cycle. Without nucleotide, NPH-II binds both ssRNA and dsRNA and the NPH-II-ssRNA complex readily alternates between two conformations. ATP binding impedes dissociation from ssRNA and changes the kinetics of bound-state transitions. In the ATP transition state, NPH-II no longer binds to dsRNA and interconversion kinetics change again. The helicase associates with dsRNA upon ATP hydrolysis and phosphate dissociation. Different shapes mark the different conformational states if NPH-II traversed during unwinding initiation. Figure adapted from reference 121.
In the absence of the helicase, the free RNA yielded a distribution centered around 0.85 FRET (Fig. 4C), as expected for the two fluorophores in close proximity (Fig. 4A). Addition of NPH-II alone induced a conformational change in the RNA-NPH-II complex that led to two distinct conformations with FRET distributions centered at ~0.15 and ~0.33 (Fig. 4D). Addition of ATP shifted the FRET distribution back to 0.85 (Fig. 4E). However, the time trajectories clearly revealed a rapid decrease in FRET followed by a loss of fluorescence, a signature indicative of helicase binding followed by duplex unwinding. To further test the mechanism, the authors replaced ATP by non-hydrolyzable analogs blocking unwinding at different states of ATP hydrolysis.149-154 In the presence of ADP-BeFX (a ground state analog), all three FRET states were observed (Fig. 4F). Addition of ADP-AlFX (a hydrolysis transition state analog) resulted in population of the two low FRET states (Fig. 4G). And in the presence of the hydrolysis product, ADP, only the high FRET peak was observed (Fig. 4H).
Based on these results and other ensemble-averaged control experiments, the authors proposed a two-state kinetic model for unwinding initiation, and explained it through alternating ssRNA/dsRNA-binding properties of the protein at different stages of the ATPase cycle (Fig. 4H): Without nucleotide, the helicase binds to both ssRNA and dsRNA, displaying intrinsic dynamics (i.e., 0.15 and 0.33 FRET states). Initial ATP binding stabilizes the bound complex preventing helicase dissociation from the ssRNA. In the hydrolysis transition state, NPH-II no longer binds the duplex. Following ATP hydrolysis, the helicase no longer binds to ssRNA. Thus, single-molecule experiments provide a mechanistic explanation for the maintenance of RNA-helicase contacts during duplex unwinding initiation, a long-standing question in the field.147,155,156 A future challenge will be to clarify why a high number of ATP molecules are often hydrolyzed prior to strand separation and which specific molecular events induce strand unwinding. This could be achieved by directly labeling the helicase in order to probe its conformational dynamics prior to unwinding, thus complementing smFRET experiments using fluorophore-labeled RNA substrates.
Single-molecule analysis of Mss116-mediated group II intron folding124
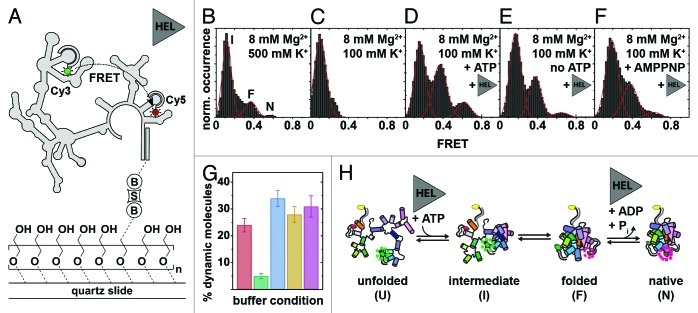
The Saccharomyces cerevisiae DEAD-box helicase Mss116 is essential for mitochondrial group I and group II intron splicing in vivo (Table 2).157 Mss116 activity has been extensively studied in ensemble-averaged experiments using the D135 ribozyme, which is a minimal model system for the S. cerevisiae group II intron ai5γ.158,159 While Mss116 helicase activity depends on ATP binding and hydrolysis, the Mss116-mediated D135 folding mechanism is still not fully understood. As an ideal tool to dissect RNA folding pathways, smFRET experiments were performed to characterize Mss116-mediated D135 folding.124
Fluorescently labeled D135 ribozymes were surface-immobilized onto PEG-coated quartz slides via a biotin-streptavidin linkage,160,161 and FRET was measured using total internal reflection fluorescence (TIRF) microscopy (Fig. 5A).162 Previous smFRET studies have shown that, in the absence of Mss116, D135 requires high Mg2+ concentrations for efficient folding in vitro.162 Moreover, the presence of Ca2+ altered the ribozyme’s structural dynamics and impeded its function.163 Building averaged FRET histograms from over 100 individual time trajectories recorded under precedented high-salt conditions suggested the presence of three distinct FRET states (Fig. 5B).162 As group II introns fold in an Mg2+ ion-dependent fashion,164 it was rationalized in an earlier smFRET study conducted at different Mg2+ ion concentrations that the three FRET states correspond to an extended folding intermediate (I, ~0.1 FRET), a folded intermediate (F, ~0.4 FRET) and the native state (N, ~0.6 FRET).162 Assignment of the native state was further supported by experiments conducted in the presence of the cleavable substrate 17/7, which significantly increased the population of N.162 smFRET trajectories confirmed that F is an obligatory folding intermediate in the D135 folding pathway, because only a small fraction of molecules (< 2%) folded directly from I to N. Under near-physiological conditions (Fig. 5C), smFRET experiments in the absence of Mss116 showed only the presence of the extended intermediate (I), confirming that the ribozyme alone cannot form the native state in low salt. Addition of Mss116 and ATP, however, restored all three FRET states (Fig. 5D), confirming that Mss116 promotes the formation of the native state. Control experiments with other, basic RNA-binding proteins showed that electrostatic interactions alone account for the initial stabilization of the folded intermediate but not the native state. Similar experiments in absence of ATP (Fig. 5E) showed that Mss116 alone can induce the formation of all three FRET states, albeit structural conversion is promoted to a lesser extent. Similar results were obtained for the non-hydrolyzable ATP analog AMPPNP (Fig. 5F). Since Mss116 can facilitate formation of N both in the presence and absence of ATP, the authors proposed that ATP hydrolysis may play a role in Mss116 dissociation and recycling to increase helicase-assisted folding efficiency.
