Abstract
In eukaryotic cells, introns are spliced from pre-mRNAs by the spliceosome. Both the composition and the structure of the spliceosome are highly dynamic, and eight DExD/H RNA helicases play essential roles in controlling conformational rearrangements. There is evidence that the various helicases are functionally and physically connected with each other and with many other factors in the spliceosome. Understanding the dynamics of those interactions is essential to comprehend the mechanism and regulation of normal as well as of pathological splicing. This review focuses on recent advances in the characterization of the splicing helicases and their interactions, and highlights the deep integration of splicing helicases in global mRNP biogenesis pathways.
Keywords: DExD/H helicase, DEAD-box, DEAH-box, protein partners, ATPase
Pre-mRNA Splicing
Most eukaryotic precursor mRNAs (pre-mRNAs) contain introns that must be removed by RNA splicing to produce mature mRNAs. This is achieved in the spliceosome, a large and extremely dynamic ribonucleoprotein (RNP) complex (for recent reviews, see refs. 1,2). The spliceosome is highly conserved from yeast to human, with 85% of yeast splicing factors having an identified human ortholog. However, the human spliceosome contains about twice as many splicing factors as does the spliceosome of the budding yeast Saccharomyces cerevisiae (approximately 170 and 90 respectively),3-5 likely reflecting the prevalence of alternative splicing mechanisms in higher eukaryotes. Thus, S. cerevisiae might be considered to have a minimal spliceosome.3 Nevertheless, splicing in budding yeast is subject to regulation.6
Introns are identified by short sequences at the 5′ splice site (5′SS), the branch site (BS) and the 3′ splice site (3′SS). In budding yeast, these adhere quite closely to consensus sequences but are more varied in metazoans, where additional cis-acting elements and trans-acting factors affect splice site choice. RNA splicing must be highly accurate in order to join the coding exons correctly, and mechanisms exist to check the fidelity of splicing and to promote the discard and degradation of aberrant intermediates and products of splicing (proofreading see below). Among the many splicing factors, RNA helicases have been identified as important regulators of splicing, implicated in promoting conformational rearrangements as well as ensuring that only appropriate substrates proceed through the splicing reactions. These roles likely involve checking the configuration of the catalytic center of the spliceosome at each stage.
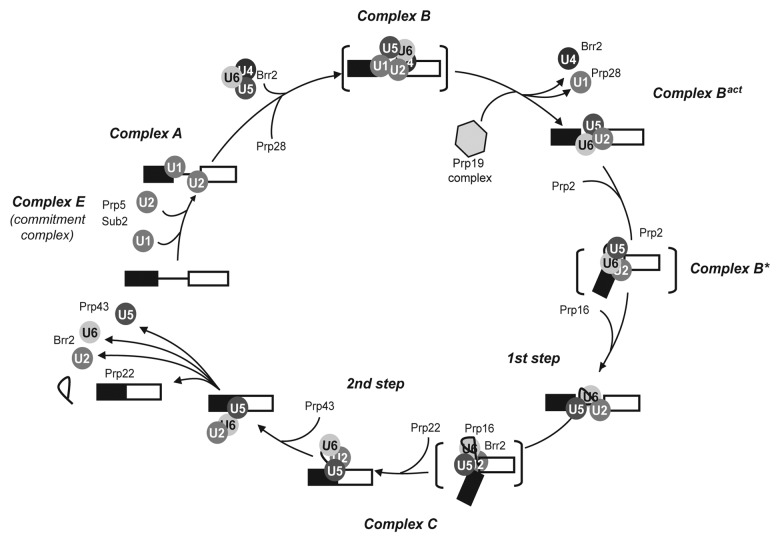
Spliceosome assembly is an ordered process in which the U1, U2, U4/U6 and U5 small nuclear RNPs (snRNPs) and non-snRNP splicing factors interact with the substrate pre-mRNA and with each other, defining the intron splice sites and the BS (reviewed in refs. 1,2,7). In the commonly accepted step-wise model of spliceosome assembly that was defined mainly from in vitro studies (Fig. 1), the 5′SS is first recognized by the U1 snRNP and the BS by the SF1/BBP and U2AF proteins (Msl5 and Mud2 in yeast) that form the commitment complex (complex E). The U2 snRNP then associates with the BS, leading to formation of the pre-spliceosome, or complex A. Complex A is converted to complex B by addition of the U4/U6 and U5 snRNPs in the form of a pre-assembled tri-snRNP particle. Within the tri-snRNP the U4 and U6 snRNAs are base-paired via two regions of sequence complementarity, but are unwound in the spliceosome during a major reorganization that displaces the U1 and U4 snRNPs. At this point, the multi-protein 19-complex (NTC) joins the spliceosome to form the almost complete, but still inactive, complex Bact. The ATP dependent activation of the spliceosome (complex B*) precedes the first catalytic step of splicing. As a consequence of the first step, complex C is formed. This is reorganized again to perform the second reaction. Finally the spliceosome is dissociated and the products of splicing, i.e., the spliced mRNA and the excised intron, are released and either processed further and exported to the cytoplasm, or degraded.
Figure 1. RNA helicases in the spliceosome assembly/disassembly pathway. The order of assembly of U snRNPs and the steps of association and activity of helicases are shown on a transcript containing a single intron. Names of complexes refer to the human nomenclature.
Although splicing has long been known to involve two trans-esterification reactions, the precise chemistry and the structural changes that occur within the spliceosome before, during and after catalysis are still the subject of intense study. During the first reaction, the 2’hydroxyl moiety of a conserved adenosine (the BS adenosine), located toward the 3′ end of the intron, attacks the 5′SS, cleaving the phosphodiester bond. This produces two intermediates, the 5′ exon and the intron-3′ exon, in which the guanosine at the 5′ end of the intron is covalently attached via a 2’-5′ phosphodiester bond to the BS adenosine, forming a branched or lariat configuration. In the second catalytic step the 3′ hydroxyl group of the 5′ exon attacks the 3′SS, joining the exons and excising the intron in lariat form. The spliceosome has only one active site for both trans-esterification reactions. Thus reorganization of the catalytic center must occur between the two reactions, such that the products of the first reaction are repositioned as substrates for the second reaction.
Most early studies of the mechanism of splicing were performed with a few model substrates in vitro or using a small number of reporter constructs in vivo,8-10 and the influence of transcription and other cellular processes on splicing was largely ignored. However, it is apparent that splicing factors can have distinct effects with different pre-mRNAs,11 and that other pathways of RNA metabolism can affect splicing.12-14 Therefore we conclude this review with a summary of links between splicing helicases and other RNA metabolic processes.
RNA Helicase Families and Mechanisms
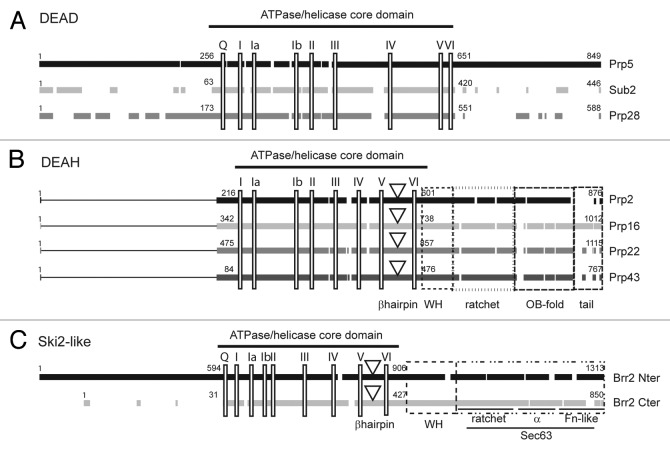
Eight RNA helicases are required for pre-mRNA splicing in all eukaryotes (Fig. 1).15 They all belong to the superfamily 2 (SF2) of helicases16 that are characterized by the presence of two RecA-like domains and variable amino and/or carboxy terminal extensions. SF2 helicases generally function as monomers but some act as homo-dimers.17,18 They share conserved motifs (Fig. 2) involved in NTP (usually ATP) binding and hydrolysis, and nucleic acid interaction. Motif III is involved in communicating between the motifs for nucleotide and nucleic acid binding. Other motifs are responsible for differences in activity observed between families. For example, among the eight spliceosomal RNA helicases, three (Prp5, Sub2 and Prp28) belong to the DEAD-box family, four (Prp2, Prp16, Prp22 and Prp43) to the DEAH-box family and one (Brr2) to the Ski-2 like family (Fig. 2) (reviewed in ref. 15).
