Abstract
CsdA is one of five E. coli DEAD-box helicases and as a cold-shock protein assists RNA structural remodeling at low temperatures. The helicase has been shown to catalyze duplex unwinding in an ATP-dependent way and accelerate annealing of complementary RNAs, but detailed kinetic analyses are missing. Therefore, we performed kinetic measurements using a coupled annealing and strand displacement assay with high temporal resolution to analyze how CsdA balances the two converse activities. We furthermore tested the hypothesis that the unwinding activity of DEAD-box helicases is largely determined by the substrate’s thermodynamic stability using full-length CsdA and a set of RNAs with constant length, but increasing GC content. The rate constants for strand displacement did indeed decrease with increasing duplex stability, with a calculated free energy between -31.3 and -40 kcal/mol being the limit for helix unwinding. Thus, our data generally support the above hypothesis, showing that for CsdA substrate thermal stability is an important rate limiting factor.
Keywords: DEAD-box helicase, CsdA, DeaD, StpA, RNA chaperone, RNA annealing, strand displacement, kinetics, FRET
Introduction
DEAD-box helicases are ubiquitous and important players in RNA metabolism that disrupt RNA duplexes in an ATP-dependent fashion. E. coli encodes five DEAD-box helicases that share nine conserved motifs located in the core of the enzyme.1 The core, formed by two structurally similar repeats, catalyzes ATP hydrolysis and RNA duplex unwinding. Additionally, all E. coli DEAD-box proteins contain highly positively charged C-terminal domains (CTD) of different lengths which might possess auxiliary functions, such as conferring RNA binding specificity, nucleic acid tethering or protein-protein interactions.2-5
Cold-shock DEAD-box protein A (CsdA) has the longest CTD of all five proteins. The 70kDa-protein has been isolated as a suppressor of a mutant of the ribosomal protein S2 gene and is considered to be essential for bacterial growth at low temperatures.6,7 Specifically, CsdA (which is also called DeaD) participates in translational regulation, ribosome biogenesis and the regulation of mRNA stability and degradation.8-12 In vitro, CsdA catalyzes the unwinding of RNA duplexes, provided they have a 5′ or 3′ single-stranded extension or a hairpin structure attached to them.13,14 While some groups provide evidence that this activity is strictly ATP-dependent,13,14 others claim that CsdA can also unwind duplexes in the absence of ATP.7,15 Interestingly, CsdA is also able to accelerate annealing of complementary RNA and DNA strands independently of nucleoside triphosphates.15 In other DEAD-box helicases capable of annealing acceleration the activity has been found to reside mainly in the basic CTD and is thus physically and functionally distinct from the helicases’ duplex unwinding activity.16-19 The balancing of the two converse activities might be facilitated through the ratio between ATP and ADP, although the exact regulation mechanism involving these nucleotides is not clear yet.18,20,21
The mechanism of duplex unwinding has been studied more intensely. As indicated by structural studies, after binding a double-stranded RNA as well as an ATP molecule, the helicase forces the RNA backbone into a bent conformation which is incompatible with its duplex architecture.22-24 This conformational strain is believed to initiate duplex opening. Subsequent ATP hydrolysis and inorganic phosphate release trigger a change in RNA affinity so that the substrate is released.25 Unlike canonical helicases, DEAD-box helicases do not translocate on the substrate. Because of this, unwinding is non-proccessive with the result that the activity is restricted to short or unstable duplexes.13,26,27
While it has been suggested that a substrate’s overall thermodynamic stability rather than only its length determines the efficiency of unwinding through DEAD-box helicases, this hypothesis has not been tested systematically. Therefore, we performed kinetic analysis of E. coli CsdA’s annealing and strand displacement activities using substrates with the same length, but different GC contents. To get insights into the regulation of annealing and unwinding activities, we further tested how the rate constant for CsdA-catalyzed annealing is influenced by different ATP concentrations and other nucleotides as well as helicase concentration.
Results
Design of RNA duplexes with increasing thermal stability
In order to test the ability of CsdA to open up double-strands with different thermal stability, we designed a set of perfectly complementary RNAs with GC-contents between 4.8 and 52.4% and a constant length of 21 base-pairs (Table 1). These artificial substrates were devoid of internal secondary structures up to a GC content of around 50%. The 5′ ends of the RNA strands were labeled with Cy5- (J1 to J4 and J6 strands) or Cy3-dyes (M1 to M4 and M6 strands), respectively, to allow for FRET upon duplex formation (Fig. 1). CsdA had been shown before to most efficiently unwind duplexes containing a helicase binding platform.14 We therefore attached an additional hairpin to the 3′-end of the + strands J1–4 and J6, separated from the duplex by a short stretch of single-stranded RNA, yielding J1h-J4h and J6h (Fig. 1). The hairpin was adopted from the sequence of hairpin 92 in 23S rRNA, a specific binding platform of the RNA helicase DbpA.28 In addition to the extended 3′ end, these RNA substrates contained a 5′-UU-overhang.