Figure 5. Mss116-mediated group II intron folding using smFRET.124,162 (A) Experimental design. The fluorophore-labeled D135 ribozyme (Cy3, green cycle; Cy5, red cycle) is immobilized on a PEG-coated quartz slide via a biotin-streptavidin linkage (“B,” “S”). Structural interconversion under different folding conditions is monitored by following FRET efficiency over time. (B–F) Averaged FRET histograms, each built from over 100 single-molecule time traces. Imaging conditions: (B) 8 mM Mg2+, 500 mM K+. Three FRET distributions are observed, termed “intermediate” (“I”), “folded” (“F”) and “native” (“N”) based on earlier results.162,163 (C)Eight mM Mg2+, 100 mM K+. Only the “I” FRET state is observed at near-physiological conditions. (D) Eight mM Mg2+, 100 mM K+, 25 nM Mss116, 1 mM ATP. Addition of Mss116 and ATP shifts the distribution of FRET states toward the folded intermediate and the native state. (E and F) Eight mM Mg2+, 100 mM K+, 25 nM Mss116 (and 1 mM AMPPNP). Effect of ATP hydrolysis on D135 folding. Prevalence of the native state is lowered in the absence of ATP (E) and in the presence of non-hydrolyzable AMPPNP (F). (G) Percentage of dynamic molecules at different imaging conditions. Red, 500 mM K+, 8 mM Mg2+; green, 8 mM Mg2+, 100 mM K+; blue, 8 mM Mg2+, 100 mM K+, 25 nM Mss116, 1 mM ATP; yellow, 8 mM Mg2+, 100 mM K+, 25 nM Mss116; purple, 8 mM Mg2+, 100 mM K+, 25 nM Mss116, 1 mM AMPPNP. (H) Proposed model of Mss116-mediated group II intron folding. D135 interconverts between four conformations referred to as “unfolded” (“U”), “intermediate” (“I”), “folded” (“F”) and “native” (“N”). Mss116 promotes the transition from U to I, even in the absence of ATP. It further catalyzes ATP-dependent conversion from F to N. Figure modified from reference 124.
As an inherent advantage, smFRET provides a direct means to characterize the ribozyme’s dynamics and to identify the presence of subpopulations. Single-molecule time traces revealed the presence of two subpopulations: static molecules that did not undergo any changes in FRET during the observation time (minutes) and dynamic molecules that displayed FRET transitions. In the absence of Mss116 and under high salt conditions, the dynamic subpopulation represented ~25% of the observed molecules (Fig. 5G, red). Decreasing the ionic strength to near physiological levels, decreased the dynamic population to below 5% (Fig. 5G, green). Under these ionic conditions, addition of Mss116 and ATP increased the dynamic population to ~35% (Fig. 5G, blue). Removal of ATP, or the presence of the non-hydrolyzable ATP analog (AMPPNP), also yielded ~25–30% of dynamic molecules. The large subpopulations of static molecules in the I state in the absence of Mss116 indicate the presence of at least one high-energy activation barrier for folding between I and F.
Traditionally, folding rate constants (k1, k-1, k2, k-2) are obtained by measuring dwell times in each FRET states using manual thresholds. But noisy data sets or species with close FRET values can make this analysis challenging.61 A powerful alternative is the use of hidden Markov Models (HMM) to analyze the smFRET trajectories, and to determine the folding rate constants and characterize the energy barriers on the folding pathway (Fig. 5H). In the absence of Mss116, high salt concentration was found to accelerate the folding reaction by ~3-fold, while slowing down the unfolding reaction by ~2-fold. Interestingly, addition of Mss116 and ATP had a similar effect. These results indicate that Mss116 functions by decreasing the activation barriers between folding intermediates and by stabilizing the native state.
