Abstract
Ski2-like RNA helicases are large multidomain proteins involved in a variety of RNA processing and degradation events. Recent structures of Mtr4, Ski2 and Brr2 provide our first view of these intricate helicases. Here we review these structures, which reveal a conserved ring-like architecture that extends beyond the canonical RecA domains to include a winged helix and ratchet domain. Comparison of apo- and RNA-bound Mtr4 structures suggests a role for the winged helix domain as a molecular hub that coordinates RNA interacting events throughout the helicase. Unique accessory domains provide expanded diversity and functionality to each Ski2-like family member. A common theme is the integration of Ski2-like RNA helicases into larger protein assemblies. We describe the central role of Mtr4 and Ski2 in formation of complexes that activate RNA decay by the eukaryotic exosome. The current structures provide clues into what promises to be a fascinating view of these dynamic assemblies.
Keywords: Brr2, Mtr4, RNA helicase, RNA processing, Ski2, structure
Ski2-like RNA helicases are a relatively small family of superfamily 2 helicases.1-5 A distinctive feature of these helicases is their large size, spanning 120–225 kDa in S. cerevisiae. Named for their founding member, Ski2,6-9 Ski2-like RNA helicases play important roles in RNA degradation, processing and splicing pathways.1,10 As helicases, they process RNAs in a 3′-5′ direction. They generally function as parts of larger complexes and in many cases appear to act as a platform for complex assembly. Several recent studies have given us our first structural view of this family. Here we highlight the common architectural themes and functional implications emerging from these structures. We also discuss the role of Ski2-like helicases in larger complexes, with a particular emphasis on complexes involved in activation of the exosome for RNA degradation and processing.
Ski2-Like Family Helicase Structures
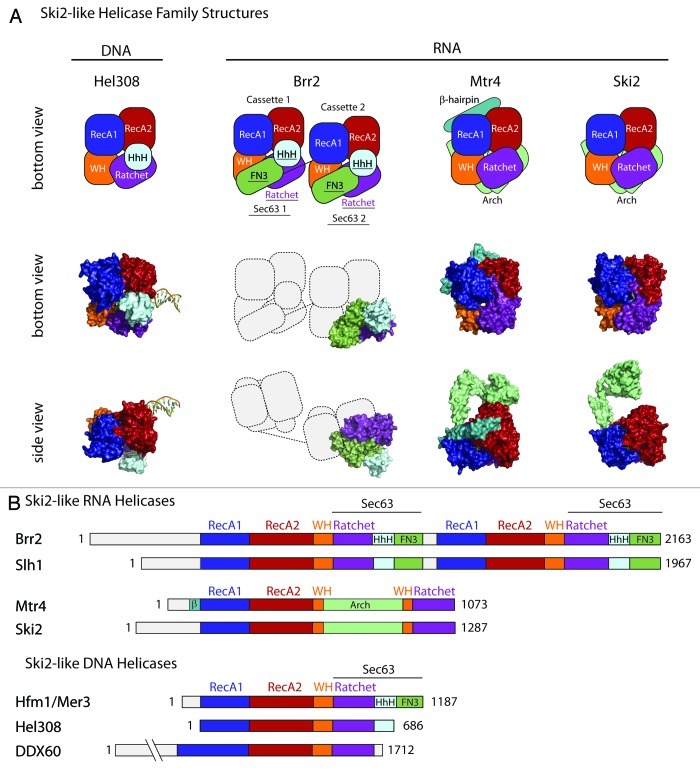
In S. cerevisiae, four Ski2-like RNA helicases have been identified, including Ski2, Mtr4, Brr2, and Slh1. (Suv3 has previously been categorized as Ski2-like, but recent studies indicate that it is phylogenetically2 and structurally11 distinct.) Human homologs exist for each protein. The biology of these helicases has been extensively reviewed.1,10,12-22 Briefly, Mtr4 and Ski2 promote RNA decay, Brr2 functions in pre-mRNA splicing, and Slh1 is involved in translation and transcription. The structural data currently available for Ski2-like helicases is summarized in Table 1 and Figure 1, and includes nearly complete structures of Mtr423,24 (75 N-terminal residues are missing) and an equivalent region for Ski2.25 Partial structures for Brr2 have also been determined26,27 that correspond to the C-terminal Sec63 domain from the second of two helicase repeats. The only structure bound to RNA substrates is an Mtr4 complex bound to ADP and a five nucleotide single stranded RNA.24 In addition to the RNA helicase structures, several related Ski2-like DNA helicase structures are also available.28-31 As will be discussed below, the structure of Hel308,28 an archaeal DNA helicase from A. fulgidus, includes a partially unwound DNA duplex and has provided valuable insight into unwinding and translocation mechanisms that are likely applicable to the RNA helicases.
Table 1. X-ray crystal structures of Ski2-like helicases.
| Protein Name | Organism | PDB | Substrate | Domains obs.* | Function | Reference |
|---|---|---|---|---|---|---|
|
Ski2-like RNA helicases (eukaryotic) | ||||||
|
Brr2, |
S. cerevisiae |
3HIB |
— |
4,5,6a |
RNA splicing |
2009 Zhang et al.27 |
| (Rss1/Slt2/Prp44/Snu246) |
|
3IM1 3IM2 |
— — |
4,5,6 4,5,6 |
|
2009 Pena et al.26 |
| U5–200KD |
H. sapiens |
2Q0Z |
— |
4,5,6 |
|
2007 NESG† |
|
Mtr4, (Dob1) |
S. cerevisiae |
3L9O |
— |
1,2,3,3ab,4 |
Nuclear RNA |
2010 Jackson et al.23 |
| |
|
2XGJ |
RNA/ADP |
1,2,3,3a,4 |
decay |
2010 Wier et al.24 |
|
Ski2 |
S. cerevisiae |
4A4Z |
ADPNP |
1,2,3,3a,4 |
Cytoplasmic |
2012 Halbach et al.25 |
| |
|
4A4K |
— |
3a |
RNA decay |
|
|
Ski2-like DNA helicases (archaeal) | ||||||
|
Hel308, |
A. fulgidus |
2P6R |
DNA |
1,2,3,4,5 |
DNA repair & |
2007Buttner et al.28 |
| (Hjm) |
|
2P6U |
— |
1,2,3,4,5 |
recombination |
|
| |
P. horikoshii |
2Z41 |
— |
1,2,3,4,5 |
|
2008 Zhang et al.31 |
| |
S. solfataricus |
2VA8 |
— |
1,2,3,4,5 |
|
2008 Richards et al.30 |
| P. furiosus | 2ZJA 2ZJ8 2ZJ5 2ZJ2 |
ADPNP — ADP — |
1,2,3,4,5 1,2,3,4,5 1,2,3,4,5 1,2,3,4,5 |
2009 Oyama et al.29 | ||
Ski2-like helicase observed domains are numbered as follows 1 = RecA1, 2 = RecA2, 3 = Winged Helix, 3a = Arch, 4 = Ratchet domain, 5 = Helix hairpin Helix and 6 = FN3. †Northeast Structural Genomics Consortium, unpublished. aDomains 4,5 and 6 form the previously named Sec63 domain. bThe Arch domain of Mtr4 and Ski2 (3a) inserts in the middle of domain 3.