Table 1. Sequences of RNA substrates with different thermal stabilities.
| RNA | GC content [%] | ∆G [kcal/mol] | Sequences |
|---|---|---|---|
| JM1 |
4.8 |
-18.4 |
5′- AAUUUAAUGUUUUAUUUAUUA -3′ 3′- UUAAAUUACAAAAUAAAUAAU -5′ |
| J1h |
|
|
5′- UUAAUUUAAUGUUUUAUUUAUUAGGGUUUAUGGCUGUUCGCCAUUU –3′ |
| JM2 |
14.3 |
-23.0 |
5′- AAUUUAAUGUUUUAGUUACUA -3′ 3′- UUAAAUUACAAAAUCAAUGAU -5′ |
| J2h |
|
|
5′- UUAAUUUAAUGUUUUAGUUACUAGGGUUUAUGGCUGUUCGCCAUUU –3′ |
| JM3 |
23.8 |
-26.4 |
5′- AAUUCAAUGUUUUAGUUACUG -3′ 3′- UUAAGUUACAAAAUCAAUGAC -5′ |
| J3h |
|
|
5′- UUAAUUCAAUGUUUUAGUUACUGGGGUUUAUGGCUGUUCGCCAUUU –3′ |
| JM4 |
33.3 |
-31.3 |
5′- AACUCAAUGUCUUAGUUACUG -3′ 3′- UUGAGUUACAGAAUCAAUGAC -5′ |
| J4h |
|
|
5′- UUAACUCAAUGUCUUAGUUACUGGGGUUUAUGGCUGUUCGCCAUUU –3′ |
| JM6 |
52.4 |
-40.0 |
5′- GACUCAGUGUCAGAGUCACUG -3′ 3′- CUGAGUCACAGUCUCAGUGAC -5′ |
| J6h | 5′- UUGACUCAGUGUCAGAGUCACUGGGGUUUAUGGCUGUUCGCCAUUU –3′ |
Cy5-labeled J strands (first line in each row) base-pair with Cy3-labeled M strands (second line in each row) to yield the duplex JM. Strands J1h-J4h and J6h were derived from strands J1–4 and J6 and thus also base-pair with the M strands. They contain an additional 3′hairpin binding-platform (separated by GGGUUU from the base-pairing region) as well as a 5′-UU overhang. Bases forming the hairpin stem are underlined. Thermodynamic stabilities were calculated using the Vienna RNA package cofold algorithm.

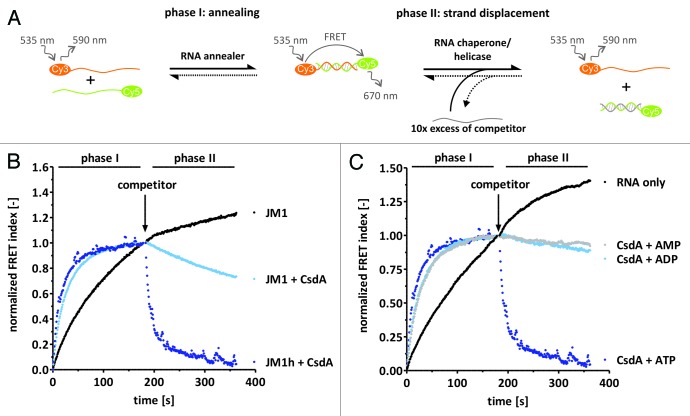
Figure 1. RNAs with a helicase binding platform. RNA substrates JM1 and JM1h were chosen exemplarily to illustrate secondary structure formation in RNAs with the helicase binding platform.