Figure 2. Splicing helicases belong to three distinct families. Primary sequence alignment of S. cerevisiae helicases from the DEAD-box (A), DEAH-box (B) and Ski2-like (C) families involved in pre-mRNA splicing. Black and gray blocks represent the conserved regions within each family. The lines in the amino-termini of DEAH-box helicases indicate the lack of conserved sequences in this region. The positions of the conserved motifs are indicated by vertical rectangles. For DEAH-box and Ski2-like helicases, a downward triangle indicates the position of the β-hairpin proposed to act as a strand separator. Dashed boxes indicate the conserved domains in the carboxy-termini of DEAH-box proteins and in the two helicase modules of the Ski2-like Brr2 helicase. For clarity of the figure we refer to the old nomenclature for conserved motifs19 although a new nomenclature has been proposed.17
DEAD-box helicases
DEAD-box RNA helicases are found in nearly every organism.19 They are exclusively ATP specific with ATP hydrolysis usually being stimulated by RNA. Although commonly referred to as helicases, DEAD-box proteins are poor unwindases and might appropriately be considered ATP-dependent RNA binding proteins. DEAD-box proteins can bind a single strand of RNA, regardless of whether it is engaged in a duplex or not. Upon ATP binding, the helicase undergoes a conformational change, resulting in local physical constraint that destabilizes the structure of the bound RNA. In some cases the substrate can be a protein bound to the RNA. Additionally, some DEAD-box helicases possess bona fide RNA annealing activity.20 These properties suggest that DEAD-box proteins could be efficient ATP-dependent switches. The three DEAD-box helicases involved in splicing share a high degree of conservation in their core domains, whereas their amino- and carboxy-termini are poorly conserved (Fig. 2A). In higher eukaryotes, the amino termini of Prp5 and Prp28 contain serine-arginine (SR) repeats. SR repeats are commonly found in RNA splicing factors involved in alternative splicing21-23 where they participate in protein or RNA binding.
DEAH-box helicases
Yeast DEAH-box helicases possess an extremely well conserved core domain that contains the common SF2 motifs, except for the Q-motif that confers ATP specificity (Fig. 2B), and conservation extends to the carboxy terminus. All possess a similar organization, that includes a conserved β-hairpin (5′HP) in their core domain, a winged helix (WH) domain, a ratchet domain involved in RNA binding and RNA translocation during duplex unwinding, and a DUF1605 domain (Domain of Unknown Function) that adopts the Oligosaccharide Binding fold (OB-fold)24-27 (Fig. 2B). Unlike DEAD-box and Ski2-like helicases, DEAH-box helicases can bind and hydrolyse any NTP (or dNTP) in vitro,15 although such substrate promiscuity may not be relevant in vivo. The conserved 5′HP and the DUF1605 domain participate in the control of the RNA binding and unwinding activities. The presence of these structures implies that DEAH-box helicases require a single-stranded region in the substrate on which to load. These domains also confer polarity and a certain degree of processivity to those helicases. Of six DEAH-box helicases in S. cerevisiae, three, Prp2, Prp16 and Prp22, participate specifically in pre-mRNA splicing, while Prp43 is necessary for both pre-mRNA splicing and rRNA processing.
Their extensive sequence conservation suggests a common mechanism of action and similar mode of regulation. In the spliceosome, the ATPase activity of both Prp2 and Prp43 is activated by G-patch proteins, Spp2 and Spp382, respectively.5,26,28,29 For its role in rRNA processing, Prp43 is stimulated by another G-patch protein, Pfa1.26 The G-patch proteins mediate regulation through interaction with the OB-fold domain of DEAH-box helicases.25 Although very similar, the primary sequences of OB-fold domains of splicing helicases show clear differences that might account for partner-specific binding. In S. cerevisiae, no G-patch protein has been found associated with Prp16 or Prp22, although human Prp16 could be a target of GPNOW30 (the ortholog of Spp2, that also interacts with hPrp2). The N-terminal domains of DEAH-box splicing helicases differ greatly in both primary sequence and length (from 84 amino acids for Prp43 to 475 amino acids for Prp22) (Fig. 2B) and little is known about their function. A PWI domain has been predicted in the N-terminus of human Prp2, but is absent in the S. cerevisiae ortholog.31 In all splicing DEAH-box helicases a large portion of the N-terminus can be deleted without altering the function of the protein in vivo.31-37
Ski2-like helicases
Brr2 is the only Ski2-like helicase involved in pre-mRNA splicing (reviewed in ref. 18). Ski2-like helicases share structural features with both DEAD- and DEAH-box helicases (reviewed in ref. 15). They possess a version of the Q-motif,15 which is also present in DEAD-box RNA helicases, and a putative strand separator, the 5′HP located between motifs V and VI, also found in DEAH-box helicases.
Brr2 is unusual, in that it possesses two Ski2-like helicase modules, each of which comprises a Ski2-like helicase domain connected to a Sec63 domain through a structurally versatile WH domain (Fig. 2C). Only the N-terminal module has ATP hydrolysis and RNA unwinding activities in vitro,38 and it alone interacts with RNA in vivo.39 The sequence of the second module is divergent and appears to have a protein interaction function rather than the canonical RNA helicase function.40,,41 The N-terminal module starts with a domain of unknown function (aa. 1–474 in budding yeast) that is essential in vivo (Turner, I.A. and Newman, A., personal communication). Interestingly it includes a PWI domain (aa. 258–338) that could participate in RNA binding.31 Determination of the structure of the C-terminal WH-Sec63 domains42,43 highlighted the presence of three conserved sub-domains40 that strongly resemble the C-terminal domain of the Ski2-like DNA helicase Hel308.44-46 The Sec63 domain itself includes a ratchet domain found in all Ski2-like and DEAH-box helicases,15 followed by a short α-helical domain, which may provide flexibility to the Sec63 domain. Finally a fibronectin-like domain, rich in β-strands, interacts strongly with the other two domains. Altogether the Sec63 domain of Brr2 is likely to function in regulation of substrate binding by the helicase domain. A crystal structure of nearly full-length human Brr2 (residues 395–2129) was recently reported.38 The structure confirmed the modular organization of Brr2 and, notably, the contribution of the first Sec63 domain to the formation of a tunnel in which RNA can bind and be translocated during unwinding. The second helicase domain and the second Sec63 domain form a similar tunnel, although negatively charged residues likely prevent RNA binding. It is proposed that the second helicase module has retained its capacity to bind ATP but not to hydrolyse it. The crystal structure reveals physical contacts between the RecA-1 domain of the N-terminal helicase module and the RecA-2 domain of the C-terminal helicase module, and a mutation within the carboxy terminal ATP-binding motif I reduced U4/U6 unwinding by the N-terminal helicase domain in vitro. Furthermore, the N-terminal ATPase activity was found to be enhanced in the presence of the C-terminal cassette.