Figure 1. Members of the Ski2-like helicase family share a common four domain helicase core. (A). Schematic representations (top row) highlight the architectural arrangement of the Ski2-like helicases for the Brr2, Mtr4 and Ski2 RNA helicases and the related Hel308 DNA helicase. Representative structures are also indicated (middle and bottom rows). The helicase core is composed of the RecA1, RecA2, Winged Helix (WH) and Ratchet domains (white lettering). Accessory domains are also indicated (black lettering). Brr2 contains two helicase core cassettes. The current Brr2 structures are limited to the second Brr2 Sec63 domain (composed of the ratchet, HhH and FN3 domains). (B) Domain organization for the Ski2-like helicases. Amino acid residue numbers are from S. cerevisiae, except for Hel308 (A. fulgidus) and DDX60 (H. sapiens). DNA or RNA helicase activity has not been demonstrated for DDX60, although it has been shown to bind both DNA and RNA.119 Uncharacterized N-terminal regions are not indicated in (A) but are included in (B) as gray boxes. Structures were rendered using PyMol.120
Common Architecture of Ski2-Like Helicases
All superfamily 2 (SF2) helicases contain a pair of RecA-like domains arranged on a single polypeptide with N- and C-terminal extensions.3,5 These two domains constitute the “core” helicase domains and provide the motor associated with helicase activity.32,33 Conserved sequence motifs found within the RecA domains are involved in ATP and nucleic acid interactions. Classification of helicase families is typically based on the precise sequence and arrangement of these motifs. In the case of Ski2-like helicases, sequence analysis has identified two RecA domains containing 12 conserved sequence motifs.1-5 The Mtr4 and Ski2 structures confirm that the RecA domains (designated here as RecA1 and RecA2) adopt structures and sequence motif arrangements similar to that observed in other SF2 helicases (Fig. 1).
Despite the strong conservation of the RecA domains between Ski2-like helicase family members, it was unclear whether any other similarities existed between family members until determination of crystal structures. The Hel308 structure28 revealed that a small winged helix domain and a larger seven helix bundle or “ratchet” domain packs against the RecA domains to form a ring-like structure that accommodates passage of a single strand of DNA. An additional C-terminal helix-hairpin-helix (HhH) domain interacts with the ratchet domain and the exiting DNA strand. Structures of the second Brr2 Sec63 domain26,27 showed that it resembles the ratchet and HhH domains of Hel308, and additionally contains a Fibronectin type III domain (FN3). Unexpectedly, the Mtr423,24 and Ski225 structures showed that they also contain a winged helix and ratchet domain similar to what is seen in the Hel308 structure. A large insertion in the middle of the winged helix domain in Mtr4 and Ski2 (the arch domain, discussed below) had prevented prior identification of this domain by sequence analysis. Likewise, although the sequence similarity between the Hel308/Brr2 seven helix bundle and the Mtr4/Ski2 eight helix bundle was not obvious (11% sequence identity/26% similarity), they are clearly structurally related (RMSD = 2.7 Å for 106 residues out of 140 for Hel308 and 162 for Mtr4, as calculated by PDBeFold34).
Combined, these structures demonstrate that the fundamental molecular “core” of all Ski2-like helicases is a ring-like four domain assembly of two RecA domains, a winged helix domain and a ratchet domain (Fig. 1). Recent structures of DEAH/RHA-box RNA helicases have revealed a similar core structure,35-37 indicating that it may be a common feature with useful characteristics for a variety of helicases. This common architecture has been referred to elsewhere as the DExH-box core.24 Here we will simply refer to it as the helicase core.
Unwinding Mechanism
Our current understanding of RNA/DNA unwinding and translocation by Ski2-like helicases has been developed primarily from structures of the Hel308 DNA helicase,28 and is detailed in recent reviews.1,32,38 Key structural features of the proposed mechanism include a β-hairpin in domain RecA2 that acts as a wedge to split the DNA duplex; nucleic acid backbone interactions with conserved motifs in RecA1 and RecA2; and stabilizing base stacking interactions opposite the RecA domains with the ratchet helix of the ratchet domain. ATP binding and hydrolysis between the RecA domains are believed to cause conformational changes that allow the helicase to step along the DNA backbone using an inchworm mechanism.39
The Mtr4 and Ski2 structures suggest that Ski2-like RNA helicases function similarly to Hel308. All of the key structural features described in Hel308 that are proposed to be important for unwinding are retained in Mtr4 and Ski2, including the β-hairpin and ratchet helix. Although no structures of the corresponding region exist for Brr2, mutagenic disruption of the predicted β-hairpin results in a slow growth phenotype in vivo and loss of helicase activity in vitro.27 No duplex RNA has been crystallized with either Mtr4 or Ski2, so strand splitting is not directly observed. However, the RNA-bound Mtr4 structure does show backbone interactions with the conserved RecA motifs Ia, Ib, IV and V. Interactions with the RNA bases are observed along the ratchet helix, although it has been noted that there are some differences in the mode of RNA binding between the two crystallographically observed molecules for Mtr4.40 The ratchet helix interaction also differs slightly in detail between Mtr4 and Hel308, but it remains to be determined whether this corresponds to any functional differences between the two proteins. What is clear is that the ratchet domain is functionally important. Deletion of the ratchet domain of Mtr4 is lethal,41 consistent with the loss of unwinding activity observed in Hel308 when the ratchet domain is removed.28 The Mtr4–1 mutation (C942Y), which arose from the initial genetic screen that identified Mtr4, is located on a loop near the ratchet helix and produces a growth phenotype in S. cerevisiae.42 Similarly, mutations along the putative ratchet helix in Brr2 result in slow growth phenotypes and also exhibit reduced unwinding activity in vitro.26,27,43