In summary, this study showed that the ribozyme folds through a series of obligatory intermediates and that Mss116 functions by facilitating formation of specific species (Fig. 2H): formation of the folded intermediate is promoted by electrostatoc interactions, even in the presence of non-specific basic proteins. However, efficient transition to the native state requires the presence of both Mss116 and ATP. In contrast to other RNA-helicase studies in singulo, the authors used a native Mss116 substrate (a large, catalytically competent group II intron ribozyme), under near-physiological ionic conditions. Such large substrates can lead to complex data analysis. For example, the free rotation assumption of the cyanine dyes may not hold true. However, the authors performed anisotropy measurements to address this issue.162 Alternatively, linearly polarized nanosecond ALEX can help monitor fluorophore rotational freedom in real time.144
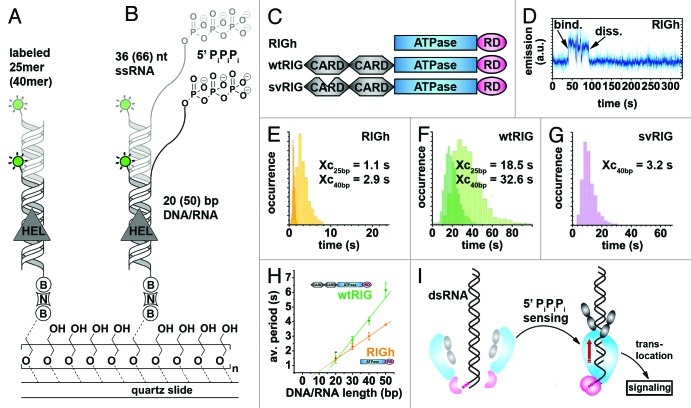
Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocation on double-stranded RNA135
During viral infections, the Retinoid acid inducible-gene protein (RIG-I) specifically recognizes 5′-triphosphate on viral RNA and subsequently initiates the antiviral immune response in the host (Fig. 6 and Table 2).165,166 RIG-I has a modular structure comprising two N-terminal caspase activation and recruitment domains (CARDs), a central ATPase domain, required for antiviral signaling and a C-terminal regulatory domain (RD).167,168 In a recent study, Myong and coworkers employed smPIFE to characterize the role of the central domain and to address whether RIG-I action is primarily induced by the presence of a 5′-triphosphate or the prevalence of dsRNA, both of which are characteristic markers of viral RNA.135 They assessed ability of the full-length protein (wtRIG), as well as two truncated versions of RIG-I (RIGh and svRIG) to recognize different DY547-labeled RNA substrates, followed by translocation along a stretch of dsRNA (Fig. 6A–C).135
Figure 6. Probing RIG-I translocation on dsRNA using PIFE.135 (A and B) Experimental design. A 25/40 bp dsRNA with blunt ends (A) or a 20–50 bp dsRNA with a 66–36 nt ssRNA overhang and a 5′ triphosphate (“PiPiPi”). RNA construct (B) is labeled with a single DY547 fluorophore (green circle) and immobilized on a PEG-passivated surface via biotin-neutravidin (“B,” “N”).161 (C) Depiction of three modular RIG-I variants used in this study. RIGh consists of the central DExH-box ATPase domain and a C-terminal regulatory domain (“RD”). wtRIG additionally has two N-terminal caspase activation and recruitment domains (“CARD”). In svRIG, one of the CARDs is non-functional. (D) Representative time trajectory recorded in the presence of RIGh and the blunt end RNA substrate shown in (A). Helicase binding is accompanied by a sudden increase of fluorophore emission. (E–G) Dwell-time analyses for time traces recorded in the presence of RIGh (E), wtRIG (F) and svRIG (G) for 25-bp and 40-bp blunt end dsRNA shown in (A). RIGh and svRIG translocate faster along dsRNA than wtRIG. In all cases, the average time required for end-to-end translocation increases with the substrate length. (H) RIG-I translocation on dsRNA in the presence of 5′-triphosphate, average time spent in the bound state vs. duplex length. (I) Proposed model for pathogen-associated molecular pattern (“PAMP”) signal integration by RIG-I.168 Binding of the RIG-I regulatory domain (pink) to RNA 5′ triphosphates induces RIG-I dimerization as described previously.174 This triggers the translocase domain (blue), followed by translocation along the dsRNA substrate (red arrow) and induction of a CARD signaling conformation (gray). Figure modified from reference 135.
In an initial set of experiments, fluorophore-labeled dsRNA devoid of a 5′-triphosphate was surface-immobilized onto a polymer-passivated quartz slide (Fig. 6A).135 RIGh binding to the dsRNA resulted in sharp fluctuations of DY547 fluorescence in the single-molecule trajectories (Fig. 6D) that became more frequent in the presence of both RIGh and ATP.135 To test whether RIGh translocates during substrate binding, the authors measured the distribution of dwell times between fluorescence bursts using 25 and 40 basepair dsRNA. As expected for a translocating enzyme, the distribution of dwell times in the bound state increased for the substrate with the longer end-to-end distance (Fig. 6E), indicating that RIGh unidirectionally translocates on dsRNA. Analogous experiments in the presence of wtRIG and ATP revealed similar, though less frequent fluctuations in fluorescence trajectories. The resulting dwell time histograms (1) confirmed the trend toward longer dwell times in the presence of the 40 bp-substrate and (2) revealed a much broader distribution with ~15-fold higher average value as compared with RIGh (Fig. 6F). Decreased translocation velocity along dsRNA lacking the 5′-triphosphate in the presence of CARDs is in agreement with earlier reports,169 and was explained by inhibition of ATP hydrolysis by conducting further ATP-dependent experiments. A splice variant of RIG-I was constructed by removing amino acids 36–80 in the first CARD domain (svRIG, Fig. 6C). The single molecule PIFE trajectories and the resulting dwell time histograms (Fig. 6C) revealed that svRIG behaves similar to RIGh, indicating that translocation activity was restored for svRIG. These results confirm that both CARD domains of RIG-I are required to suppress translocation activity in the absence of the 5′-triphosphate.169
Next, the authors evaluated the effect of a 5′ triphosphate attached to a single-stranded overhang on RIG-I translocation (Fig. 6B). Single-molecule PIFE trajectories and the resulting dwell time histograms show that wtRIG translocates ~20-fold faster on the 5′-triphosphate substrate than on standard dsRNA. This suggests that the 5′-triphosphate prevents ATPase inhibition by the two CARDs, and the authors proposed that RIG-I translocation is preceded by 5′-triphosphate recognition. To rule out scanning on the single-stranded overhang prior to translocation along the dsRNA, the authors varied the ratio of ssRNA and dsRNA stretches and observed linear increase of dwell times with increasing length of the duplex for both RIGh and wtRIG (Fig. 6H). These results pointed the authors to proposing a model where RIG-I first recognizes the 5′-triphosphate of viral RNA, which, in turn, prevents ATPase inhibition by the two CARDs, thus enabling RIG-I unidirectional translocation and the associated immune response cascade (Fig. 6I). However, the way in which the immune response is linked to the unidirectional translocation of RIG-I remains unclear. The authors speculated that repetitive shuttling might inhibit binding of pathogenic proteins playing a role in the viral replication cycle. This does not explain, however, why ATPase-deficient RIG-I mutants capable of unidirectional translocation do not elicit an immune response in vivo. Future studies should be performed with native RNA substrates, as these may form higher-order structures that can be recognized by RIG-I, thus modulating its activity, and in the presence of viral RNA-binding proteins known to play a role in replication.