Helicases in the Spliceosome
Sub2
Two RNA helicases, Sub2 (yeast)/UAP56 (human) and Prp5 participate in the recognition of the BS sequence by U2 snRNP during the formation of the pre-spliceosome (Fig. 1). UAP56 (Fig. 3A) was originally identified in humans as an interactor of U2AF65.47 In vivo, Mud2 and Msl5 form a heterodimer and physically interact with U1 snRNP proteins, but not with U2 snRNP proteins, linking the recognition of the 5′SS by the U1 snRNP with recognition of the branch-site by Msl5.48 The heterodimer Mud2/Msl5 is proposed to recruit ATP-bound Sub2/UAP56 at, or close to, the BS. Subsequently ATP hydrolysis by Sub2 triggers release of Msl5, leaving Mud2 associated with Sub2/UAP56, and allowing access for U2 snRNP factors and U2 snRNA.49 The mechanism by which Sub2/UAP56 exerts its function is unclear; whether displacement of protein from an RNA or modulation of the Mud2/Msl5 protein interaction. The effects of mutations in motifs I and II of Sub2/UAP56 highlighted the need for ATP binding/hydrolysis but not unwinding for pre-spliceosome formation.49 Although Sub2/UAP56 is essential for yeast viability in normal conditions, it is dispensable when MUD2 or MSL5 is deleted.50 Therefore biochemical and genetic results are in good agreement.49
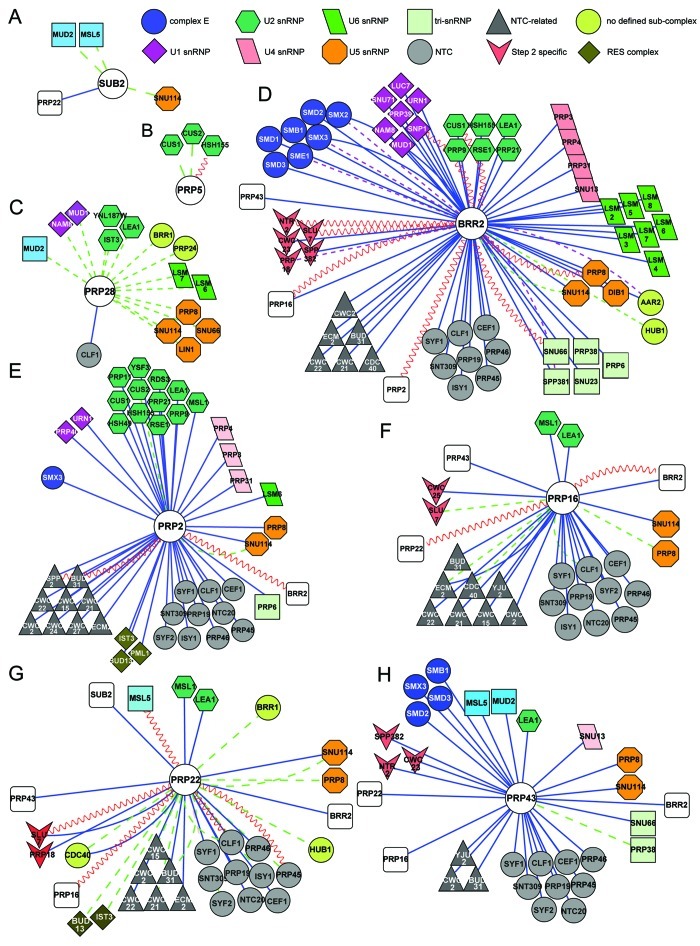
Figure 3. Cytoscape (http://www.cytoscape.org) representation of the interactome of S. cerevisiae splicing helicases. Splicing interactomes of (A) Sub2; (B) Prp5; (C) Prp28; (D) Brr2; (E) Prp2; (F) Prp16; (G) Prp22; (H) Prp43. The list of interactors was obtained from biogrid (http://thebiogrid.org). Only splicing factors are shown, grouped as in ref. 1. Colored shapes indicate sub-complex associations of splicing factors. Connectors are colored according to the experimental system used: blue lines represent affinity-capture followed by identification of the prey by mass spectrometry or western blotting, dashed purple lines represent co-fractionation or co-purification experiments, dashed green lines represent genetic interactions and sinusoidal red lines represent yeast two-hybrid interactions.
Recombinant Sub2/UAP56 can interact with U4 and U6 snRNAs in vivo and in vitro, and was proposed to play a role in unwinding the U4/U6 duplex in HeLa cell nuclear extracts.49 However, the significance of this is unclear, as Brr2 was shown to perform this function (see below, and refs. 40, 51, 52). In addition to a role in pre-mRNA splicing, the Sub2/UAP56 DEAD-box RNA helicase is implicated in Pol II transcription regulation, mRNA transport and localization, and the control of cancer and virus expression.49,53-58
Prp5
The ATPase activity of Prp5 is necessary to facilitate and proof-read the interaction of U2 snRNP with the BS. Prp5 interacts genetically with several U2 snRNP factors, including Hsh155, Cus1 and Cus2 (Fig. 3B)59-61 and several U2 snRNA mutations are suppressed by mutations in PRP5.62 The U2 snRNP-associated factor Cus2 was proposed to promote or stabilize a conformation in the U2 snRNA that is favorable for association of the SF3a and SF3b proteins prior to interaction of the U2 snRNA with pre-mRNA.62 Interestingly, when Cus2 is deleted, pre-spliceosome formation can proceed in the absence of ATP in vitro, although the presence of Prp5 is still required. Conversely, when recombinant Cus2 is added back to the extract, the ATP dependence is restored.59 Prp5 could promote the displacement of Cus2 from the U2 snRNA, while helping to stabilize U2 snRNA in the stem IIa conformation.59,63
In Schizosaccharomyces pombe, SpPrp5 associates with the U1 snRNP by directly interacting with Rsd1, mediated by the SR-like domains of each protein.64 The U1 snRNP proteins Snu71 and U1A, and the SF3b subunits of the U2 snRNP also co-purified with SpPrp5.64 In contrast, the budding yeast Prp5 does not possess SR repeats and no Rsd1 ortholog is known. Thus, although the splicing machinery is highly conserved, organism-specific interactions are also found.
Prp28
Following the selection and recognition of the 5′SS and the BS, the tri-snRNP joins the pre-spliceosome to form the transient complex B.1 Unwinding of U4/U6 leads to a major reorganization of the spliceosome, with the displacement of the U1 and U4 snRNPs and addition of spliceosome activation factors converting it to the Bact complex (Fig. 1). Displacing U4 allows U6 snRNA to base pair with the 5′SS (following U1 snRNP displacement) and with the U2 snRNA, contributing to the formation of the catalytic center.
Two RNA helicases, Prp28p (Fig. 3C) and Brr2 (Fig. 3D), play crucial roles in these rearrangements.1 Prp28, the third consecutive DEAD-box helicase to participate in spliceosome formation, was identified in a screen for cold-sensitive splicing mutants. Prp28 was initially proposed to be necessary for U4/U6 unwinding,65,66 but was later shown to be required for dissociation of the U1 snRNA/5′SS base-pairing interaction.67 The requirement for Prp28 can be bypassed by mutations in the U1 snRNP proteins U1C, Prp42 or Snu71, the cap-binding protein Cbp80 or Ynl187 that weakens the U1/5′SS interaction,68,69 suggesting that Prp28 may destabilize the U1 snRNA/5′SS interaction indirectly, by displacing proteins that stabilize it.68 Although Prp28 interacts genetically with two other U1 snRNP proteins, Nam8 and Mud1, mutation or deletion of these factors does not bypass Prp28.69 Therefore, the destabilizing effect of Prp28 may be limited to a fraction of the U1 snRNP that contacts the 5′SS.69
The replacement of U1 snRNA by U6 snRNA at the 5′SS is tightly coupled. Mutations that strengthen the U6:5′SS interaction relieve the defect caused by mutations that hyper-stabilize U1:5′SS,67 indicating an equilibrium between a “pre-spliceosome-5′SS” and a “complex B-5′SS.” Compatible with this, Prp28 interacted in large scale genetic screens with components of the U6 snRNP as well as with U5 snRNP (Fig. 3C).70-72 Thus Prp28 may proof-read the 5′SS based on the relative stability of its interactions with the U1 and U6 snRNAs.67
The N-terminal extension of the human ortholog of Prp28 has SR repeats that are targets of the SRPK2 kinase,73 and the phosphorylation status of Prp28 impacts the stable recruitment of the tri-snRNP and complex B formation, suggesting a potential regulatory mechanism.73
Brr2
Brr2 is generally accepted to be responsible for U4/U6 unwinding,40 although this activity of Brr2 appears to be functionally linked to Prp28 and Sub2/UAP5649 in vivo (see above). Brr2 is a component of the U5 snRNP, within which it contacts Prp8 and the GTPase Snu114, as visualized by cryo-electronmicroscopy.74 Brr2 associates with the U5 snRNP in a late maturation event of this particle75 that is coupled with the release of the chaperone-like protein, Aar2.75 Two recent studies showed that Brr2 loads onto the single stranded region of U4 located upstream of U4/U6 helix I and it was proposed to translocate 3′ to 5′ along the single stranded RNA to reach its duplexed target.39,76 Interestingly the RNase H domain of Prp8 binds the same region of U4 and prevents Brr2 loading there.76 Unwinding of the U4/U6 duplex by Brr2 in vitro is stimulated by a C-terminal region of Prp8 that contains the conserved RNase H and the ubiquitin-binding Jab1/MPN domains.42,43,77 The ubiquitination status of Prp8 and the GDP/GTP bound state of Snu114 can also regulate the unwinding of U4/U6 by Brr2 in vivo.71,78-80 Mutations that alter the interaction between Brr2 and Prp8 also reduce U4/U6 unwinding in vivo and in vitro.71 Interestingly, in humans some of these mutations cause retinal degeneration and blindness.81,82 It is unclear how essential and ubiquitous splicing factors can be responsible for a tissue-specific disorder.