It has been proposed that the base stacking interactions mediated by the ratchet helix provide a mechanism for enforcing directionality during translocation.28,32 These interactions, combined with the ring-like shape of the helicase core, may also confer some degree of processivity to the helicase. However, while Hel308 has been described as a processive helicase,28,44 the processivity of the RNA helicases has proven difficult to determine.26,45
Accessory Domains
The unique characteristics of Ski2-like family members are derived in large measure from accessory domains that decorate the helicase core and provide expanded functionality. Indeed, this appears to be a common theme throughout SF2 helicase families.2,13
Arch Domain
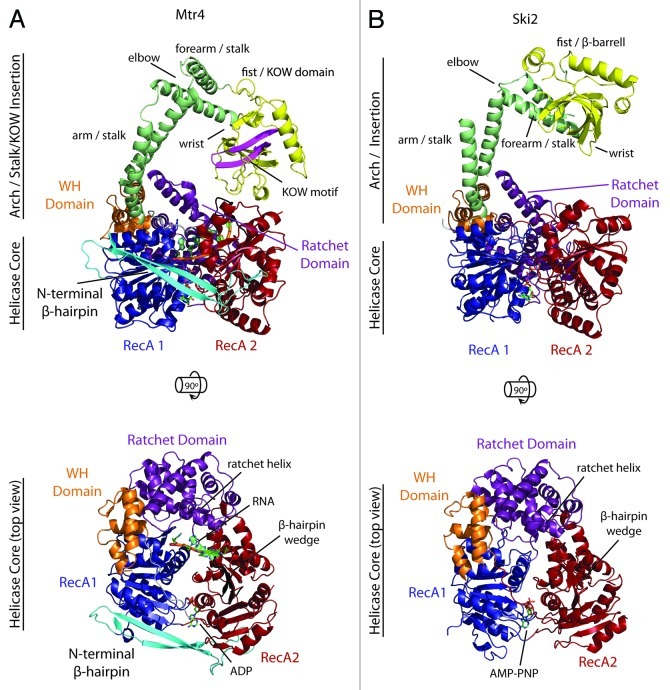
The most distinctive Ski2-like accessory domain described to date is the Arch domain of Mtr4 and a related domain in Ski2. The domain is a prominent 256–266 amino acid insertion within the winged helix domain that is unique to Mtr4 and Ski2 (Fig. 1). Originating from the back side of the helicase core, two anti-parallel coiled coils (arm and forearm), each composed of an ascending and descending helix, extend over the helicase core and terminate in a globular region called the fist that in Mtr4 is positioned directly above the RNA entry site. A dramatic bend between the arm and forearm creates the arch-like appearance for which the domain was named. In Ski2, this bend is stabilized by a zinc-binding CCCH-type motif.25 We note that multiple terms have been used in the literature to describe the same structural features throughout this domain, as indicated in Figure 2.
Figure 2. Comparison of Mtr4 and Ski2. The structures of Mtr4 (A) and Ski2 (B) are shown from a side-view and top-view (PDB ID 2XGJ and 4A4Z, respectively). For clarity, the arch domain has been removed in the top-view. Domain names and important structural features are indicated, including alternative nomenclature used in the literature.
While the arch domain is common both to Mtr4 and Ski2, significant sequence and structural differences exist between the two helicases (Fig. 2). In contrast to the helicase core, the sequence similarity between Mtr4 and Ski2 in the arch is limited to a few residues23,25 and provides a useful basis for distinguishing between the two proteins. Structurally, the largest differences are observed in the fist, as described below.
The fist of Mtr4 adopts a β-barrel fold that contains a KOW motif46 and has therefore been described as a KOW domain.24 KOW domains are often associated with proteins that interact with structured RNAs,47 including the bacterial L24 protein, the archaeal L14 protein and the eukaryotic L26 and L27 proteins from the large ribosomal subunit.46,48-50 (The L14 protein was actually used to model the fist in the original low resolution maps of Mtr4.23) Although sequence conservation is limited on the surface of the fist, and generally throughout the arch, a few conserved positively charged and aromatic residues are found on the surface of the fist which may facilitate RNA interactions.23 In vitro gel shift assays confirm that the fist binds hypomodified tRNA24 and pre-rRNA (Johnson lab, unpublished data). However, the fist does not bind single stranded poly(A) RNA,25 indicating a preference for structured RNA.
Like Mtr4, the Ski2 fist contains a β-barrel, but it lacks a KOW motif and adopts a more rigid conformation by associating directly with the arch forearm via hydrophobic interactions.25 The Ski2 fist appears to be less selective than that of Mtr4, binding to both structured and single stranded RNAs. The surface of the Ski2 fist is more positively charged than Mtr4. Presumably, the different RNA binding properties reflect differences in the types of substrates processed by Mtr4 and Ski2.25
While the arch domain is poised to interact with an incoming substrate and may play a role in substrate specificity, it is not required for helicase activity.23 Nor has it been shown to be directly involved in protein-protein interactions. It does, however, play an important functional role. Archless-mtr4 mutants have a slow growth phenotype in S. cerevisiae and exhibit rRNA processing defects.23 Further studies will be required to elucidate the role of the arch domain in Mtr4/Ski2 function.
Other Accessory Domains
Mtr4 and Ski2 both contain N-terminal extensions off of the helicase core that may play functional roles. The N-terminus of Ski2 is significantly longer than Mtr4. No structural information is available for this region, with the exception of a 50 amino acid extended β-hairpin in Mtr4 that spans both RecA domains and appears to stabilize the RecA1-RecA2 structure23,24 (Fig. 1). It is unclear whether a similar structure exists in Ski2 since the corresponding sequence was removed to facilitate crystallization. However, the N-terminus of Ski2 is required for Ski complex formation (see discussion below), suggesting a role in mediating protein-protein interactions.
In Brr2, a helix-hairpin-helix (HhH) domain and Fibronectin type III (FN3) domain flank the C-terminus of the ratchet domain. These three domains are jointly referred to as a Sec63 domain,51 although the ratchet domain may be more appropriately considered part of the helicase core.21 A similar HhH domain is observed in Hel308 where it interacts with DNA emerging from the helicase core28 and acts as an autoinhibitory “molecular brake” to repress helicase activity.30 FN3 domains are generally associated with protein-protein interactions.52
Brr2 and Slh1 both contain two copies of the core helicase-Sec63 module, although only the first module is believed to have functional helicase activity.53 The role of the second module is uncertain, but has been suggested to play a role in protein-protein interactions26,54,55 and was shown to regulate Brr2 helicase activity,26 and may therefore be considered an accessory domain to the Brr2/Slh1 helicase core.