Conclusions
It is increasingly clear that RNA helicases play numerous crucial and diverse roles in essentially all aspects of RNA metabolism. Here, we have reviewed recent single-molecule studies that have provided unprecedented information on helicase mechanisms. Force-based methods have allowed for the observation of individual mechanistic cycles of different helicases with near-basepair resolution, unveiling unwinding forces and processivity. In turn, fluorescence-based methods have been used to observe and characterize helicase-mediated strand exchange, helicase-RNA interaction, helicase conformational dynamics, as well as helicase translocation along a double helix in real time. The resulting models did not only expand our understanding of helicase mechanisms, but they also underpinned the mechanical and functional diversity between different helicases that are highly related with regard to the conservation of specific sequential motifs. Such detailed characterization would have been hardly accessible with conventional ensemble-averaged techniques, making single-molecule spectroscopy a powerful tool to study these key biological reactions.
In turn, some of the studies reviewed make use of artificially designed RNA model systems (such as RNA hairpins), which may simplify data interpretation, but my also fair to reveal the biological function of the enzyme. Other aspects mimicking a living cell were also missing. Therefore, future studies should not only include other biologically important RNA helicases such as DICER, but also known protein cofactors, for example eIF4G that has been demonstrated to increase eIF4A activity, as well as proteins known to bind to the natural RNA substrate, such as viral cofactors in the case of RIG-I. A possibly interesting approach to simulate the crowded environments inside living cells would be imaging in the presence of molecular crowding agents that have been successfully employed in other research fields.170 Methodological efforts have already resolved mysteries associated with single-molecule data (see the section on NS3). We believe that technical advances will continue to contribute to making single-molecule spectroscopy more robust. Current efforts aim at the development of multidimensional FRET to simultaneously map multiple domains, a new generation of fluorophores with improved photostability and higher spatial and temporal resolution.65,78,131,171-173 We anticipate that such technical improvements will ultimately lead to imaging folding of single RNA molecules inside live cells, which will provide further fascinating insights into the function of RNA helicases.
Acknowledgments
Work in the Rueda and Sigel labs is supported by the NIH (GM085116, D.R.), the NSF (MCB-075068, D.R.), the University of Zürich (R.K.O.S.), the ERC (MIRNA N° 259092, R.K.O.S.) and the Forschungskredit of the University of Zürich (57010302, S.L.B.K.). S.L.B.K. thanks Dr. O. Otto (Technical University of Dresden) for advice on optical tweezers.
Glossary
Abbreviations:
- mRNA
messenger RNA
- rRNA
ribosomal RNA
- tRNA
transfer RNA
- snRNA
small nuclear RNA
- snoRNA
small nucleolar RNA
- miRNA
microRNA
- siRNA
small interfering RNA
- aRNA
antisense RNA
- ATP
adenosine-5′-triphosphate
- SF
superfamiliy
- AFM
atomic force microscopy
- TPM
tethered-particle microscopy
- UTR
untranslated region
- HCV
hepatitis C virus
- NS3
non-structural protein 3
- smFRET
single-molecule Förster resonance energy transfer
- Cy3/Cy5
cyanine dye 3/5
- PIFE
protein-induced fluorescence enhancement
- ALEX
alternating laser excitation
- ADP
adenosine-5′-diphosphate
- ssRNA
single-stranded RNA
- dsRNA
double-stranded RNA
- TIRF
total internal reflection fluorescence
- AMPPNP
5′-adenylyl-β-γ-imidodiphosphate
- PAMP
pathogen-associated molecular pattern
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23507
References
- 1.Crick FHC. Central dogma of molecular biology. Nature. 1970;227:561–3. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 3.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 4.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–60. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–12. doi: 10.1016/S0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 6.Yusupov MM, Yusupova GZ, Baucom Å, Lieberman K, Earnest TN, Cate JHD, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–96. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 7.Bachellerie J-P, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–90. doi: 10.1016/S0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 8.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–37. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 9.Guo Z, Karunatilaka KS, Rueda D. Single-molecule analysis of protein-free U2-U6 snRNAs. Nat Struct Mol Biol. 2009;16:1154–9. doi: 10.1038/nsmb.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37:179–88. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Feltz C, Anthony K, Brilot A, Pomeranz Krummel DA. Architecture of the spliceosome. Biochemistry. 2012;51:3321–33. doi: 10.1021/bi201215r. [DOI] [PubMed] [Google Scholar]
- 12.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12:84–8. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 13.Feng J, Funk WD, Wang S-S, Weinrich SL, Avilion AA, Chiu C-P, et al. The RNA component of human telomerase. Science. 1995;269:1236–41. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10:102–9. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Ferré-D’Amaré AR, Winkler WC. The roles of metal ions in regulation by riboswitches. Met Ions Life Sci. 2011;9:141–73. doi: 10.1039/9781849732512-00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–2. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 18.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–33. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 19.Krupovic M, Cvirkaite-Krupovic V. Virophages or satellite viruses? Nat Rev Microbiol. 2011;9:762–3. doi: 10.1038/nrmicro2676. [DOI] [PubMed] [Google Scholar]