A large body of results recently shed light on Brr2 structure and function. In addition to its association with U5 snRNP, Brr2 interacts genetically and physically with most spliceosomal sub-complexes (Fig. 3D), including components of U4/U6 snRNP, U1 snRNP and U2 snRNP.71,83-88 Brr2 was also proposed to be responsible for U2:U6 dissociation during spliceosome disassembly.79 In two-hybrid experiments the C-terminal domains of Brr2 interact with Prp2 and Prp16, and Brr2 was proposed to act as a receptor for these helicases at the catalytic center of the spliceosome.41 Being present at the heart of the spliceosome, Brr2 could exert the dual functions of a helicase and a protein-protein interaction platform, thereby participating in the control of the progression of the splicing reaction through the sequential interaction of its partners with its C-terminal domain.41-43,77 Furthermore, the use of a brr2 mutant, brr2-G858R, in combination with UV cross-linking and sequencing has recently revealed a new function for Brr2, driving conformational rearrangements at the catalytic center of the spliceosome that lead to competence for the second step of splicing.39
Prp2
Prp2 (Fig. 3E) joins the Bact complex along with Spp2 and is required for activation of the spliceosome prior to the first transesterification reaction. Spp2 is a member of the G-patch protein family that contains a glycine-rich domain. Spp2 is necessary for the recruitment and the function of Prp2,28,29 although it is not known how Spp2 affects Prp2 activity. The exact role of Prp2 is also unclear and its direct target is unknown, although an ATP-dependant conformational rearrangement promoted by Prp2 leads to destabilization of SF3a and SF3b proteins from the BS.5,89 Prp2 activity may therefore contribute to positioning the branch-site adenosine for nucleophilic attack on the 5′SS.
Cwc22, which is loosely associated with the NTC, contributes to destabilization of the SF3a/b proteins.90 In absence of Cwc22, Prp2 binds the spliceosome, hydrolyzes ATP and dissociates from the spliceosome, but fails to trigger SF3a/b release. It was suggested that Cwc22 could assist in positioning Prp2 close to the SF3-bound BS, thereby making use of the ATPase dependant 5′ to 3′ translocation of Prp2 to dissociate (directly or not) SF3a/b proteins from the BS. Indeed Prp2 cross-links beside the BS.91 Another target of Prp2 activity could be the connection between Cwc24 and the NTC.92,93
Using a dual-color fluorescence cross-correlation spectroscopy approach to measure the affinity of several splicing factors in the activated spliceosome, Ohrt et al.92 showed that Prp2 initiates a cascade of rearrangements, including displacement of the NTC-associated proteins Cwc24, Cwc27 and the RES complex protein Bud13 and reduced association of the SF3a/b proteins. Additionally, the two essential 1st step factors, Yju2 and Cwc25, become more strongly bound to the spliceosome. It was suggested that Cwc25 shifts the equilibrium from an inactive, step 1 incompetent catalytic center to an active, step 1 competent catalytic center.5 In this way, Prp2 may not only expose the BS adenosine but also promote the correct alignment of the 5′SS and the BS prior to the first trans-esterification reaction.
A large-scale two-hybrid screen of human splicing factors highlighted the profound integration of the various sub-complexes that form the spliceosome.94 This screen illustrated the dynamics of interactions and confirmed the conservation in human of the interaction between Prp2 and Spp2 (GPNOW in human). Moreover, the authors suggested that two molecules of Prp2 could interact with Spp2 prior to the first step, with only one copy remaining after the first step. More strikingly the post-first step Prp2-Spp2 dimer might recruit Prp16 to the catalytic center of the spliceosome. It is still unclear whether these Prp2 interactions are direct or whether the interactions between Prp2, Spp2 and Prp16 impact the enzymatic activity of the helicases.
Prp16
Prp16 (Fig. 3F) was originally identified in a screen for suppressors of the C259 branch site mutation of an ACT1-HIS4 gene fusion.95 For a long time, Prp16 was believed to associate with the spliceosome only transiently, for the duration of the second catalytic step.96 However, Prp16 was subsequently shown to be recruited to a suboptimal pre-mRNA after Cwc25 but before the first step of splicing.97,98 At this stage, Prp16 could stabilize the interaction of Cwc25 with the BS sequence in an ATP independent fashion. Thus, during the first step of splicing, Prp16 may play a facilitator role (see below). During splicing of optimum pre-mRNAs, the ATPase activity of Prp16 is required for the transition between the first and second step of splicing, and is proposed to trigger the release of Yju2 and Cwc25.97 Mutations in ISY1, PRP8 or the U6 snRNA were shown to suppress prp16 mutations, suggesting that those factors are also possible targets for Prp16.99-101 The activity of Prp16 could “terminate” the first step of splicing and “initiate” the formation of a step 2 competent spliceosome. In the step 2 spliceosome, the 3′exon becomes resistant to RNase H cleavage, suggesting that it may be bound within the active site of the spliceosome.102 Several factors (Cwc23, Slu7, Prp18, Prp22, Prp43, Ntr1 and Ntr2)3 join the complex C spliceosome before the second catalytic step. However, besides Prp16, only Slu7, Prp18 and Prp22 are necessary for second step catalysis in vitro.5,103
Prp22
The precise mechanisms that govern disassembly of the spliceosome are not well understood. Prp22 is the first helicase to participate in spliceosome disassembly, triggering release of the spliced mRNA . The ATPase activity of Prp22 is dispensable for the second step itself, but is necessary for release of U5 snRNP proteins and the spliced mRNA after the second step. 104-107
Based on yeast two-hybrid interactions and co-precipitation assays, Prp16 was proposed to serve as a receptor for Prp22 (Fig. 3G) in the spliceosome.41 Moreover, both these proteins were observed to interact with the substrate near the 3′SS; Prp16 was found to crosslink from -4 to +13 relative to the 3′SS prior the second step, whereas Prp22 bound in a Prp16 dependent manner at positions -8 to -4 upstream of the 3′SS at this stage.108,109 Prp22 also binds downstream of the splice junction in the spliced mRNA when the 3′ exon is longer than 13 nucleotides.110 Prp8 has a footprint of at least 13 nucleotides in the 3′ exon,111,112 and is thought to stabilize the duplex formed between the U5 snRNA and the first bases of the 3′ exon during the second step. In vitro, Prp22 can unwind RNA duplex with a 3′ to 5′ polarity.32 It is therefore tempting to speculate that Prp22 could bind the 3′ exon just behind Prp8 and track along in a 3′ to 5′ direction, stripping away Prp8 and U5 snRNA loop I from the spliced RNA.110
Prp43
Release of the excised intron lariat and disassembly of the associated post-splicing complex necessitate ATP hydrolysis by Prp43.37,113-116 Prp43 co-precipitates U2, U5 and U6 snRNAs,117-119 although in a CRAC (UV crosslinking and cloning) analysis Prp43 was only crosslinked to the U6 snRNA (positions 18–43 and 76–83). In vivo, Prp43 (Fig. 3H) associates with Spp382120-122 (also called Ntr1) and Ntr2.113,121 Spp382 is a G-patch protein that functions as a receptor for Prp43 and also stimulates the otherwise weak RNA unwinding activity of Prp43 in vitro.113 Prp43 was implicated not only in the disassembly of spliceosomes following the splicing of optimal pre-mRNA but also in the dissociation of spliceosomes that become stalled with sub-optimal substrates.98 Curiously, Prp43 also participates in ribosome biogenesis (see below). Thus, Prp43 could be a general disassembly factor.