Helicase Dynamics in Mtr4
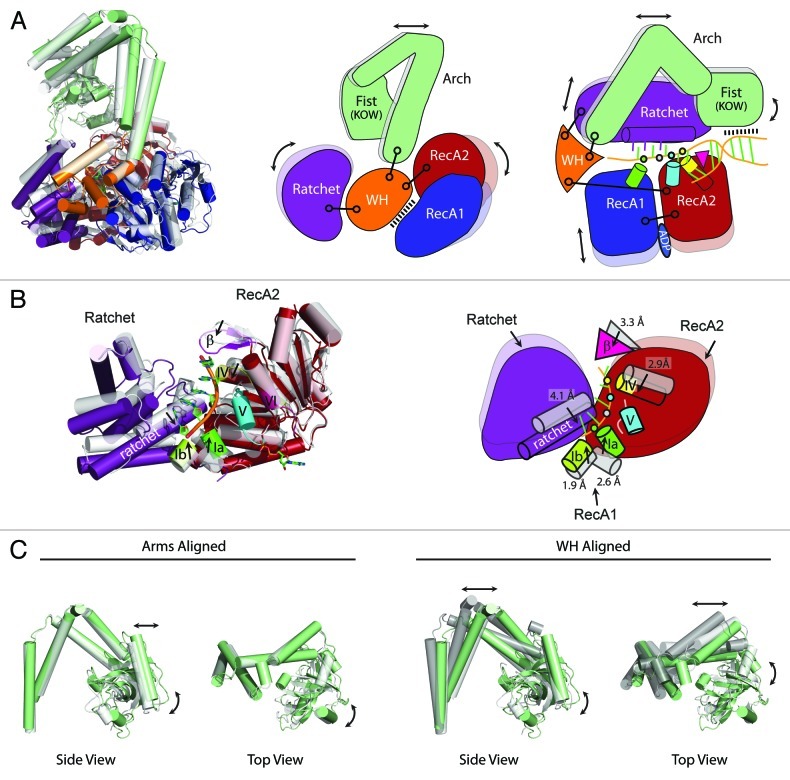
Because multiple structures of Mtr4 are available (apo and ADP/RNA-bound), it is possible to get a sense for the range of motions accessible to the helicase. Since both structures were determined at about the same time, no comparison of the structures has been published to date. Here we describe the major differences between the two structures.
The first observation is that while extensive contacts place restraints on the helicase core, the core domains retain moderate flexibility. The net effect of RNA binding to Mtr4 is a collapse of the entire helicase core around the RNA (Fig. 3). The RecA domains engage the RNA backbone through the conserved sequence motifs Ia, Ib, IV and V. This binding is accompanied by shifts that bring the RecA domains closer together. On the opposite side of the RNA, the ratchet helix shifts ~4.1 Å to interact directly with the RNA bases. Larger motions (up to 5.5 Å) and helical rearrangements are observed throughout the ratchet domain in response to RNA binding (Fig. 3).
Figure 3. Substrate binding induces conformational changes in Mtr4. (A) Comparison of the apo23 (gray) and RNA/ADP bound24 (color) Mtr4 structures (left panel). Alignment was performed by superimposing the winged helix domains. The cartoon rendition (middle panel) highlights domain motions with respect to the winged helix. Covalent linkages connecting the winged helix to the RecA2, arch and ratchet domains are indicated (black lines). The winged helix also stacks with RecA1. The right panel depicts RNA interactions with the RecA (observed), arch (predicted) and ratchet domains (observed), demonstrating the ability of Mtr4 to interact with every region of the unwinding RNA (i.e., duplex, fork, ssRNA backbone and bases). The winged helix is physically linked to each of the other domains (considering the RecA domains as a single module), suggesting a potential role as a molecular hub that coordinates motions between the other domains. (B) Comparison of the RecA2 and ratchet domains in the apo and substrate-bound structures. Structures were aligned as in (A). The Arch, WH and most of RecA1 are removed for clarity. The ratchet helix, motifs Ia, Ib, IV, and V and the β-hairpin collapse toward the RNA substrate upon binding. Conformational shifts are depicted with black arrows and labeled with observed distances. Phosphates are colored the same as interacting helicase motifs. (C) Superposition of the arch domains (left) indicates that the arms adopt a rigid structure. Comparison of the arch domain when the winged helix domains are aligned (right) highlights conformational differences upon substrate binding. The right view includes apo (white) and both molecules from the RNA-bound structure (green and dark gray).
The arch domain is also mobile. This is particularly true for the fist. Compared with the apo structure, rotation occurs in the RNA-bound structure at the “wrist” between the arms and the fist. In one of the molecules observed in the RNA-bound crystal, significant disorder is observed in the electron density which prevented modeling of the fist, suggesting substantial flexibility in this region.24 In contrast, the “elbow” position where the arms make a sharp bend appears to be quite fixed and the arms move as a rigid body (Fig. 3C). Rotation of the arms with respect to the helicase core occurs at the junction with the winged helix domain. The different arch conformations observed in the apo and RNA bound states are not a function of direct contact with RNA since no interaction is observed in the structure, but may be a result of indirect long-range interactions with the helicase core.
An important conclusion from comparison of the Mtr4 structures is that each domain appears to rotate independently with respect to the other domains (Fig. 3A). Furthermore, the winged helix domain is strategically positioned to act as the central hub around which all of the other domains rotate. The winged helix domain is physically linked to each of the other domains, providing a three-way junction with the RecA domains attached at the N-terminal end, the arch domain inserted in the middle, and the ratchet domain attached to the C-terminus. Superposition of the winged helix reveals hinge-like motions that occur between each domain and the winged helix (Fig. 3A).
Winged helix domains are commonly utilized to bind DNA and have also been observed in protein-protein interactions.56 Among Ski2-like helicases, the domain appears to be structurally conserved. However, no general function has been ascribed to this domain other than maintaining the structural integrity of the helicase core.38 We note that an extended 13 amino acid linker located in Mtr4, Ski2 and Hel308 between RecA2 and the winged helix traverses the RecA domains and places the winged helix on the opposite side of the helicase from the RNA entry point. Consequently, the winged helix is not expected to interact with RNA in a significant manner. (Minimal interactions are observed between the winged helix and DNA in the Hel308 structure.28) Instead, it is poised to coordinate and potentially regulate motions between the other domains, each of which interact extensively with RNA.