- 20.Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat Rev Microbiol. 2011;9:617–26. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 22.König SLB, Evans AC, Huppert JL. Seven essential questions on G-quadruplexes. BioMol Concepts. 2010;1:197–213. doi: 10.1515/bmc.2010.011. [DOI] [PubMed] [Google Scholar]
- 23.Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl. 1999;38:2326–43. doi: 10.1002/(SICI)1521-3773(19990816)38:16<2326::AID-ANIE2326>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Sigel H, Sigel RKO. Metal ion interactions with nucleic acids and their constituents. In: Reedijk J, Poepelmeier K, eds. Comprehensive Inorganic Chemistry. Oxford, United Kingdom: Elsevier Ltd., 2012:in press. [Google Scholar]
- 25.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res. 2011;44:1302–11. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 26.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–8. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyle AM. Metal ions in the structure and function of RNA. J Biol Inorg Chem. 2002;7:679–90. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 28.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–45. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 29.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol. 2001;11:309–14. doi: 10.1016/S0959-440X(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 30.Williamson JR. Cooperativity in macromolecular assembly. Nat Chem Biol. 2008;4:458–65. doi: 10.1038/nchembio.102. [DOI] [PubMed] [Google Scholar]
- 31.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr Opin Chem Biol. 2005;9:104–9. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Zhao R, Rueda D. Memory effects in RNA folding dynamics revealed by single-molecule fluorescence. In: Russell R, ed. Biophysics of RNA folding. New York, NY: Springer, 2012. [Google Scholar]
- 33.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–4. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 34.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 35.Gingras A-C, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–26. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 36.Parsyan A, Shahbazian D, Martineau Y, Petroulakis E, Alain T, Larsson O, et al. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc Natl Acad Sci USA. 2009;106:22217–22. doi: 10.1073/pnas.0909773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/S0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 38.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–91. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 39.Kar A, Fushimi K, Zhou X, Ray P, Shi C, Chen X, et al. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol Cell Biol. 2011;31:1812–21. doi: 10.1128/MCB.01149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z-R. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol Cell Biol. 2002;22:5443–50. doi: 10.1128/MCB.22.15.5443-5450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 42.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–92. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 44.Gong Z, Dong C-H, Lee H, Zhu J, Xiong L, Gong D, et al. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005;17:256–67. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai M-C, Lee YH, Tarn W-Y. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–58. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranji A, Boris-Lawrie K. RNA helicases: emerging roles in viral replication and the host innate response. RNA Biol. 2010;7:775–87. doi: 10.4161/rna.7.6.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lattmann S, Stadler MB, Vaughn JP, Akman SA, Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011;39:9390–404. doi: 10.1093/nar/gkr630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henn A, Bradley MJ, De La Cruz EM. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu Rev Biophys. 2012;41:247–67. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao W, Coman MM, Ding S, Henn A, Middleton ER, Bradley MJ, et al. Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins. J Mol Biol. 2011;409:399–414. doi: 10.1016/j.jmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henn A, Cao W, Licciardello N, Heitkamp SE, Hackney DD, De La Cruz EM. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc Natl Acad Sci USA. 2010;107:4046–50. doi: 10.1073/pnas.0913081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 52.Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol. 2007;17:316–24. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 53.Pyle AM. RNA helicases and remodeling proteins. Curr Opin Chem Biol. 2011;15:636–42. doi: 10.1016/j.cbpa.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andreou AZ, Klostermeier D. Conformational changes of DEAD-box helicases monitored by single molecule fluorescence resonance energy transfer. Methods Enzymol. 2012;511:75–109. doi: 10.1016/B978-0-12-396546-2.00004-8. [DOI] [PubMed] [Google Scholar]
- 55.Tai C-L, Chi W-K, Chen D-S, Hwang L-H. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–84. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enukashvily N, Donev R, Sheer D, Podgornaya O. Satellite DNA binding and cellular localisation of RNA helicase P68. J Cell Sci. 2005;118:611–22. doi: 10.1242/jcs.01605. [DOI] [PubMed] [Google Scholar]
- 57.Eggleston AK, Rahim NA, Kowalczykowski SC. A helicase assay based on the displacement of fluorescent, nucleic acid-binding ligands. Nucleic Acids Res. 1996;24:1179–86. doi: 10.1093/nar/24.7.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tseng-Rogenski SS, Chang T-H. RNA unwinding assay for DExD/H-box RNA helicases. Methods Mol Biol. 2004;257:93–102. doi: 10.1385/1-59259-750-5:093. [DOI] [PubMed] [Google Scholar]