Proofreading in splicing
An increasing body of evidence links several helicases with proofreading and discard of defective substrates (reviewed in refs. 124–126). Prp5 is proposed to proofread the BS during pre-spliceosome formation,127,128 Prp16 proofreads the 5′SS and the BS for the first step of splicing98,129,130 and Prp22131 appears to proofread the 3′SS and the BS for the second step. As previously mentioned, Prp28 was suggested to proofread the U1 snRNA:5′SS and/or U6 snRNA:5′SS interaction.67
The mechanism of proofreading is incompletely understood. Burgess and Guthrie129 proposed a version of kinetic proofreading in which the rate of ATP hydrolysis by helicases determines the fate of pre-mRNAs and splicing reactions. The kinetic proofreading model is based on the equilibrium between rejection and acceptance of a substrate for the next step of splicing. In the case of a suboptimal pre-mRNA, the rate of rejection is normally higher than the rate of acceptance and the defective pre-mRNA is discarded. In a possible mechanism (“timer model”124), the activation of a helicase ATPase activity would restrict the time allotted for a given splicing event to occur. For example, in the case of a pre-mRNA with a normal BS, the recognition and the association of U2 snRNP would happen quickly. Activation of Prp5 ATPase activity would “validate” the U2:BS interaction and promote pre-spliceosome formation. In the case of a mutated or suboptimal BS, the establishment of the U2:BS interaction would be slow and Prp5 activation would “reject” the defective spliceosome and promote its dissociation. Thus the helicase could play a dual function of stabilizer of correct interactions and de-stabilizer of impaired interactions. Studies performed on Prp5, Prp16 and Prp22 showed that mutations that reduce the level of ATPase activity (not necessarily the rate of the ATP hydrolysis) allow the splicing of reporter constructs in which the 5′SS, BS or 3′SS is not optimal.127-131
Prp43 also plays a crucial role in the quality control of splicing. Prp43 is responsible for entry into the non-reversible discard pathway (reviewed in refs. 2, 124, 125, 132). Whereas pre-mRNAs rejected by Prp16 or Prp22 can re-enter the splicing cycle, activation of Prp43 seals the fate of discarded RNAs.2,98,126,129,130,133
Possible Links Between Splicing Helicases and Other RNA Maturation Processes
The biogenesis of mRNPs involves a complex and highly integrated series of events that begins with initiation of transcription, quickly followed by addition of a monomethyl guanosine cap at the 5′ end of the nascent transcript, co-transcriptional spliceosome assembly (if an intron exists), 3′ end formation (cleavage and polyadenylation), association with nuclear pores and export to the cytoplasm,134 and these processing events can influence each other. There is evidence that Sub2/UAP56 participates in several mRNA processing pathways in the nucleus.53,134 The roles of other helicases have not been addressed so far. However, below we review physical and genetic interactions of splicing helicases that may suggest their participation in the regulation and/or integration of various processing events.
Links to transcription
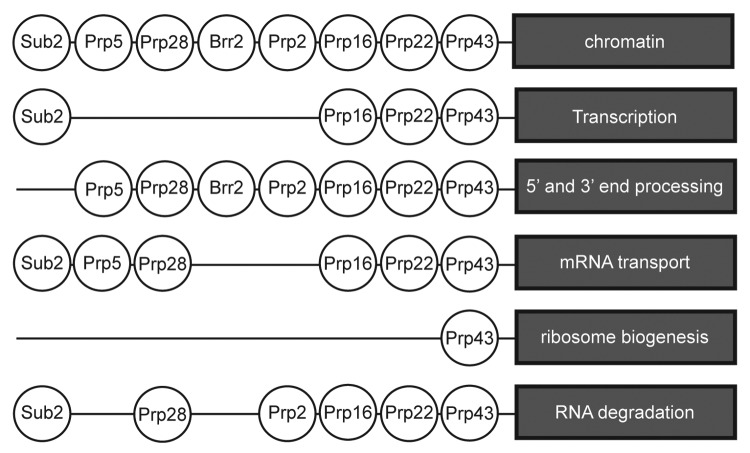
There is considerable evidence for the coupling of transcription and pre-mRNA splicing.135-140 Intriguingly, Chanarat et al.141 showed that the yeast Prp19 complex is necessary for the recruitment of THO /TREX (transcription and export) complex during transcription of both intron-containing and intronless genes. They also showed that a mutant of SYF1 (syf1–37), that encodes an NTC factor, slows transcription by Pol II while pre-mRNA splicing is unaffected. However, although Sub2, Prp16 and Prp22 associate with the THO (transcription elongation) complex in large scale proteomic and genetic screens,70,89 to date there is no information regarding the direct participation of splicing RNA helicases in the coupling process. Most splicing helicases show functional and/or physical interaction with chromatin and chromatin remodelling complexes (Fig. 4) such as the Swi2/Snf2 complex (Prp5, Prp16, Prp22, Prp43), the Swr1 complex (Prp22, Prp28) or with the RNA polymerase machinery. The Swi/Snf complex is an ATP-dependent chromatin remodelling complex that can displace/remodel/modify nucleosomes.142,143 The Swr1 complex catalyzes the replacement of histone H2A by histone H2Az at or around transcription start sites, thereby shifting the position of the reprogrammed nucleosome. This action can regulate transcription positively or negatively depending on the new position of the modified nucleosome. Somewhat confusingly, in addition to its role in splicing, U1 snRNA has been implicated in a splicing independent role in transcriptional activation.144
Figure 4. Splicing helicases are connected with other pathways of RNP metabolism. Based on large scale physical and genetic screens (see text), most splicing helicases are connected with several RNP biogenesis events. Only in few cases (see text) has the biological relevance of the proteome data been validated.
Among splicing helicases, Brr2 shows the least connections with proteins involved in chromatin remodelling or transcription (only Spt2 and Yta741). This may underline a splicing-only role for Brr2 and suggest that trans-acting helicases are more likely candidates for co-regulating splicing and transcription.
Links to 5′ and 3′ end processing
Links between pre-mRNA splicing and 5′ end capping or 3′ end cleavage/polyadenylation of transcripts have long been known.12,145,146 For example, in higher eukaryotes the 3′ end processing machinery plays an important role in the definition of the last exon and, conversely, splicing influences the choice of cleavage/polyadenylation site (reviewed in ref. 12). In particular, U1 snRNPs inhibit premature cleavage and polyadenylation.147
In yeast, where the spliceosome appears to recognize introns rather than exons, a direct effect of the 3′ end processing machinery on splicing is not clear. Nevertheless, most splicing helicases interact genetically and/or physically with factors involved in 3′end-processing such as Pab1 (Brr2, Prp43),83,148 or Nab2 (Prp5, Prp28).149
Capping at the 5′ end of transcripts occurs shortly (20 to 30 nucleotides) after initiation, as the nascent transcript emerges from the Pol II complex.150 The cap binding proteins Cbp80 and Cbp20 (Sto1 and Cbc2/Mud13 in yeast) promote splicing of cap-proximal introns by stabilizing the 5′SS:U1 snRNA interaction151-155 and deletion of the Cap Binding Complex (CBC) abolishes the co-transcriptional recruitment of the U1 snRNP to intron-containing transcripts.156 In Arabidopsis thaliana, mutations in AtCbp80 or AtCbp20 affect alternative splicing and 5′SS selection preferentially in the first intron.14 The cap binding protein Cbp20 co-purifies only with Brr2, Prp2, Prp16 and Prp2289,157 while the regulatory subunit Cbp80 co-purifies with every splicing helicase69,83,89,158 except Prp43. The biological significance of this difference is not clear. However, it may suggest that Cbp80 participates in the control of the early splicing helicases and formation of the commitment complex while the fully assembled cap binding complex would associate with the spliceosome from complex B onwards. The absence of interaction between the CBC and Prp43 could imply that the dissociation of the CBC from the spliceosome precedes Prp43 recruitment and Prp43-dependent spliceosome disassembly.