A detailed understanding of the conformational dynamics employed by the helicase during substrate recognition and processing will require additional structural information. Based on the differential affinity of the Mtr4 arch for various substrates, it seems likely that substrate specific information could be communicated throughout the helicase. For example, given the length and positioning of the arch domain, subtle changes in RNA sequence or structure detected by the fist could have significant conformational and functional effects on the helicase core. Conversely, specific features sensed by the ratchet helix or RecA domains could be easily communicated to the protein surface. However, in the absence of relevant structural and biochemical data, such models remain speculative.
RNA Processing and Degradation
Activation of the exosome by Mtr4 and Ski2
A recent review describes the role of RNA helicases in pre-mRNA splicing, including the integral role of Brr2 in the spliceosome.12 Here we discuss the role of Mtr4 and Ski2 in exosome-mediated RNA decay. The exosome is a large multi-protein ring-like structure57-63 involved in the processing and degradation of a wide variety of nuclear and cytosolic RNAs, which has been extensively reviewed.14,20,64-67 Multiple nucleases are associated with the exosome, including Rrp44 (3′-5′ exonuclease and endonuclease)58,68-71 and Rrp6 (3′-5′ exonuclease, associated with the nuclear exosome).58,72-74 Exosome activation requires Mtr4 in the nucleus and Ski2 in the cytosol.8,75-79 Activation generally occurs in the context of larger assemblies, including the Mtr4-mediated TRAMP and NEXT complexes and the Ski2-mediated Ski complex.18,19,80-87 Mtr4 may also independently activate the exosome for processing of some substrates, such as the 5.8S rRNA.40,77,78,83
How do Mtr4 and Ski2 stimulate exosome activity? One simple explanation is that unwinding of RNA secondary structures and resolution of ribonucleoprotein complexes by Mtr4/Ski2 makes the 3′ end of the RNA accessible to the exosome.16,19 The exosome may also utilize the translocation activity provided by the helicase motor to effectively degrade substrates.19 Extensive conservation is observed along the base of Mtr4 and Ski2 where RNA exits the helicase core,23-25 whereas the base of Hel308 is much less conserved. This suggests that the conservation is related to a function common to Mtr4 and Ski2, but distinct from canonical helicase activity. One possibility is that Mtr4 and Ski2 interact directly with the exosome core, which is roughly similar in dimensions to the base of Mtr4 and Ski2.23 A second possibility is that other proteins directly interact with the base of Mtr4 and Ski2 and mediate association with the exosome. Mtr4 and Ski2 association with the exosome has been demonstrated in vivo through a variety of co-immunoprecipitation experiments in yeast88,89 and human.80,90-92 Although direct interactions with the exosome core have yet to be demonstrated in vitro, human Mtr4 appears to directly interact with the exosome associated components Mpp6 and Rrp6.80,91 Importantly, Ski2 association with the exosome requires Ski7, although the mode of interaction remains to be determined.88
Maturation of the 5.8S rRNA suggests that the role of Mtr4 in exosome activation may be more complex than simply providing an accessible 3′ end. Conversion of the 7S precursor rRNA to the mature 5.8S rRNA involves exonucleolytic processing by Rrp44, followed by further processing by Rrp6.40,78 Both of these steps require Mtr4. Remarkably, when the arch domain is removed from Mtr4 (“mtr4-archless”), the 7S rRNA is effectively degraded to the 5.8+30S rRNA precursor (a step associated with Rrp44 activity) at which point processing stalls, similar to what is observed in a Rrp6 knockout.23 This indicates that Rrp6 activity is impaired and further suggests that Mtr4 is able to partition substrates between Rrp44 and Rrp6 in an arch-dependent manner. Recombinant Rrp6 completely degrades a naked 7S rRNA substrate in vitro.93 However, a separate study showed that TAP-tag purified Rrp6 from S. cerevisiae cannot degrade 7S rRNA or tRNA substrates unless Mtr4 is present. Furthermore, removal of the Mtr4 fist or the entire arch inhibits RNA degradation.41 These data suggest that Rrp6 association with the core exosome (which co-purified with TAP-tagged Rrp6) has an inhibitory effect on Rrp6 that is overcome by the Mtr4 arch domain. Notably, the arch domain is only observed in helicases that are associated with exosome function (Mtr4 and Ski2).25 Additional studies are needed to elucidate the precise mechanisms involved in exosome activation.
Substrate recognition by Mtr4/Ski2-mediated macromolecular complexes
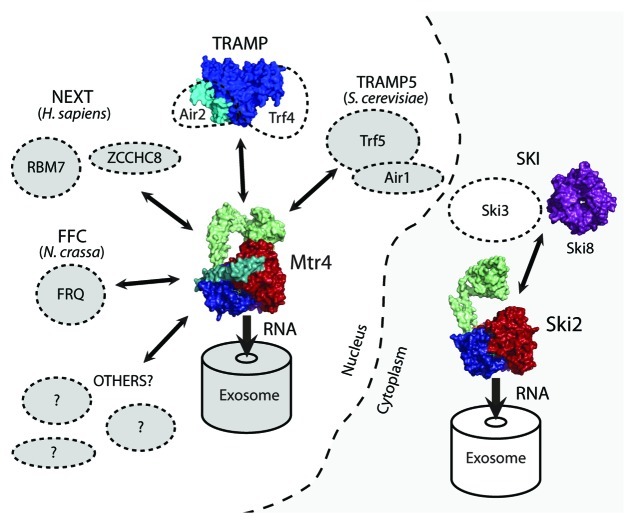
The fundamental biological problem for exosome dependent decay pathways is distinguishing the correct RNA substrate from all of the other cellular RNAs. Inadvertent removal of needed RNAs or retention of unwanted RNAs is likely to have deleterious consequences for cellular function. Consequently, substrate recognition must be specific and carefully regulated. It is therefore not surprising that a variety of multi-protein complexes have been identified that play a critical role in targeting substrates to the exosome. Significantly, Mtr4 or Ski2 are vital components of these RNA targeting complexes (Fig. 4). For the most part, a detailed molecular description for these complexes is severely lacking. Our current understanding of these assemblies is summarized below. In each case, important structural questions include: What are the structures of the individual components and how do they assemble to form a larger complex? How does complex formation affect the activity/function of individual components? How are RNA substrates recognized by the complex? What is the trajectory of the RNA through the complex? How are RNAs ultimately delivered to the exosome?
Figure 4. Exosome activating complexes are mediated by Mtr4 and Ski2. Existing crystal structures are indicated (see text for details). Exosome structures also exist,58-61,63 but are not shown.