Kinetics of CsdA-catalyzed annealing and helix unwinding
CsdA’s RNA unwinding and annealing activities have so far mainly been tested in assays measuring the equilibrium between double- and single-stranded RNA but detailed kinetic measurements are still lacking.13-15 We therefore employed a FRET-based annealing and strand displacement assay which allows for monitoring the reactions every second, thus providing high resolution and accuracy (Fig. 2A).29 The observed annealing rates of all substrates in the absence of protein were between 0.005 and 0.006 sec−1. Strand displacement did not occur in the absence of protein. Kinetic constants obtained with native gel electrophoresis were identical within error with the FRET measurements (data not shown). Consistent with earlier reports, CsdA catalyzed RNA duplex unwinding efficiently only in the presence of a helicase binding platform. In contrast, blunt-ended substrates were not efficiently unwound (Fig. 2B, Table 2; Fig. S1). Using either the unlabelled J or the M RNA as the competitor strand yielded the same strand displacement rate constants (data not shown). Furthermore, strand displacement activity was strictly dependent on the presence of ATP. In the absence of ATP, the FRET index curves after competitor injection (phase II) could not be fitted to a mono-exponential decrease equation (Fig. 2C). In contrast to RNA unwinding, CsdA accelerated annealing also in the absence of nucleoside triphosphates. ATP stimulated CsdA’s acceleration activity almost 2-fold while ADP and AMP did not have such an effect. Furthermore, CsdA accelerated annealing of blunt-ended substrates as well as of RNAs containing the helicase binding platform (Table 2, Fig. 2B).
Figure 2. Kinetics of CsdA’s RNA annealing and helicase activities. (A) Scheme of the FRET-based annealing and strand displacement assay used to determine RNA annealing, helicase and chaperone activity. Briefly, two complementary 21mers that are 5′-labeled with Cy5 and Cy3, respectively, are annealed with each other which results in a FRET signal (phase I). Proteins that accelerate this reaction are referred to as RNA annealer proteins. Phase II is started through the addition of a 10-molar excess of an unlabeled competitor RNA which will replace one strand if an RNA helicase or chaperone is present to catalyze the reaction, resulting in a decrease of the FRET signal. In the absence of such a protein the FRET index either remains constant or increases further, indicating annealing of the residual single-strands. (B) CsdA-dependent annealing and strand displacement of a blunt-ended RNA (JM1) and a substrate with a helicase binding platform (JM1h). The JM1h ‘RNA only’ curves were identical with the JM1 ‘RNA only’ curves and are thus not shown. (C) Annealing and strand displacement activities of CsdA were tested in the presence of 2 mM AMP, ADP, or ATP and using the JM1h substrate. Observed annealing reaction rates were as follows: kobs, ann (CsdA+ATP) = (0.067 ± 0.026) s−1; kobs, ann (CsdA+ADP) = (0.042 ± 0.006) s−1; kobs, ann (CsdA+AMP) = (0.045 ± 0.026) s−1; kobs, ann (CsdA) = (0.030 ± 0.004) s−1.
Table 2. CsdA-dependent reaction rates of annealing and strand displacement of RNAs with increasing thermal stability as measured with the FRET-based chaperone assay.
| RNA | kobs, ann [s−1] | kobs, SD [s−1] | Duplex displaced [%] |
|---|---|---|---|
| JM1h |
0.078 ± 0.017 |
0.053 ± 0.004 |
75 ± 28 |
| JM1 |
0.045 ± 0.017 |
0.005 ± 0.002 |
n.d. |
| JM2h |
0.091 ± 0.008 |
0.045 ± 0.009 |
94 ± 20 |
| JM3h |
0.088 ± 0.011 |
0.011 ± 0.003 |
39 ± 21 |
| JM4h |
0.073 ± 0.009 |
0.012 ± 0.001 |
45 ± 21 |
| JM4 |
0.061 ± 0.021 |
0.008 ± 0.002 |
n.d. |
| JM6h | No ann acc | No SD | No SD |
The RNA substrates all contain a hairpin linked to their 3′ end to allow for helicase loading. Values are means ± standard deviation of at least three measurements. No SD, no strand displacement; No ann acc, no annealing acceleration; n.d., not determined.
Regulation of CsdA’s RNA annealing and duplex unwinding activities
It has been suggested that in DEAD-box helicases with annealing activity, the two converse activities of duplex unwinding and annealing acceleration are balanced against each other based on the concentration ratio of ATP and ADP.18,20,30 Specifically, high ATP concentrations might inhibit annealing while high ADP concentrations might decrease unwinding activity. We therefore tested the influence of ATP on annealing and strand displacement activities of CsdA using the JM1h substrate and saturating Mg2+ concentrations (Fig. 3A). RNA unwinding catalyzed by CsdA became faster with increasing ATP concentrations, reaching a plateau at ATP concentrations of around 1–2 mM. Annealing acceleration was not inhibited by ATP. On the contrary, ATP concentrations above 0.5 mM even increased the observed rate constant of annealing.