Links to mRNA transport
Splicing and mRNA transport are tightly coupled in eukaryotes,159 with spliced mRNAs being more efficiently exported to the cytoplasm than intronless RNAs.160 Several splicing helicases (Sub2, Prp16, Prp22, Prp43) display both genetic and physical links to the THO/TREX complex that connects transcription and mRNA transport.161-163 However, Sub2/UAP56 is the only splicing RNA helicase whose function in RNA transport is clearly established.53,164 In yeast, a sub2 mutation leads to nuclear accumulation of polyadenylated, spliced mRNA.56 In higher eukaryotes, UAP56 is part of the Exon Junction Complex that is deposited on spliced mRNA 24 to 26 bases upstream of exon-exon junctions and plays a role in mRNP transport.165-168 UAP56 also appears to bind intronless mRNAs prior to export.167,169 The signals for binding to intronless RNAs are not currently known, but they may depend on the presence of non-splicing factors such as the CBC.170 Sub2/UAP56 was shown to recruit the mRNA export factor Aly co-transcriptionally to both spliced and intronless mRNAs,160,171 and was proposed to act as an ATP-dependent chaperone of the Aly-RNA interaction.169 In yeast, ATP binding to Sub2 is necessary for mRNA transport172 and in higher eukaryotes the association of UAP56, Aly and CIP29 with the TREX complex is also dependent on the presence of ATP.173 CIP29 stimulates the helicase activity of UAP56,174 while Aly stimulates ATP hydrolysis by UAP56.169 UAP56/Sub2 is released prior to the export of mRNA to the cytoplasm.175
Links to ribosome biogenesis
During ribosome biogenesis two G-patch proteins, Gno1 and Pfa1, associate with Prp43.26,117,176 Pfa1 stimulates Prp43 activity in vitro, and in vivo their interaction seems to be required for maturation of the 20S pre-rRNA (pre-rRNA).117,177 Several Prp43 binding sites that were identified in 18S and 25S rRNA precursors mapped close to cleavage sites in the pre-rRNA, or close to snoRNA-rRNA base-pairing sites.178 Those data support the likely involvement of Prp43 in remodelling snoRNAs and/or in displacing snoRNAs from pre-rRNA complexes. Thus, it appears that Prp43 functions as a disassembly factor during ribosome biogenesis and during pre-mRNA splicing, subject to control by different G-patch proteins.
Links to RNA degradation
Several RNA helicases co-purify or interact genetically with proteins involved in RNA degradation (Fig. 4) but the functional significance is currently not known. The mutant sub2–20155 is synthetic sick when combined with depletion of the nuclear degradation factor Rrp6, but not with loss of the cytoplasmic degradation factor Xrn1.179 Additionally, overexpression of SUB2 leads to a synthetic growth defect when combined with mutations affecting the nuclear 5′ to 3′ exonuclease Rat1 or the TRAMP complex factor Mtr4.180 Furthermore, Egecioglu et al.125 found that the nuclear exosome and Rat1 participate in a quality control pathway that allows discard of aberrantly spliced mRNAs or incompletely spliced transcripts. For example the level of pre-mRNAs in a prp2–1 mutant increased when components of the nuclear exosome or Rat1 were depleted or mutated.181 The interaction of Prp43 with Xrn1 is likely linked to its role during pre-rRNA processing rather than its role in splicing.117
Conclusions
The fact that splicing helicases function within large and highly dynamic RNP complexes has greatly complicated the identification of their targets and characterization of their modes of action and regulation. Genetic studies, mainly in budding yeast, have been a rich source of information about interacting partners and potential targets. However, genetics may not distinguish functional interactions (which may be indirect) from direct physical interactions. In vitro analyses of splicing helicase activities using purified components have yielded limited information, and because pre-mRNA splicing is intricately intertwined with transcription and other RNA maturation events, in vitro studies of splicing factors will lack their influences unless coupled in vitro systems are developed. As discussed above, splicing RNA helicases are not only necessary for the progression of the spliceosome through the splicing cycle, they also function as proofreaders of the splicing process. Proofreading by helicases is only one level of quality control, which also involves the degradation of aberrant RNA molecules in the nucleus or in the cytoplasm.124-126 Therefore, a full understanding of the contribution of helicases to this process will require detailed kinetic studies in vivo.
As RNA helicases make highly transient interactions with their targets, small molecule inhibitors, including substrate or cofactor analogs, may prove useful to capture transient complexes for structural studies. Similarly, mutant helicases in combination with cross-linking approaches may permit global “snapshots” to be obtained of normally transient protein-protein and protein-RNA interactions,15 as mentioned above for Brr2. At the other end of the scale, combining chemical biology methodologies with single molecule fluorescence techniques now allows kinetic studies of spliceosome assembly and the splicing reactions to be performed on single transcripts in real time (reviewed in ref. 132).
Clearly, much remains to be unravelled about the precise role(s) played by splicing helicases, and how their activities are modulated and timed by their multiple partners, whether proteins, nucleic acids or small molecules. Deciphering the splicing helicase code remains an exciting challenge with profound repercussions in the understanding of normal and pathogenic pre-mRNA splicing.
Acknowledgments
We are grateful to Keerthi Chathoth and Daniela Hahn for helpful suggestions. This work was supported by Wellcome Trust grant 087551. The Wellcome Trust Centre for Cell Biology is supported by Wellcome Trust core funding [092076]. JDB is the Royal Society Darwin Trust Research Professor.
Glossary
Abbreviations:
- 3′SS
3′ splice site
- 5′SS
5′ splice site
- aa
amino acid
- BS
branch site
- CBC
Cap Binding Complex
- DUF
Domain of Unknown Function
- NTC
NineTeen Complex
- OB-fold
oligosaccharide binding fold
- pre-mRNA
precursor messenger RNA
- pre-rRNA
pre-ribosomal RNA
- RNP
ribonucleoprotein complex
- SF2
superfamily 2
- snRNP
small nuclear RNP
- snoRNA small nucleolar RNP
- SR
serine-arginine
- WH
winged helix
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22547
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:0003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37:179–88. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 4.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/S1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 5.Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol. 2009;16:1237–43. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 6.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–37. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staley JP, Woolford JL., Jr. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21:109–18. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens SW, Abelson J. Yeast pre-mRNA splicing: methods, mechanisms, and machinery. Methods Enzymol. 2002;351:200–20. doi: 10.1016/S0076-6879(02)51849-8. [DOI] [PubMed] [Google Scholar]
- 9.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–63. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–40. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinson HG. An active role for splicing in 3′-end formation. Wiley Interdiscip Rev RNA. 2011;2:459–70. doi: 10.1002/wrna.68. [DOI] [PubMed] [Google Scholar]
- 13.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–80. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 14.Raczynska KD, Simpson CG, Ciesiolka A, Szewc L, Lewandowska D, McNicol J, et al. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010;38:265–78. doi: 10.1093/nar/gkp869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordin O, Hahn D, Beggs JD. Structure, function and regulation of spliceosomal RNA helicases. Curr Opin Cell Biol. 2012;24:431–8. doi: 10.1016/j.ceb.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Koonin EV. Similarities in RNA helicases. Nature. 1991;352:290. doi: 10.1038/352290c0. [DOI] [PubMed] [Google Scholar]
- 17.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 21.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Björk P, Jin S, Zhao J, Singh OP, Persson JO, Hellman U, et al. Specific combinations of SR proteins associate with single pre-messenger RNAs in vivo and contribute different functions. J Cell Biol. 2009;184:555–68. doi: 10.1083/jcb.200806156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 24.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–7. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebaron S, Papin C, Capeyrou R, Chen YL, Froment C, Monsarrat B, et al. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009;28:3808–19. doi: 10.1038/emboj.2009.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–6. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol. 2004;24:10101–10. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL., Jr. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA. 1995;1:375–90. [PMC free article] [PubMed] [Google Scholar]
- 30.Aksaas AK, Larsen AC, Rogne M, Rosendal K, Kvissel AK, Skålhegg BS. G-patch domain and KOW motifs-containing protein, GPKOW; a nuclear RNA-binding protein regulated by protein kinase A. J Mol Signal. 2011;6:10. doi: 10.1186/1750-2187-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korneta I, Magnus M, Bujnicki JM. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res. 2012;40:7046–65. doi: 10.1093/nar/gks347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider S, Schwer B. Functional domains of the yeast splicing factor Prp22p. J Biol Chem. 2001;276:21184–91. doi: 10.1074/jbc.M101964200. [DOI] [PubMed] [Google Scholar]
- 33.Schneider S, Hotz HR, Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J Biol Chem. 2002;277:15452–8. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- 34.Edwalds-Gilbert G, Kim DH, Silverman E, Lin RJ. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA. 2004;10:210–20. doi: 10.1261/rna.5151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–29. doi: 10.1017/S1355838298980992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotz HR, Schwer B. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics. 1998;149:807–15. doi: 10.1093/genetics/149.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem. 2002;277:17743–50. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 38.Santosa KS, Mozaffari-Jovin S, Webera G, Pena V, Lührmann R, Wahl MC. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci USA. 2012;••• doi: 10.1073/pnas.1208098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hahn D, Kudla G, Tollervey D, Beggs MC. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 2012;26 doi: 10.1101/gad.199307.112. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn D, Beggs JD. Brr2p RNA helicase with a split personality: insights into structure and function. Biochem Soc Trans. 2010;38:1105–9. doi: 10.1042/BST0381105. [DOI] [PubMed] [Google Scholar]