TRAMP (Trf4/5-Air1/2-Mtr4 polyadenylation complex)
The best characterized exosome activating complex is the nuclear TRAMP complex.19,94,95 TRAMP is a 3-protein complex composed of Mtr4, a poly(A) polymerase (Trf4 or Trf5), and an RNA binding protein containing five zinc knuckles (Air1 or Air2).82-85 The complex has been described in yeast83-85,96 and human,80 and individual protein components are found throughout eukaryotic species,19,97-101 suggesting that the complex is found throughout eukaryotes. In S. cerevisiae, two forms of TRAMP have been described, TRAMP4 (Trf4, Air2 and Mtr4) and TRAMP5 (Trf5, Air1 and Mtr4).82,83 These TRAMP complexes localize differentially in the nucleoplasm (TRAMP4) and nucleolus (TRAMP5),102-104 but they appear to be functionally redundant.85 Deletion of Air1 or Air2 results in only a slight growth defect, whereas the Air1/Air2 double deletion is either synthetically lethal or results in a slow growth phenotype.83,105 Similarly, no growth defects are observed upon deletion of Trf4 or Trf5, but the double deletion is lethal.85 Given the high sequence identity (55% identity for Trf4/5106; 45% identity for Air1/2105) and functional redundancy for each TRAMP homolog, it is expected that the TRAMP4 and TRAMP5 complexes will also be similar.
TRAMP adds a short (~5-nt) poly(A) tail to the 3′ end of RNA substrates,107,108 which is preferentially unwound by Mtr445 and subsequently degraded by the exosome. Remarkably, Mtr4 directly modulates the polyadenylation activity of Trf4. In the presence of Mtr4, Trf4 polymerase activity is enhanced until the RNA has 5 adenosines at the 3′ end, at which point Trf4 activity is severely inhibited.107 Similarly, Mtr4 helicase activity is enhanced simply by binding to Trf4-Air2.45 In the context of TRAMP, Mtr4 has the ability to detect the length of a poly(A) tail, which directly influences Trf4 and Mtr4 activity.45,107 A point mutation in the ratchet domain (E947A) disrupts modulation of Trf4 activity by Mtr4.107 In the RNA-bound crystal structure,24 this residue is located near one of the RNA bases and potentially helps recognize the poly(A) tail. Whether Mtr4 is directly reading the sequence in this context is unclear, although Mtr4 does exhibit an affinity for polyadenylated RNAs over non-polyadenylated substrates in in vitro binding and unwinding assays.45,109,110
Understanding the assembly of the TRAMP complex is an active focus of investigation. Trf4 and Air2 are tightly associated in vitro, remaining in complex above 1M NaCl, while Mtr4 interaction is more dynamic and dissociates from Trf4-Air2 around 500 mM NaCl.83,84,96 A recent crystal structure of the catalytic domain of Trf4 bound to Air2 zinc knuckles 4–5 reveals that these two proteins interact primarily through zinc knuckle 5 and a loop between zinc knuckles 4 and 5.111 (An NMR structure of Air2 zinc knuckles 1–5 was also recently reported.41) This interaction has been confirmed by mutagenic analysis.41,111,112 The site of Mtr4 interactions is less well understood. In vitro pull-down assays from the Conti lab24 and our lab (unpublished data) indicate that the helicase core is sufficient to form a TRAMP-like complex (i.e., the N-terminus and arch domains are not required). Similarly, a domain deletion study shows that TRAMP fails to pull-down with Mtr4 when any of the individual helicase core domains are removed,41 although it isn’t clear what effect deletion of individual core domains has on the overall structure or localization. Presumably, the Trf4/5-Air1/2 docking site would be located near the RNA entry site in Mtr4. A few conserved patches on the surface of Mtr4 that are not found in Ski2 suggest potential interaction regions,24 but the location of TRAMP assembly on Mtr4 remains obscure. Recent studies, however, show that the N-terminus of Air1/2 interacts directly with Mtr4 and may provide a bridge between Mtr4 and Trf4/5.41 Earlier yeast 2-hybrid studies indicated that an N-terminal region of Trf4/5 is required for Mtr4 interaction.83 No structures of the relevant regions in Air1/2 or Trf4/5 are currently available.
NEXT (Mtr4-Rbm7-ZCCHC8)
Another Mtr4-mediated activator of the exosome is the human nuclear exosome targeting complex (NEXT), which was recently identified by co-immunoprecipitation studies and shown to target promoter upstream transcripts (PROMPTs) for degradation by the exosome.80 Like TRAMP, the NEXT complex contains 3 components including Mtr4, a zinc knuckle protein called ZCCHC8, and a putative RNA binding protein, Rbm7. However, no poly(A) activity is associated with NEXT, indicating that polyadenylation is not required for degradation in this context. In humans, NEXT is localized in the nucleus while TRAMP appears to be confined to the nucleolus. Since Mtr4 is shared by both complexes, it has been suggested that Mtr4 potentially acts as a common scaffold for a variety of protein complexes aimed at shuttling substrates to the exosome.80 It will be interesting to learn whether Mtr4 interacts with each complex (and other yet to be discovered complexes) in a similar manner. For example, does the putative Mtr4-ZCCHC8 interaction resemble that of the TRAMP Mtr4-Air2 interaction?
FFC (FRQ-FRH complex)
The FRH (FRQ-interacting RNA helicase) protein from N. crassa is an indispensable component of the circadian clock.113 Sequence alignments indicate that FRH is best described as a homolog of S. cerevisae Mtr4 (56% sequence identity; 73% similarity overall).101 Alignment of the arch domain sequence reveals strong similarity with Mtr4, but fails to align with Ski2.23 FRH forms a complex with the Frequency protein (FRQ) that regulates oscillating gene expression.101 The FRQ-FRH complex (FFC) acts within a negative feedback loop that represses transcription of specific genes, including the frq allele, by binding and promoting phosphorylation of the white collar complex (WCC) transcription promoter.113 FRH is required for the direct interaction between FFC and WCC.101 Moreover, FFC negatively regulates FRQ expression by specifically binding frq mRNA and mediating its exosomal decay.114
Mutation of the conserved arginine R806 to histidine allows the formation of the FFC but abolishes interactions with WCC.115 Interestingly, the equivalent residue in S. cerevisiae Mtr4, R774, is a conserved, solvent exposed residue positioned on the upper region of the fist/KOW domain in the arch.23 Thus, in this system the arch domain appears to play an important role in complex formation.