- 59.Yodh JG, Schlierf M, Ha T. Insight into helicase mechanism and function revealed through single-molecule approaches. Q Rev Biophys. 2010;43:185–217. doi: 10.1017/S0033583510000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gordon MP, Ha T, Selvin PR. Single-molecule high-resolution imaging with photobleaching. Proc Natl Acad Sci USA. 2004;101:6462–5. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanco M, Walter NG. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 2010;472:153–78. doi: 10.1016/S0076-6879(10)72011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diao J, Su Z, Lu X, Yoon T-Y, Shin Y-K, Ha T. Single-vesicle fusion assay reveals Munc18-1 binding to the SNARE core is sufficient for stimulating membrane fusion. ACS Chem Neurosci. 2010;1:168–74. doi: 10.1021/cn900034p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rasnik I, McKinney SA, Ha T. Surfaces and orientations: much to FRET about? Acc Chem Res. 2005;38:542–8. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- 64.Walter NG, Huang C-Y, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 2008;5:475–89. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uphoff S, Holden SJ, Le Reste L, Periz J, van de Linde S, Heilemann M, et al. Monitoring multiple distances within a single molecule using switchable FRET. Nat Methods. 2010;7:831–6. doi: 10.1038/nmeth.1502. [DOI] [PubMed] [Google Scholar]
- 66.Chung HS, Gopich IV, McHale K, Cellmer T, Louis JM, Eaton WA. Extracting rate coefficients from single-molecule photon trajectories and FRET efficiency histograms for a fast-folding protein. J Phys Chem A. 2011;115:3642–56. doi: 10.1021/jp1009669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rueda D, Walter NG. Single molecule fluorescence control for nanotechnology. J Nanosci Nanotechnol. 2005;5:1990–2000. doi: 10.1166/jnn.2005.505. [DOI] [PubMed] [Google Scholar]
- 68.Karunatilaka KS, Rueda D. Single-molecule fluorescence studies of RNA: a decade's progress. Chem Phys Lett. 2009;476:1–10. doi: 10.1016/j.cplett.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao R, Rueda D. RNA folding dynamics by single-molecule fluorescence resonance energy transfer. Methods. 2009;49:112–7. doi: 10.1016/j.ymeth.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 72.Cardo L, Karunatilaka KS, Rueda D, Sigel RKO. Single molecule FRET characterization of large ribozyme folding. In: Sigel A, Sigel H, Sigel RKO, eds. Ribozymes: methods and protocols. Totowa, NJ: Humana Press Inc., 2012:227-51. [DOI] [PubMed] [Google Scholar]
- 73.Andreou AZ, Klostermeier D. The DEAD-box helicase eIF4A: Paradigm or the odd one out? RNA Biol. 2012;10 doi: 10.4161/rna.21966. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hopfner K-P, Michaelis J. Mechanisms of nucleic acid translocases: lessons from structural biology and single-molecule biophysics. Curr Opin Struct Biol. 2007;17:87–95. doi: 10.1016/j.sbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 75.Rasnik I, Myong S, Ha T. Unraveling helicase mechanisms one molecule at a time. Nucleic Acids Res. 2006;34:4225–31. doi: 10.1093/nar/gkl452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steimer L, Klostermeier D. RNA helicases in infection and disease. RNA Biol. 2012;9:751–71. doi: 10.4161/rna.20090. [DOI] [PubMed] [Google Scholar]
- 77.Tinoco I, Chen G, Qu X. RNA Worlds: From Life’s Origins to Diversity in Gene Regulation. In: Atkins JF, Gesteland RF, Cech TR, eds. Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 2011. [Google Scholar]
- 78.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–90. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Henn A, Medalia O, Shi S-P, Steinberg M, Franceschi F, Sagi I. Visualization of unwinding activity of duplex RNA by DbpA, a DEAD box helicase, at single-molecule resolution by atomic force microscopy. Proc Natl Acad Sci USA. 2001;98:5007–12. doi: 10.1073/pnas.071372498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marsden S, Nardelli M, Linder P, McCarthy JEG. Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J Mol Biol. 2006;361:327–35. doi: 10.1016/j.jmb.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 81.Dohoney KM, Gelles J. X-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2000;409:370–4. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 82.Fan H-F, Li H-W. Studying RecBCD helicase translocation along Chi-DNA using tethered particle motion with a stretching force. Biophys J. 2009;96:1875–83. doi: 10.1016/j.bpj.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dessinges M-N, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci USA. 2004;101:6439–44. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun B, Wei K-J, Zhang B, Zhang X-H, Dou S-X, Li M, et al. Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO J. 2008;27:3279–87. doi: 10.1038/emboj.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56:930–3. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 86.Cohen SR, Bitler A. Use of AFM in bio-related systems. Curr Opin Colloid Interface Sci. 2008;13:316–25. doi: 10.1016/j.cocis.2008.02.002. [DOI] [Google Scholar]
- 87.Heus HA, Puchner EM, van Vugt-Jonker AJ, Zimmermann JL, Gaub HE. Atomic force microscope-based single-molecule force spectroscopy of RNA unfolding. Anal Biochem. 2011;414:1–6. doi: 10.1016/j.ab.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 88.Santos NC, Castanho MARB. An overview of the biophysical applications of atomic force microscopy. Biophys Chem. 2004;107:133–49. doi: 10.1016/j.bpc.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 89.Vilfan ID, Kamping W, van den Hout M, Candelli A, Hage S, Dekker NH. An RNA toolbox for single-molecule force spectroscopy studies. Nucleic Acids Res. 2007;35:6625–39. doi: 10.1093/nar/gkm585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Katan AJ, Dekker C. High-speed AFM reveals the dynamics of single biomolecules at the nanometer scale. Cell. 2011;147:979–82. doi: 10.1016/j.cell.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 91.Neaves KJ, Huppert JL, Henderson RM, Edwardson JM. Direct visualization of G-quadruplexes in DNA using atomic force microscopy. Nucleic Acids Res. 2009;37:6269–75. doi: 10.1093/nar/gkp679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Picco LM, Bozec L, Ulcinas A, Engledew DJ, Antognozzi M, Horton MA, et al. Breakingthe speed limit with atomic force microscopy. Nanotechnology. 2007;18:044030. doi: 10.1088/0957-4484/18/4/044030. [DOI] [Google Scholar]
- 93.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt Lett. 1986;11:288–90. doi: 10.1364/OL.11.000288. [DOI] [PubMed] [Google Scholar]