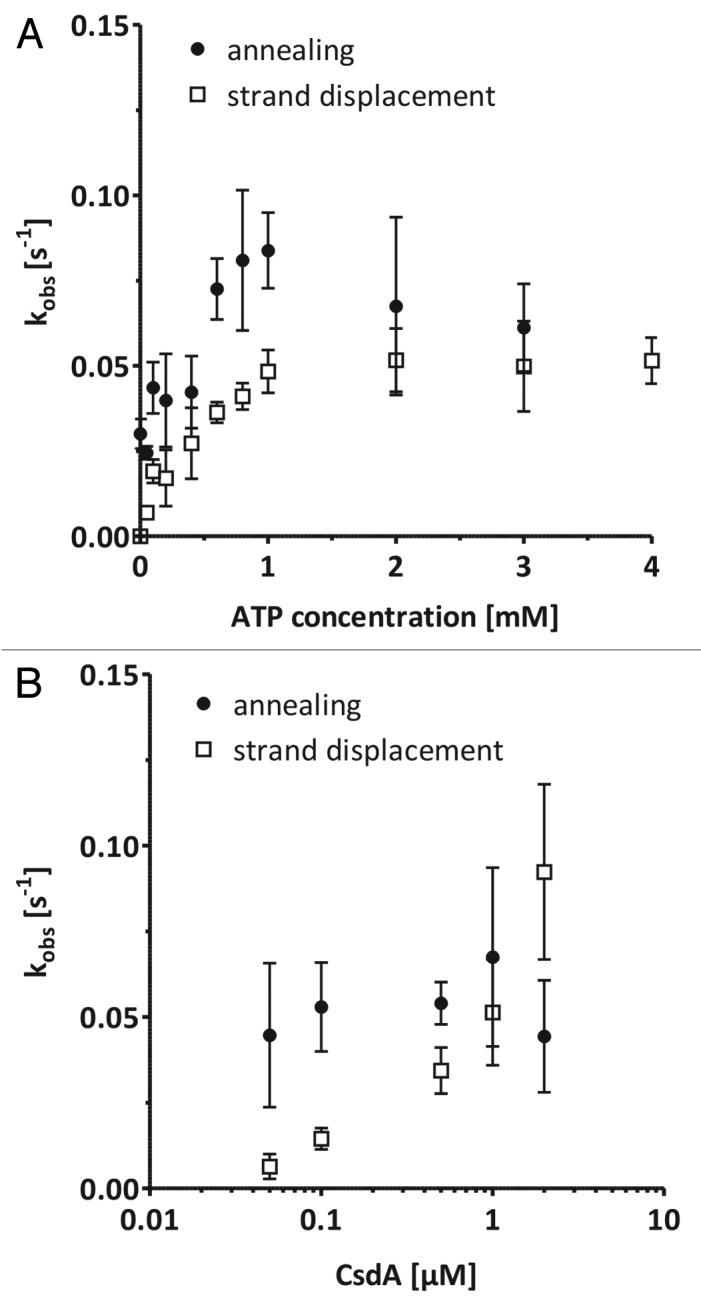
Figure 3. Balancing of annealing and helicase activities. (A) FRET-based annealing and strand displacement assays were performed using the JM1h substrate and different ATP concentrations. (B) Dependence of the observed rate constants kobs, ann and kobs, SD on CsdA concentrations determined with the FRET-based annealing and strand displacement assay. Measurements were performed using the JM1h substrate. Values are means ± standard deviation of at least three measurements.
Besides the ATP concentration, the concentration of the helicase itself influenced the unwinding efficiency strongly as was tested with the JM1h substrate (Fig. 3B). Unwinding was barely detectable at 50 nM CsdA and increased only moderately with increasing CsdA concentration. The highest CsdA concentration we tested was 2 µM which did not confer the maximum unwinding activity. These results are in accordance with Bizebard et al. (2004) who also reported that a high CsdA: RNA ratio was necessary to obtain similar unwinding rates as with lower concentrations of other helicases, e.g., E. coli RhlE. Interestingly, annealing was already efficient at CsdA concentrations of 50 nM corresponding to a protein: RNA molar ratio of 4.5 (RNA length = 21 base-pairs, Fig. 3B). The observed rate constant for annealing did not increase, nor decrease above this CsdA concentration. Thus, the protein concentration might be an important factor to balance annealing and unwinding activities of CsdA
CsdA’s helicase activity is strongly dependent on the substrate’s thermodynamic stability
To test the influence of substrate stability on CsdA’s unwinding activity, we performed our coupled annealing and strand displacement assay using RNA substrates with different thermodynamic stabilities (Table 1). Interestingly, CsdA accelerated annealing of the RNAs JM1h to JM4h with very similar observed rate constants kobs, ann (Table 2). Annealing of JM6h, however, was not accelerated. The observed rate constant for strand displacement decreased strongly with increasing GC content (Table 2, Fig. 4). The most stable of the tested duplexes, JM6h, was not destabilized by CsdA at all. Additionally, the amplitudes of CsdA-catalyzed strand displacement decreased with increasing duplex stability. These data confirm the hypothesis that the duplex stability is a determining factor for unwinding activity.