Ski complex (Ski2-Ski3-Ski8)
The Ski complex components are conserved in most eukaryotes including human, and promotes degradation of mRNAs by the cytosolic exosome.18 It is composed of Ski2, a tetratricopeptide protein (Ski3), and a WD-40 repeat protein (Ski8).86 The complex was originally described as a trimer,86 but subsequent mass spec analysis indicates that it is a tetramer containing two copies of Ski8.87 Activation of the exosome also requires an accessory protein, Ski7, that has been shown to physically link the two protein machineries.88 A crystal structure of Ski8 reveals a seven-bladed β-propeller that is proposed to function as a scaffold protein.116,117 Yeast 2-hybrid screens and co-immunoprecipitation studies indicate that the N-terminus of Ski2 associates directly with Ski3, while direct interactions with Ski8 or Ski7 are not observed.118 While the Ski2 crystal structure provides little information regarding complex assembly,25 forthcoming structures are expected to significantly clarify the architecture of this complex.
Outlook
As the above complexes demonstrate, the assembly of different proteins on Mtr4 and Ski2 allow for targeting of a variety of RNA substrates for exosome processing and degradation. The FCC complex also highlights the potential of some complexes to affect protein post-translational modifications. It would not be surprising to discover more complexes that expand the versatility of these helicases. Despite the valuable insight provided by the current structures, we are just beginning to understand how RNA substrates are recognized and interact with Ski2-like RNA helicases. The interaction of these helicases in larger complex assemblies is a critical question that promises to be an active focus of continued research.
Acknowledgments
We thank Jeremy Bakelar, Lindsey Lott, Lacy Taylor and Joan Hevel for helpful discussions. This work was supported by NSF grant MCB 0952920.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22101
References
- 1.Caprara MG. Chapter 6 Ski2-Like Proteins: Biology and Mechanism. RNA Helicases: The Royal Society of Chemistry, 2010:149-67. [Google Scholar]
- 2.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–29. doi: 10.1016/S0959-440X(05)80116-2. [DOI] [Google Scholar]
- 4.Jankowsky E, Fairman-Williams ME. Chapter 1 An Introduction to RNA Helicases: Superfamilies, Families, and Major Themes. RNA Helicases: The Royal Society of Chemistry, 2010:1-31. [Google Scholar]
- 5.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 6.Widner WR, Wickner RB. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–41. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AW, Kolodner RD. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–27. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toh-E A, Guerry P, Wickner RB. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J Bacteriol. 1978;136:1002–7. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–52. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Jedrzejczak R, Wang J, Dauter M, Szczesny RJ, Stepien PP, Dauter Z. Human Suv3 protein reveals unique features among SF2 helicases. Acta Crystallogr D Biol Crystallogr. 2011;67:988–96. doi: 10.1107/S0907444911040248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordin O, Hahn D, Beggs JD. Structure, function and regulation of spliceosomal RNA helicases. Curr Opin Cell Biol. 2012;24:431–8. doi: 10.1016/j.ceb.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Pyle AM. RNA helicases and remodeling proteins. Curr Opin Chem Biol. 2011;15:636–42. doi: 10.1016/j.cbpa.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebreton A, Séraphin B. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim Biophys Acta. 2008;1779:558–65. doi: 10.1016/j.bbagrm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 15.de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–8. doi: 10.1016/S0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 16.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–76. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Norbury CJ. Regional specialization: the NEXT big thing in nuclear RNA turnover. Mol Cell. 2011;43:502–4. doi: 10.1016/j.molcel.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Schaeffer D, Clark A, Klauer AA, Tsanova B, van Hoof A. Functions of the cytoplasmic exosome. Adv Exp Med Biol. 2010;702:79–90. doi: 10.1007/978-1-4419-7841-7_7. [DOI] [PubMed] [Google Scholar]
- 19.Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 20.Butler JS. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–6. doi: 10.1016/S0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- 21.Hahn D, Beggs JD. Brr2p RNA helicase with a split personality: insights into structure and function. Biochem Soc Trans. 2010;38:1105–9. doi: 10.1042/BST0381105. [DOI] [PubMed] [Google Scholar]
- 22.Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–9. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–16. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci USA. 2010;107:12139–44. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–34. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena V, Jovin SM, Fabrizio P, Orlowski J, Bujnicki JM, Lührmann R, et al. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell. 2009;35:454–66. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, et al. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009;16:731–9. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–52. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 29.Oyama T, Oka H, Mayanagi K, Shirai T, Matoba K, Fujikane R, et al. Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm. BMC Struct Biol. 2009;9:2. doi: 10.1186/1472-6807-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards JD, Johnson KA, Liu H, McRobbie AM, McMahon S, Oke M, et al. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008;283:5118–26. doi: 10.1074/jbc.M707548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Nakashima T, Kakuta Y, Yao M, Tanaka I, Kimura M. Crystal structure of an archaeal Ski2p-like protein from Pyrococcus horikoshii OT3. Protein Sci. 2008;17:136–45. doi: 10.1110/ps.073107008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 33.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–68. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 35.He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–6. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudlinzki D, Schmitt A, Christian H, Ficner R. Structural analysis of the C-terminal domain of the spliceosomal helicase Prp22. Biol Chem. 2012 doi: 10.1515/hsz-2012-0158. In press. [DOI] [PubMed] [Google Scholar]
- 38.Woodman IL, Bolt EL. Winged helix domains with unknown function in Hel308 and related helicases. Biochem Soc Trans. 2011;39:140–4. doi: 10.1042/BST0390140. [DOI] [PubMed] [Google Scholar]
- 39.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–62. doi: 10.1016/S1097-2765(01)00329-X. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein J, Toth EA. Yeast nuclear RNA processing. World J Biol Chem. 2012;3:7–26. doi: 10.4331/wjbc.v3.i1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holub P, Lalakova J, Cerna H, Pasulka J, Sarazova M, Hrazdilova K, et al. Air2p is critical for the assembly and RNA-binding of the TRAMP complex and the KOW domain of Mtr4p is crucial for exosome activation. Nucleic Acids Res. 2012;40:5679–93. doi: 10.1093/nar/gks223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang S, Hitomi M, Hu YH, Liu Y, Tartakoff AM. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–46. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell. 2006;23:389–99. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guy CP, Bolt EL. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 2005;33:3678–90. doi: 10.1093/nar/gki685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia H, Wang X, Anderson JT, Jankowsky E. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc Natl Acad Sci USA. 2012;109:7292–7. doi: 10.1073/pnas.1201085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kyrpides NC, Woese CR, Ouzounis CA. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci. 1996;21:425–6. doi: 10.1016/S0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- 47.Steiner T, Kaiser JT, Marinkoviç S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–53. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–7. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–33. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponting CP. Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem J. 2000;351:527–35. doi: 10.1042/0264-6021:3510527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bloom L, Calabro V. FN3: a new protein scaffold reaches the clinic. Drug Discov Today. 2009;14:949–55. doi: 10.1016/j.drudis.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 53.Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5:959–71. doi: 10.1017/S135583829999012X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Lührmann R, et al. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–20. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 55.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–67. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–6. doi: 10.1016/S0959-440X(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 57.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–59. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 58.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 59.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–6. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005;12:575–81. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 61.Navarro MV, Oliveira CC, Zanchin NI, Guimarães BG. Insights into the mechanism of progressive RNA degradation by the archaeal exosome. J Biol Chem. 2008;283:14120–31. doi: 10.1074/jbc.M801005200. [DOI] [PubMed] [Google Scholar]