- 94.Dholakia K, Reece P, Gu M. Optical micromanipulation. Chem Soc Rev. 2008;37:42–55. doi: 10.1039/b512471a. [DOI] [PubMed] [Google Scholar]
- 95.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–5. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–94. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rohrbach A. Switching and measuring a force of 25 femtoNewtons with an optical trap. Opt Express. 2005;13:9695–701. doi: 10.1364/OPEX.13.009695. [DOI] [PubMed] [Google Scholar]
- 98.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–7. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–8. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophys J. 1997;72:1335–46. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–9. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 102.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–7. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 103.Hendricks AG, Holzbaur ELF, Goldman YE. Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc Natl Acad Sci USA. 2012;109:18447–52. doi: 10.1073/pnas.1215462109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Block SM. Making light work with optical tweezers. Nature. 1992;360:493–5. doi: 10.1038/360493a0. [DOI] [PubMed] [Google Scholar]
- 105.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–4. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 107.Vega Laso MR, Zhu D, Sagliocco F, Brown AJP, Tuite MF, McCarthy JEG. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J Biol Chem. 1993;268:6453–62. [PubMed] [Google Scholar]
- 108.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3:218–21. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rogers GW, Jr., Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–44. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 110.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–83. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 111.Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011;39:2260–70. doi: 10.1093/nar/gkq1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr., Pyle AM, et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–8. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dimitrova M, Imbert I, Kieny MP, Schuster C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J Virol. 2003;77:5401–14. doi: 10.1128/JVI.77.9.5401-5414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–76. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cheng W, Dumont S, Tinoco I, Jr., Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc Natl Acad Sci USA. 2007;104:13954–9. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 117.Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–80. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 118.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–6. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cheng W, Arunajadai SG, Moffitt JR, Tinoco IJ, Jr., Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–9. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Appleby TC, Anderson R, Fedorova O, Pyle AM, Wang R, Liu X, et al. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J Mol Biol. 2011;405:1139–53. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fairman-Williams ME, Jankowsky E. Unwinding initiation by the viral RNA helicase NPH-II. J Mol Biol. 2012;415:819–32. doi: 10.1016/j.jmb.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kalinin S, Peulen T, Sindbert S, Rothwell PJ, Berger S, Restle T, et al. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat Methods. 2012;9:1218–25. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- 123.Orte A, Clarke R, Klenerman D. Single-molecule two-colour coincidence detection to probe biomolecular associations. Biochem Soc Trans. 2010;38:914–8. doi: 10.1042/BST0380914. [DOI] [PubMed] [Google Scholar]
- 124.Karunatilaka KS, Solem A, Pyle AM, Rueda D. Single-molecule analysis of Mss116-mediated group II intron folding. Nature. 2010;467:935–9. doi: 10.1038/nature09422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–6. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 126.Aregger R, Klostermeier D. The DEAD box helicase YxiN maintains a closed conformation during ATP hydrolysis. Biochemistry. 2009;48:10679–81. doi: 10.1021/bi901278p. [DOI] [PubMed] [Google Scholar]
- 127.Karow AR, Klostermeier D. A conformational change in the helicase core is necessary but not sufficient for RNA unwinding by the DEAD box helicase YxiN. Nucleic Acids Res. 2009;37:4464–71. doi: 10.1093/nar/gkp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Linden MH, Hartmann RK, Klostermeier D. The putative RNase P motif in the DEAD box helicase Hera is dispensable for efficient interaction with RNA and helicase activity. Nucleic Acids Res. 2008;36:5800–11. doi: 10.1093/nar/gkn581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Theissen B, Karow AR, Köhler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci USA. 2008;105:548–53. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Holden SJ, Uphoff S, Hohlbein J, Yadin D, Le Reste L, Britton OJ, et al. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys J. 2010;99:3102–11. doi: 10.1016/j.bpj.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ha T, Tinnefeld P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu Rev Phys Chem. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–3. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 133.Cordes T, Vogelsang J, Tinnefeld P. On the mechanism of Trolox as antiblinking and antibleaching reagent. J Am Chem Soc. 2009;131:5018–9. doi: 10.1021/ja809117z. [DOI] [PubMed] [Google Scholar]
- 134.Vogelsang J, Kasper R, Steinhauer C, Person B, Heilemann M, Sauer M, et al. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew Chem Int Ed Engl. 2008;47:5465–9. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- 135.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–4. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108:7414–8. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang Z, Ji D, Xia A, Koberling F, Patting M, Erdmann R. Direct observation of delayed fluorescence from a remarkable back-isomerization in Cy5. J Am Chem Soc. 2005;127:8064–6. doi: 10.1021/ja050050+. [DOI] [PubMed] [Google Scholar]