Figure 4. Strand displacement activities of CsdA and StpA depending on the thermodynamic stability of the RNA duplex. (A) CsdA-catalyzed annealing and strand displacement rates were measured with our combined FRET-based chaperone assay and the substrates JM1h-JM4h and JM6h. (B) Comparison of StpA- and CsdA-catalyzed strand displacement rates kobs, SD depending on the substrate’s GC content. Values are means ± standard deviation of at least three measurements.
As was shown recently for several DEAD-box helicases,25 ATP hydrolysis is not necessary for base-pair disruption, but for efficient helicase recycling. Therefore, we wanted to know how similar the helicases’ mechanism of base-pair destabilization is in comparison to the strand displacement activity of ATP-independent RNA chaperones- another group of proteins responsible for RNA structural remodeling.31 The E. coli protein StpA is one of the best-studied RNA chaperones, catalyzing both annealing and strand displacement.32 We compared StpA’s annealing and strand displacement activities using the same set of RNAs with varying GC contents, although the used RNAs JM1 to 4 and JM6 were blunt-ended (Table 3, Fig. 4). For StpA, the observed annealing rates kobs, ann as well as strand displacement rates kobs, SD were identical within the error range for all seven RNA substrates. Notably, StpA also disrupted the most stable duplex with a GC content of 52.4%. Attachment of the helicase binding platform did not influence StpA-catalyzed annealing or strand displacement rates as tested on the substrate JM3h (Table 3).
Table 3. StpA-catalyzed annealing and strand displacement reactions of RNAs with different GC contents.
| RNA | kobs, ann [s−1] | kobs, SD [s−1] |
|---|---|---|
| JM1 |
0.025 ± 0.004 |
0.016 ± 0.004 |
| JM1h |
0.020 ± 0.006 |
0.014 ± 0.002 |
| JM2 |
0.024 ± 0.004 |
0.012 ± 0.005 |
| JM2h |
0.024 ± 0.004 |
0.007 ± 0.001 |
| JM3 |
0.027 ± 0.004 |
0.019 ± 0.004 |
| JM3h |
0.026 ± 0.003 |
0.019 ± 0.008 |
| JM4 |
0.027 ± 0.005 |
0.016 ± 0.003 |
| JM4h |
0.021 ± 0.003 |
0.014 ± 0.003 |
| JM6 |
0.025 ± 0.006 |
0.015 ± 0.006 |
| JM6h | 0.010 ± 0.002 | 0.014 ± 0.003 |
Observed rate constants were determined with our FRET-based annealing and strand displacement assay. Values are means ± standard deviation of at least three measurements.
Therefore, in contrast to CsdA, StpA’s unwinding activity is completely independent of thermodynamic stability of the RNA substrate. CsdA-catalyzed strand displacement of the two substrates of the lowest GC content was faster than the StpA-catalyzed reaction (Fig. 4). However, because helix disruption through CsdA was inefficient at higher GC contents, the ATP-independent RNA chaperone StpA catalyzed strand displacement more efficiently than the helicase at higher GC contents. CsdA also accelerated annealing significantly more efficient than StpA (compare Tables 2 and 3). In accordance with structural studies,22,23 these results suggest that the mechanisms of base-pair disruption catalyzed by RNA chaperones and helicases are fundamentally different from each other.
Discussion
CsdA’s strand displacement activity
Our data support the hypothesis that in DEAD-box helicases, besides the duplex length, the overall stability of the substrate is limiting for duplex unwinding. The RNAs used in this study formed 21 base-pair duplexes which exceeds the substrate length of 15 base-pairs which other groups suggested to be the upper limit for duplex unwinding by CsdA14 and other DEAD-box helicases.27 The free energy we determined to be the limit for helix CsdA-catalyzed disruption is between -31.3 to -40 kcal/mol, corresponding to substrates JM4h and JM6h. This seems to be the free energy that must be provided through the formation of interactions between the helicase and the RNA substrate, the ATP molecule bound as well as internal protein-protein interactions. Structural studies show that DEAD-box helicase cores can accommodate RNA helices of between six and ten base pairs in length.22,23,33 A question that remains to be addressed is how the helicase ‘senses’ the overall stability of the duplex while contacting only a part of the substrate. The enzyme might need a nucleation site to start unwinding, which needs to be rather unstable.