- 41.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–67. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pena V, Jovin SM, Fabrizio P, Orlowski J, Bujnicki JM, Lührmann R, et al. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell. 2009;35:454–66. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, et al. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009;16:731–9. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodman IL, Bolt EL. Molecular biology of Hel308 helicase in archaea. Biochem Soc Trans. 2009;37:74–8. doi: 10.1042/BST0370074. [DOI] [PubMed] [Google Scholar]
- 45.Richards JD, Johnson KA, Liu H, McRobbie AM, McMahon S, Oke M, et al. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008;283:5118–26. doi: 10.1074/jbc.M707548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–52. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 47.Fleckner J, Zhang M, Valcárcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–72. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 48.Chang J, Schwer B, Shuman S. Structure-function analysis and genetic interactions of the yeast branchpoint binding protein Msl5. Nucleic Acids Res. 2012;40:4539–52. doi: 10.1093/nar/gks049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 2008;22:1796–803. doi: 10.1101/gad.1657308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–9. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–55. doi: 10.1016/S0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 52.Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5:959–71. doi: 10.1017/S135583829999012X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen H. UAP56- a key player with surprisingly diverse roles in pre-mRNA splicing and nuclear export. BMB Rep. 2009;42:185–8. doi: 10.5483/BMBRep.2009.42.4.185. [DOI] [PubMed] [Google Scholar]
- 54.Meignin C, Davis I. UAP56 RNA helicase is required for axis specification and cytoplasmic mRNA localization in Drosophila. Dev Biol. 2008;315:89–98. doi: 10.1016/j.ydbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jensen TH, Boulay J, Rosbash M, Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr Biol. 2001;11:1711–5. doi: 10.1016/S0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- 57.Wisskirchen C, Ludersdorfer TH, Müller DA, Moritz E, Pavlovic J. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J Virol. 2011;85:8646–55. doi: 10.1128/JVI.02559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Wang X, Zhang X, Goodrich DW. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol Cell Biol. 2005;25:4023–33. doi: 10.1128/MCB.25.10.4023-4033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perriman R, Barta I, Voeltz GK, Abelson J, Ares M., Jr. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci U S A. 2003;100:13857–62. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33:5053–62. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wells SE, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr. CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev. 1996;10:220–32. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 62.Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–9. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perriman RJ, Ares M., Jr. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–20. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao W, Kim HS, Cao Y, Xu YZ, Query CCA. A U1-U2 snRNP interaction network during intron definition. Mol Cell Biol. 2012;32:470–8. doi: 10.1128/MCB.06234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strauss EJ, Guthrie C. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes Dev. 1991;5:629–41. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- 66.Strauss EJ, Guthrie C. PRP28, a ‘DEAD-box’ protein, is required for the first step of mRNA splicing in vitro. Nucleic Acids Res. 1994;22:3187–93. doi: 10.1093/nar/22.15.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/S1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 68.Chen JY, Stands L, Staley JP, Jackups RR, Jr., Latus LJ, Chang TH. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell. 2001;7:227–32. doi: 10.1016/S1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 69.Hage R, Tung L, Du H, Stands L, Rosbash M, Chang TH. A targeted bypass screen identifies Ynl187p, Prp42p, Snu71p, and Cbp80p for stable U1 snRNP/Pre-mRNA interaction. Mol Cell Biol. 2009;29:3941–52. doi: 10.1128/MCB.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 2010;327:425–31. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brenner TJ, Guthrie C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–80. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuhn AN, Reichl EM, Brow DA. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc Natl Acad Sci U S A. 2002;99:9145–9. doi: 10.1073/pnas.102304299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mathew R, Hartmuth K, Möhlmann S, Urlaub H, Ficner R, Lührmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15:435–43. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 74.Häcker I, Sander B, Golas MM, Wolf E, Karagöz E, Kastner B, et al. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol. 2008;15:1206–12. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- 75.Weber G, Cristão VF, de L Alves F, Santos KF, Holton N, Rappsilber J, et al. Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev. 2011;25:1601–12. doi: 10.1101/gad.635911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mozaffari-Jovin S, Santos KF, Hsiao H-H, Will CL, Urlaub H, Wahl MC, et al. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 2012;26 doi: 10.1101/gad.200949.112. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol. 2009;16:42–8. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ. A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol. 2008;15:444–51. doi: 10.1038/nsmb.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell. 2006;23:389–99. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartels C, Klatt C, Lührmann R, Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3:875–80. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boon KL, Grainger RJ, Ehsani P, Barrass JD, Auchynnikava T, Inglehearn CF, et al. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol. 2007;14:1077–83. doi: 10.1038/nsmb1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao C, Bellur DL, Lu S, Zhao F, Grassi MA, Bowne SJ, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009;85:617–27. doi: 10.1016/j.ajhg.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 84.Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, et al. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA. 2001;7:1543–53. [PMC free article] [PubMed] [Google Scholar]
- 85.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–71. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 87.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–10. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 88.Ren L, McLean JR, Hazbun TR, Fields S, Vander Kooi C, Ohi MD, et al. Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS One. 2011;6:e16719. doi: 10.1371/journal.pone.0016719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lardelli RM, Thompson JX, Yates JR, 3rd, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16:516–28. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yeh TC, Liu HL, Chung CS, Wu NY, Liu YC, Cheng SC. Splicing factor Cwc22 is required for the function of Prp2 and for the spliceosome to escape from a futile pathway. Mol Cell Biol. 2011;31:43–53. doi: 10.1128/MCB.00801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teigelkamp S, McGarvey M, Plumpton M, Beggs JD. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994;13:888–97. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ohrt T, Prior M, Dannenberg J, Odenwälder P, Dybkov O, Rasche N, et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012;18:1244–56. doi: 10.1261/rna.033316.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goldfeder MB, Oliveira CC. Cwc24p, a novel Saccharomyces cerevisiae nuclear ring finger protein, affects pre-snoRNA U3 splicing. J Biol Chem. 2008;283:2644–53. doi: 10.1074/jbc.M707885200. [DOI] [PubMed] [Google Scholar]
- 94.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, et al. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell. 2012;45:567–80. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 95.Couto JR, Tamm J, Parker R, Guthrie C. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987;1:445–55. doi: 10.1101/gad.1.5.445. [DOI] [PubMed] [Google Scholar]
- 96.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–9. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 97.Tseng CK, Liu HL, Cheng SC. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011;17:145–54. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell. 2010;39:385–95. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–54. doi: 10.1016/S1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 101.Madhani HD, Guthrie C. Genetic interactions between the yeast RNA helicase homolog Prp16 and spliceosomal snRNAs identify candidate ligands for the Prp16 RNA-dependent ATPase. Genetics. 1994;137:677–87. doi: 10.1093/genetics/137.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–9. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–94. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schneider S, Campodonico E, Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J Biol Chem. 2004;279:8617–26. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- 105.Aronova A, Bacíková D, Crotti LB, Horowitz DS, Schwer B. Functional interactions between Prp8, Prp18, Slu7, and U5 snRNA during the second step of pre-mRNA splicing. RNA. 2007;13:1437–44. doi: 10.1261/rna.572807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–93. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 107.James SA, Turner W, Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–77. doi: 10.1017/S1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McPheeters DS, Schwer B, Muhlenkamp P. Interaction of the yeast DExH-box RNA helicase prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res. 2000;28:1313–21. doi: 10.1093/nar/28.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McPheeters DS, Muhlenkamp P. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol Cell Biol. 2003;23:4174–86. doi: 10.1128/MCB.23.12.4174-4186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–54. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grainger RJ, Barrass JD, Jacquier A, Rain JC, Beggs JD. Physical and genetic interactions of yeast Cwc21p, an ortholog of human SRm300/SRRM2, suggest a role at the catalytic center of the spliceosome. RNA. 2009;15:2161–73. doi: 10.1261/rna.1908309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Teigelkamp S, Whittaker E, Beggs JD. Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic Acids Res. 1995;23:320–6. doi: 10.1093/nar/23.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, et al. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–25. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arenas JE, Abelson JN. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci U S A. 1997;94:11798–802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pandit S, Lynn B, Rymond BC. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci U S A. 2006;103:13700–5. doi: 10.1073/pnas.0603188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, et al. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol. 2005;25:9269–82. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Combs DJ, Nagel RJ, Ares M, Jr., Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–34. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol. 2006;26:513–22. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pandit S, Paul S, Zhang L, Chen M, Durbin N, Harrison SM, et al. Spp382p interacts with multiple yeast splicing factors, including possible regulators of Prp43 DExD/H-Box protein function. Genetics. 2009;183:195–206. doi: 10.1534/genetics.109.106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsai RT, Tseng CK, Lee PJ, Chen HC, Fu RH, Chang KJ, et al. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol Cell Biol. 2007;27:8027–37. doi: 10.1128/MCB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Herrmann G, Kais S, Hoffbauer J, Shah-Hosseini K, Brüggenolte N, Schober H, et al. Conserved interactions of the splicing factor Ntr1/Spp382 with proteins involved in DNA double-strand break repair and telomere metabolism. Nucleic Acids Res. 2007;35:2321–32. doi: 10.1093/nar/gkm127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boon KL, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, et al. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol Cell Biol. 2006;26:6016–23. doi: 10.1128/MCB.02347-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci. 2012;37:263–73. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Egecioglu DE, Chanfreau G. Proofreading and spellchecking: a two-tier strategy for pre-mRNA splicing quality control. RNA. 2011;17:383–9. doi: 10.1261/rna.2454711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Horowitz DS. The splice is right: guarantors of fidelity in pre-mRNA splicing. RNA. 2011;17:551–4. doi: 10.1261/rna.2577511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–49. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Perriman R, Ares M., Jr. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol Cell. 2010;38:416–27. doi: 10.1016/j.molcel.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–91. doi: 10.1016/0092-8674(93)90363-U. [DOI] [PubMed] [Google Scholar]
- 130.Burgess S, Couto JR, Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–17. doi: 10.1016/0092-8674(90)90086-T. [DOI] [PubMed] [Google Scholar]
- 131.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–90. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hoskins AA, Gelles J, Moore MJ. New insights into the spliceosome by single molecule fluorescence microscopy. Curr Opin Chem Biol. 2011;15:864–70. doi: 10.1016/j.cbpa.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Perriman RJ, Ares M., Jr. Alternative splicing variability: exactly how similar are two identical cells? Mol Syst Biol. 2011;7:505. doi: 10.1038/msb.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luna R, Gaillard H, González-Aguilera C, Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–31. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- 135.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–93. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Alexander R, Beggs JD. Cross-talk in transcription, splicing and chromatin: who makes the first call? Biochem Soc Trans. 2010;38:1251–6. doi: 10.1042/BST0381251. [DOI] [PubMed] [Google Scholar]
- 137.Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol. 2003;23:5768–79. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–15. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 139.Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–66. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–6. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 141.Chanarat S, Seizl M, Strässer K. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 2011;25:1147–58. doi: 10.1101/gad.623411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tang L, Nogales E, Ciferri C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol. 2010;102:122–8. doi: 10.1016/j.pbiomolbio.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [Review] [DOI] [PubMed] [Google Scholar]
- 144.Kwek KY, Murphy S, Furger A, Thomas B, O’Gorman W, Kimura H, et al. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol. 2002;9:800–5. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 145.Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep. 2002;3:869–74. doi: 10.1093/embo-reports/kvf173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Albulescu LO, Sabet N, Gudipati M, Stepankiw N, Bergman ZJ, Huffaker TC, et al. A quantitative, high-throughput reverse genetic screen reveals novel connections between Pre-mRNA splicing and 5′ and 3′ end transcript determinants. PLoS Genet. 2012;8:e1002530. doi: 10.1371/journal.pgen.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–8. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–50. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 149.Batisse J, Batisse C, Budd A, Böttcher B, Hurt E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem. 2009;284:34911–7. doi: 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nat Struct Biol. 2000;7:838–42. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 151.Lewis JD, Görlich D, Mattaj IW. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 1996;24:3332–6. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–98. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 153.Cooke C, Alwine JC. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol. 1996;16:2579–84. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Colot HV, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 155.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, et al. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol. 1999;19:6543–53. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 157.Schwer B, Erdjument-Bromage H, Shuman S. Composition of yeast snRNPs and snoRNPs in the absence of trimethylguanosine caps reveals nuclear cap binding protein as a gained U1 component implicated in the cold-sensitivity of tgs1Δ cells. Nucleic Acids Res. 2011;39:6715–28. doi: 10.1093/nar/gkr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, et al. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods. 2007;4:951–6. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 159.Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–31. doi: 10.1016/S0092-8674(02)00627-X. [DOI] [PubMed] [Google Scholar]
- 160.Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, et al. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–7. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 161.Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–8. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 162.Strässer K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–52. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 163.Jimeno S, Rondón AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–35. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Linder P, Stutz F. mRNA export: travelling with DEAD box proteins. Curr Biol. 2001;11:R961–3. doi: 10.1016/S0960-9822(01)00574-7. [DOI] [PubMed] [Google Scholar]
- 165.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–9. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–108. [PMC free article] [PubMed] [Google Scholar]
- 167.Gatfield D, Le Hir H, Schmitt C, Braun IC, Köcher T, Wilm M, et al. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001;11:1716–21. doi: 10.1016/S0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- 168.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–7. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Taniguchi I, Ohno M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol Cell Biol. 2008;28:601–8. doi: 10.1128/MCB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nojima T, Hirose T, Kimura H, Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J Biol Chem. 2007;282:15645–51. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- 171.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–31. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kota KP, Wagner SR, Huerta E, Underwood JM, Nickerson JA. Binding of ATP to UAP56 is necessary for mRNA export. J Cell Sci. 2008;121:1526–37. doi: 10.1242/jcs.021055. [DOI] [PubMed] [Google Scholar]
- 173.Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 2010;24:2043–53. doi: 10.1101/gad.1898610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sugiura T, Sakurai K, Nagano Y. Intracellular characterization of DDX39, a novel growth-associated RNA helicase. Exp Cell Res. 2007;313:782–90. doi: 10.1016/j.yexcr.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 175.Stewart M. Nuclear export of mRNA. Trends Biochem Sci. 2010;35:609–17. doi: 10.1016/j.tibs.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 176.Guglielmi B, Werner M. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J Biol Chem. 2002;277:35712–9. doi: 10.1074/jbc.M205526200. [DOI] [PubMed] [Google Scholar]
- 177.Pertschy B, Schneider C, Gnädig M, Schäfer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284:35079–91. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–92. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol. 2002;22:8254–66. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.González-Aguilera C, Tous C, Gómez-González B, Huertas P, Luna R, Aguilera A. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell. 2008;19:4310–8. doi: 10.1091/mbc.E08-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–75. doi: 10.1016/S0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]