- 138.Markiewicz RP, Vrtis KB, Rueda D, Romano LJ. Single-molecule microscopy reveals new insights into nucleotide selection by DNA polymerase I. Nucleic Acids Res. 2012;40:7975–84. doi: 10.1093/nar/gks523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Deniz AA, Dahan M, Grunwell JR, Ha T, Faulhaber AE, Chemla DS, et al. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Förster distance dependence and subpopulations. Proc Natl Acad Sci USA. 1999;96:3670–5. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang Q, Fairman ME, Jankowsky E. DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J Mol Biol. 2007;368:1087–100. doi: 10.1016/j.jmb.2007.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–26. doi: 10.1016/S0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 142.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–19. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 143.Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101:8936–41. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kapanidis AN, Laurence TA, Lee NK, Margeat E, Kong X, Weiss S. Alternating-laser excitation of single molecules. Acc Chem Res. 2005;38:523–33. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 145.Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–6. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 146.Gross CH, Shuman S. Vaccinia virions lacking the RNA helicase nucleoside triphosphate phosphohydrolase II are defective in early transcription. J Virol. 1996;70:8549–57. doi: 10.1128/jvi.70.12.8549-8557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–51. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 148.Shuman S. Vaccinia virus RNA helicase. Directionality and substrate specificity. J Biol Chem. 1993;268:11798–802. [PubMed] [Google Scholar]
- 149.Wong I, Lohman TM. A two-site mechanism for ATP hydrolysis by the asymmetric Rep dimer P2S as revealed by site-specific inhibition with ADP-A1F4. Biochemistry. 1997;36:3115–25. doi: 10.1021/bi9621977. [DOI] [PubMed] [Google Scholar]
- 150.Levin MK, Gurjar MM, Patel SS. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J Biol Chem. 2003;278:23311–6. doi: 10.1074/jbc.M301283200. [DOI] [PubMed] [Google Scholar]
- 151.Chen B, Doucleff M, Wemmer DE, De Carlo S, Huang HH, Nogales E, et al. ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure. 2007;15:429–40. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. 2008;105:20209–14. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107:521–8. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 155.Kawaoka J, Jankowsky E, Pyle AM. Backbone tracking by the SF2 helicase NPH-II. Nat Struct Mol Biol. 2004;11:526–30. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 156.Kawaoka J, Pyle AM. Choosing between DNA and RNA: the polymer specificity of RNA helicase NPH-II. Nucleic Acids Res. 2005;33:644–9. doi: 10.1093/nar/gki208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–8. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nat Struct Mol Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- 159.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–55. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–41. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 161.Lamichhane R, Solem A, Black W, Rueda D. Single-molecule FRET of protein-nucleic acid and protein-protein complexes: surface passivation and immobilization. Methods. 2010;52:192–200. doi: 10.1016/j.ymeth.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Steiner M, Karunatilaka KS, Sigel RKO, Rueda D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci USA. 2008;105:13853–8. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Steiner M, Rueda D, Sigel RKO. Ca2+ induces the formation of two distinct subpopulations of group II intron molecules. Angew Chem Int Ed Engl. 2009;48:9739–42. doi: 10.1002/anie.200903809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM. Productive folding to the native state by a group II intron ribozyme. J Mol Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 165.Binder M, Eberle F, Seitz S, Mücke N, Hüber CM, Kiani N, et al. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286:27278–87. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 167.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–20. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 168.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 169.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–7. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ellis RJ, Minton AP. Cell biology: join the crowd. Nature. 2003;425:27–8. doi: 10.1038/425027a. [DOI] [PubMed] [Google Scholar]
- 171.Lee S, Lee J, Hohng S. Single-molecule three-color FRET with both negligible spectral overlap and long observation time. PLoS One. 2010;5:e12270. doi: 10.1371/journal.pone.0012270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sakatshii SK, Shubeita GT, Dietler G. On the possibility of observation of single quadrupoles by fluorescence resonance energy transfer scanning near-field optical microscopy. Opt Commun. 2000;188:41–5. doi: 10.1016/S0030-4018(00)01154-8. [DOI] [Google Scholar]
- 173.Selvin PR. Lanthanide-based resonance energy transfer. J Sel Top Quant Electron. 1996;2:1077–87. doi: 10.1109/2944.577339. [DOI] [Google Scholar]
- 174.Gack MU, Kirchhofer A, Shin YC, Inn K-S, Liang C, Cui S, et al. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci USA. 2008;105:16743–8. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Campos LA, Liu J, Wang X, Ramanathan R, English DS, Muñoz V. A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy. Nat Methods. 2011;8:143–6. doi: 10.1038/nmeth.1553. [DOI] [PubMed] [Google Scholar]
- 176.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–62. doi: 10.1016/S1097-2765(01)00329-X. [DOI] [PubMed] [Google Scholar]
- 177.Kossen K, Uhlenbeck OC. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 1999;27:3811–20. doi: 10.1093/nar/27.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J Mol Biol. 2002;324:625–36. doi: 10.1016/S0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]