In contrast to earlier studies,7,15 we did not find CsdA to possess ATP-independent strand displacement activity (Fig. 2C). We think that co-purified ribosomal proteins with RNA chaperone activity might have been responsible for the ATP-independent unwinding activity reported by Jones et al. (1996). Zhao and Jain (2011) used 12 base-pairs short RNA duplexes for their strand displacement assays which are not stable enough to exclude spontaneous base-pair opening.34 Thus, they probably measured accelerated annealing of the competitor strand to the breathing duplex. Another feature differentiates CsdA from ATP-independent RNA chaperones. While the latter protein group usually prefers binding of a single-stranded substrate over a double-stranded RNA,35 no such preference was found for DEAD-box helicases so far.36
CsdA’s annealing activity
Consistent with a recent report,15 CsdA stimulated annealing of most of our RNA substrates independently of the presence of a helicase binding platform or the substrate’s stability (Table 2, Fig. 2B). The lack of acceleration of JM6h annealing might be due to low binding affinity for RNAs J6h and M6. The annealing acceleration of blunt-ended RNAs as well as the dispensability of ATP suggest that CsdA’s annealing acceleration activity resides in a different part of the protein than its helix unwinding activity. The annealing activity of other DEAD-box helicases was localized mainly to their CTDs.16-19 Indeed, CsdA has a long C-terminal extension with a locally high basicity.13 Positively charged protein domains or stretches often facilitate annealing, e.g., through co-localization of complementary RNA strands, stabilization of the annealing transition state or the selection of an RNA conformation most suitable for annealing.37-40 Therefore, the CTD of CsdA is a likely candidate for the helicase’s annealing activity.
Other groups had suggested a nucleotide-conferred regulation of helicase’s annealing activity that would help balancing the two activities, annealing stimulation and duplex unwinding.18,20 We did not observe a negative (or positive) influence of ADP or AMP on observed rate constants of annealing kobs, ann (Fig. 2C). ATP even increased the observed rate constants about 1.8-fold (Fig. 3A), which might paradoxically be due to a higher unwinding activity at these ATP concentrations. During phase I of the FRET-based assay, structures can form that do not resemble the expected 21 base-pair duplex and yield a lower FRET efficiency. Through the ATP-dependent destabilization of these, more molecules arrive at the thermodynamically most stable structure within a shorter time frame so that the apparent annealing rate increases. Without decreasing overall annealing rates, the ATP concentration can, however, shift the equilibrium between single-strands and double-strands, which is simply due to an increased unwinding rate. For example, Halls et al.21 found that ATP decreased the amplitude of annealing while the rate constant for annealing was constant. Besides the concentration of nucleotides, the CsdA concentration might play a role in balancing the two RNA remodeling activities. We found that annealing acceleration was already efficient at CsdA concentrations as low as 50 nM while unwinding was significant only above 100 nM CsdA and exceeded annealing at CsdA concentrations above 1 µM.
Whether the annealing activity of CsdA and other helicases plays a role in RNA metabolism remains to be investigated. The CTD of CsdA has been found to be important for bacterial survival under cold-shock conditions,14 but unidentified functions other than annealing stimulation might be responsible for this effect. Generally, acceleration of annealing and duplex unwinding might support the remodeling of RNA structure in a synergistic fashion rather than a competitive one.
Material and Methods
Reagents and RNAs
Reagents were ordered from AppliChem, Merck and Sigma. All RNAs were ordered from Eurogentec, dissolved in H2O and stored at -20 or -80°C (for long-term storage). Sequences are listed in Table 1.
Protein purification
Purification of the E. coli protein StpA was performed as described elsewhere.41 The purified protein ran as a single band on a silver-stained SDS-PAGE. Protein concentrations were determined using the BioRad Protein assay.