- 62.Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, et al. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci USA. 2007;104:16844–9. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Büttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–71. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 64.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–8. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 65.Januszyk K, Lima CD. Structural components and architectures of RNA exosomes. Adv Exp Med Biol. 2010;702:9–28. doi: 10.1007/978-1-4419-7841-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. J Cell Sci. 2009;122:1487–94. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- 67.Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 2011;8:61–6. doi: 10.4161/rna.8.1.14237. [DOI] [PubMed] [Google Scholar]
- 68.Lebreton A, Tomecki R, Dziembowski A, Séraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–6. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 69.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 70.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–40. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA. 2011;17:1566–77. doi: 10.1261/rna.2763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci USA. 2006;103:11898–903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phillips S, Butler JS. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003;9:1098–107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–4. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′-->5′ degradation. Mol Cell. 2003;11:1405–13. doi: 10.1016/S1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 77.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–40. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–52. doi: 10.1128/MCB.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–37. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 81.Wolin SL, Sim S, Chen X. Nuclear noncoding RNA surveillance: is the end in sight? Trends Genet. 2012;28:306–13. doi: 10.1016/j.tig.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–11. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–24. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 84.Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–37. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 86.Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–57. doi: 10.1017/S1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Synowsky SA, Heck AJ. The yeast Ski complex is a hetero-tetramer. Protein Sci. 2008;17:119–25. doi: 10.1110/ps.073155908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–93. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–33. doi: 10.1016/S0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- 90.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–64. doi: 10.1016/S0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 91.Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–72. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–57. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–7. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bayne EH, White SA, Allshire RC. DegrAAAded into silence. Cell. 2007;129:651–3. doi: 10.1016/j.cell.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 95.Jensen TH, Moore C. Reviving the exosome. Cell. 2005;121:660–2. doi: 10.1016/j.cell.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 96.Keller C, Woolcock K, Hess D, Bühler M. Proteomic and functional analysis of the noncanonical poly(A) polymerase Cid14. RNA. 2010;16:1124–9. doi: 10.1261/rna.2053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schmid M, Küchler B, Eckmann CR. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev. 2009;23:824–36. doi: 10.1101/gad.494009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Etheridge RD, Clemens DM, Gershon PD, Aphasizhev R. Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei. Mol Biochem Parasitol. 2009;164:66–73. doi: 10.1016/j.molbiopara.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakamura R, Takeuchi R, Takata K, Shimanouchi K, Abe Y, Kanai Y, et al. TRF4 is involved in polyadenylation of snRNAs in Drosophila melanogaster. Mol Cell Biol. 2008;28:6620–31. doi: 10.1128/MCB.00448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lange H, Sement FM, Gagliardi D. MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 2011;68:51–63. doi: 10.1111/j.1365-313X.2011.04675.x. [DOI] [PubMed] [Google Scholar]
- 101.Cheng P, He Q, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–41. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dez C, Dlakić M, Tollervey D. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA. 2007;13:1516–27. doi: 10.1261/rna.609807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, et al. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–26. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wery M, Ruidant S, Schillewaert S, Leporé N, Lafontaine DL. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA. 2009;15:406–19. doi: 10.1261/rna.1402709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Inoue K, Mizuno T, Wada K, Hagiwara M. Novel RING finger proteins, Air1p and Air2p, interact with Hmt1p and inhibit the arginine methylation of Npl3p. J Biol Chem. 2000;275:32793–9. doi: 10.1074/jbc.M004560200. [DOI] [PubMed] [Google Scholar]
- 106.Castaño IB, Heath-Pagliuso S, Sadoff BU, Fitzhugh DJ, Christman MF. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 1996;24:2404–10. doi: 10.1093/nar/24.12.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, et al. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011;30:1790–803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bernstein J, Ballin JD, Patterson DN, Wilson GM, Toth EA. Unique properties of the Mtr4p-poly(A) complex suggest a role in substrate targeting. Biochemistry. 2010;49:10357–70. doi: 10.1021/bi101518x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bernstein J, Patterson DN, Wilson GM, Toth EA. Characterization of the essential activities of Saccharomyces cerevisiae Mtr4p, a 3′->5′ helicase partner of the nuclear exosome. J Biol Chem. 2008;283:4930–42. doi: 10.1074/jbc.M706677200. [DOI] [PubMed] [Google Scholar]
- 111.Hamill S, Wolin SL, Reinisch KM. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci USA. 2010;107:15045–50. doi: 10.1073/pnas.1003505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fasken MB, Leung SW, Banerjee A, Kodani MO, Chavez R, Bowman EA, et al. Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. J Biol Chem. 2011;286:37429–45. doi: 10.1074/jbc.M111.271494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baker CL, Loros JJ, Dunlap JC. The circadian clock of Neurospora crassa. FEMS Microbiol Rev. 2012;36:95–110. doi: 10.1111/j.1574-6976.2011.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell. 2009;138:1236–46. doi: 10.1016/j.cell.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi M, Collett M, Loros JJ, Dunlap JC. FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics. 2010;184:351–61. doi: 10.1534/genetics.109.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheng Z, Liu Y, Wang C, Parker R, Song H. Crystal structure of Ski8p, a WD-repeat protein with dual roles in mRNA metabolism and meiotic recombination. Protein Sci. 2004;13:2673–84. doi: 10.1110/ps.04856504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Madrona AY, Wilson DK. The structure of Ski8p, a protein regulating mRNA degradation: Implications for WD protein structure. Protein Sci. 2004;13:1557–65. doi: 10.1110/ps.04704704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang L, Lewis MS, Johnson AW. Domain interactions within the Ski2/3/8 complex and between the Ski complex and Ski7p. RNA. 2005;11:1291–302. doi: 10.1261/rna.2060405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–19. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.The PyMOL Molecular Graphics System, Schrödinger, LLC.