Full-length CsdA was produced in E. coli BL21 DE3 (Novagen) using the plasmid pCsdA13 and 1 L Luria-Bertani medium supplemented with 100 μg/ml of ampicillin. The cells were grown to an OD600 of 0.6 - 0.8 and the expression was induced through the addition of 0.5 mM IPTG. After 4 h, the cells were harvested by centrifugation, resuspended in lysis buffer (20 mM HEPES pH 7.5, 1 M NaCl, 0.1 mM EDTA, 1 mM β-mercaptoethanol, 1 mM PMSF and 10 μg/ml DNase I) with 1 g cells/5 ml buffer and lysed using a French press. After 30 min of incubation on ice, cell debris were removed by centrifugation (45,000 × g, 20 min). Ni2+-affinity purification was performed on an Äkta FPLC (GE Healthcare) using a 1 ml His-Trap FF column (GE Healthcare), charged with 0.1 M NiSO4 and equilibrated with buffer A (20 mM HEPES pH 7.5, 1 M NaCl, 0.1 mM EDTA, 1 mM β-mercaptoethanol). The bound protein was washed extensively with 50 column volumes of buffer A and eluted with buffer A plus 500 mM imidazole. The purified protein was concentrated using a centrifugal filter unit (Amicon Ultracell 25 kDa cut-off) to a volume applicable to size-exclusion chromatography (below 5 ml). Size exclusion chromatography was performed on a GE Healthcare, Tricorn 16/600 Superdex 200 column, prior equilibrated with buffer A. Fractions collected from size-exclusion were concentrated under stirring using a 10 ml Amicon nitrogen pressure cell equipped with a Millipore Ultrafiltration membrane (25 kDa cut-off) to ~5 mg protein / ml in the same buffer. Protein concentration was determined using the Bradford assay.
FRET assays
Combined annealing and strand-displacement assays were performed and analyzed as described by Rajkowitsch and Schroeder29 using a GENios Pro or Infinite F500 microplate reader (both from TECAN) and applying the following changes. Concentrations of donor and acceptor dye-labeled RNAs were 11 nM in a total volume of 45 µl (phase I). To start the strand displacement reaction after 180 sec, 5 µl of the unlabeled competitor strand were added to reach a final concentration of 100 nM in a final volume of 50 µl (phase II). Five µl was the minimal volume that could be injected by the microplate reader while still ensuring high accuracy. The strand concentrations of the labeled strands were thus reduced to 10 nM in phase II. The basic FRET-buffer contained 50 mM Tris-Cl pH 7.5, 3 mM MgCl2 and 1 mM DTT. If not stated otherwise, 2 mM ATP and additional 2 mM MgCl2 (final concentration 5 mM MgCl2) were added to samples containing CsdA to allow for helicase activity. The ATP-dependence of the CsdA-catalyzed rate constants for annealing and strand displacement were measured at total MgCl2 concentrations of 5 mM. Higher MgCl2 concentrations did not increase unwinding, indicating that 5 mM MgCl2 were saturating. Measurements were performed at 30°C and protein concentrations of 1 µM CsdA or 1 µM StpA if not stated otherwise. Annealing data were fitted to the following second-order reaction equation using Prism 5 (GraphPad Software, San Diego, CA):
| (1) |
(with Y- FRET index, Aann- amplitude of annealing, kobs, ann- observed rate constant for annealing, t-time).
FRET signals describing strand displacement were fit to a mono-exponential decay:
| (2) |
(with kobs, SD- observed rate constant for strand displacement, ASD- amplitude of strand displacement).
The extent of duplex displacement was calculated with
For better visualization in figures, FRET indexes were normalized to 1 (phase II) or to 0 and 1 (phase I).
RNA stability calculations
Free energies of RNAs were calculated using the Vienna RNA package cofold routine42,43 and the default settings (Turner model 1999, 37°C). This algorithm yielded very similar results as mfold44 and thus the values are comparable to the stabilities of RNA substrates used in other studies.
Supplementary Material
Acknowledgments
This work was funded by the University of Vienna and the Austrian Science Fund FWF through the SpecialResearch Program (SFB17) on ‘Modulators of RNA fate and function’, grant F1703 to RS. We thank Bob Zimmermann for proof-reading the manuscript.
Glossary
Abbreviations:
- CsdA
Cold-shock DEAD-box protein A
- CTD
C-terminal domain
- FRET
Förster resonance energy transfer
- StpA
suppressor of the td- phenotype A
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/rnabiology/article/23475
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23475
References
- 1.Iost I, Dreyfus M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006;34:4189–97. doi: 10.1093/nar/gkl500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J Mol Biol. 2002;324:625–36. doi: 10.1016/S0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 3.Klostermeier D, Rudolph MG. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res. 2009;37:421–30. doi: 10.1093/nar/gkn947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, et al. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci U S A. 2011;108:12254–9. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J Biol Chem. 2005;280:35499–505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 6.Toone WM, Rudd KE, Friesen JD. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173:3291–302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones PG, Mitta M, Kim Y, Jiang W, Inouye M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Aoki H, Ganoza MC. Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. Int J Biochem Cell Biol. 1999;31:215–29. doi: 10.1016/S1357-2725(98)00142-3. [DOI] [PubMed] [Google Scholar]
- 9.Moll I, Grill S, Gründling A, Bläsi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol Microbiol. 2002;44:1387–96. doi: 10.1046/j.1365-2958.2002.02971.x. [DOI] [PubMed] [Google Scholar]
- 10.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–9. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peil L, Virumäe K, Remme J. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 2008;275:3772–82. doi: 10.1111/j.1742-4658.2008.06523.x. [DOI] [PubMed] [Google Scholar]
- 12.Palonen E, Lindström M, Somervuo P, Johansson P, Björkroth J, Korkeala H. Requirement for RNA helicase CsdA for growth of Yersinia pseudotuberculosis IP32953 at low temperatures. Appl Environ Microbiol. 2012;78:1298–301. doi: 10.1128/AEM.07278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–66. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 14.Turner AM, Love CF, Alexander RW, Jones PG. Mutational analysis of the Escherichia coli DEAD box protein CsdA. J Bacteriol. 2007;189:2769–76. doi: 10.1128/JB.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao XL, Jain C. DEAD-box proteins from Escherichia coli exhibit multiple ATP-independent activities. J Bacteriol. 2011;193:2236–41. doi: 10.1128/JB.01488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valdez BC. Structural domains involved in the RNA folding activity of RNA helicase II/Gu protein. Eur J Biochem. 2000;267:6395–402. doi: 10.1046/j.1432-1327.2000.01727.x. [DOI] [PubMed] [Google Scholar]
- 17.Valdez BC, Henning D, Perumal K, Busch H. RNA-unwinding and RNA-folding activities of RNA helicase II/Gu--two activities in separate domains of the same protein. Eur J Biochem. 1997;250:800–7. doi: 10.1111/j.1432-1033.1997.00800.x. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 19.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, et al. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375:1344–64. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlmann-Schiffler H, Jalal C, Stahl H. Ddx42p--a human DEAD box protein with RNA chaperone activities. Nucleic Acids Res. 2006;34:10–22. doi: 10.1093/nar/gkj403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–55. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 23.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–14. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers GW, Jr., Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–44. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 27.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–12. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–9. doi: 10.1017/S1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajkowitsch L, Schroeder R. Coupling RNA annealing and strand displacement: a FRET-based microplate reader assay for RNA chaperone activity. Biotechniques. 2007;43:304–10, 306, 308 passim. doi: 10.2144/000112530. [DOI] [PubMed] [Google Scholar]
- 30.Flores-Rozas H, Hurwitz J. Characterization of a new RNA helicase from nuclear extracts of HeLa cells which translocates in the 5′ to 3′ direction. J Biol Chem. 1993;268:21372–83. [PubMed] [Google Scholar]
- 31.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–30. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 32.Doetsch M, Gstrein T, Schroeder R, Fürtig B. Mechanisms of StpA-mediated RNA remodeling. RNA Biol. 2010;7:735–43. doi: 10.4161/rna.7.6.13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–54. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 34.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–4. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 35.Doetsch M, Stampfl S, Furtig B, Beich-Frandsen M, Saxena K, Lybecker M, et al. Study of E. coli Hfq's RNA annealing acceleration and duplex destabilization activities using substrates with different GC-contents. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Müller UF, Göringer HU. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res. 2002;30:447–55. doi: 10.1093/nar/30.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doetsch M, Fürtig B, Gstrein T, Stampfl S, Schroeder R. The RNA annealing mechanism of the HIV-1 Tat peptide: conversion of the RNA into an annealing-competent conformation. Nucleic Acids Res. 2011;39:4405–18. doi: 10.1093/nar/gkq1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hargittai MR, Mangla AT, Gorelick RJ, Musier-Forsyth K. HIV-1 nucleocapsid protein zinc finger structures induce tRNA(Lys,3) structural changes but are not critical for primer/template annealing. J Mol Biol. 2001;312:985–97. doi: 10.1006/jmbi.2001.5021. [DOI] [PubMed] [Google Scholar]
- 40.Nedbal W, Frey M, Willemann B, Zentgraf H, Sczakiel G. Mechanistic insights into p53-promoted RNA-RNA annealing. J Mol Biol. 1997;266:677–87. doi: 10.1006/jmbi.1996.0813. [DOI] [PubMed] [Google Scholar]
- 41.Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35:1257–69. doi: 10.1093/nar/gkl1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70-4. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernhart SH, Tafer H, Mückstein U, Flamm C, Stadler PF, Hofacker IL. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol Biol. 2006;1:3. doi: 10.1186/1748-7188-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.