Abstract
Similar to proteins, RNA molecules must fold into the correct conformation and associate with protein complexes in order to be functional within a cell. RNA helicases rearrange RNA secondary structure and RNA-protein interactions in an ATP-dependent reaction, performing crucial functions in all aspects of RNA metabolism. In prokaryotes, RNA helicase activity is associated with roles in housekeeping functions including RNA turnover, ribosome biogenesis, translation and small RNA metabolism. In addition, RNA helicase expression and/or activity are frequently altered during cellular response to abiotic stress, implying they perform defined roles during cellular adaptation to changes in the growth environment. Specifically, RNA helicases contribute to the formation of cold-adapted ribosomes and RNA degradosomes, implying a role in alleviation of RNA secondary structure stabilization at low temperature. A common emerging theme involves RNA helicases acting as scaffolds for protein-protein interaction and functioning as molecular clamps, holding RNA-protein complexes in specific conformations. This review highlights recent advances in DEAD-box RNA helicase association with cellular response to abiotic stress in prokaryotes.
Keywords: RNA helicase, DEAD-box proteins, prokaryotes, abiotic stress, cold-adapted degradosome, ribosome biogenesis, cytoskeleton compartmentalization
RNA Helicases: Structure and Function
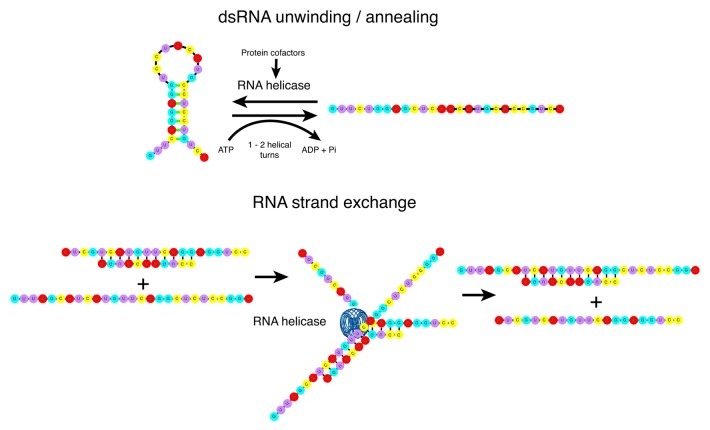
RNA helicases are molecular motors that rearrange RNA secondary structure or ribonucleoprotein (RNP) complexes.1-4 Genes encoding RNA helicases have been identified in all three kingdoms of life as well as in many viral genomes.5 RNA helicases are potentially associated with the entire lifespan of an RNA molecule, from transcription to degradation, functioning in housekeeping pathways including translation initiation, ribosome biogenesis and mRNA splicing.6-8 In addition, RNA helicase activity is associated with specialized processes involving cell growth, differentiation, development, small RNA (sRNA) metabolism and response to abiotic stress.5,9-11 The importance of RNA helicases and RNA metabolism in cell physiology is underscored by their role in viral infection, aging, neurological disorders and human diseases including cancer.12,13 Nucleic acid helicases are broadly classified into six families with the majority of RNA helicases belonging to superfamily 2 (SF2) comprised of five subfamilies, three of which are termed the DEAD, DEAH and DExH protein subfamilies after variations in the signature amino acid domain, Asp-Glu-Ala-Asp.14 The DEAD-box family is the largest subfamily, characterized by an RNA helicase core containing a minimum of 12 conserved amino acid motifs comprising two tandemly repeated RecA-like domains that structurally resemble the bacterial recombinase A protein (RecA) (Fig. 1).15 Importantly, the majority of RNA helicases also possess N- and/or C-terminal extensions that provide RNA or protein cofactor specificity.4 RNA helicases frequently function in multi-subunit complexes, prime examples being translation initiation and the mRNA spliceosomal complex in eukaryotes.4 Recently it has become apparent that RNA helicase function may not be limited to alteration of RNA structure but may include nucleation of protein-protein interactions and maintenance of the RNP complex in a long-lived, stable configuration. This process is referred to as RNA clamping16 and is an enticing model for RNA processing pathways that require numerous sequential steps, each requiring a different RNA helicase.4,9 It is also becoming evident that RNA helicases potentially link transcriptional and post-transcriptional regulation of RNA metabolism with different cellular processes.17-22 The ability of some RNA helicases to both unwind and anneal dsRNA duplexes and combine the two processes to catalyze RNA strand exchange expands the repertoire of RNA structural alterations catalyzed by these enzymes (Fig. 2).23-26
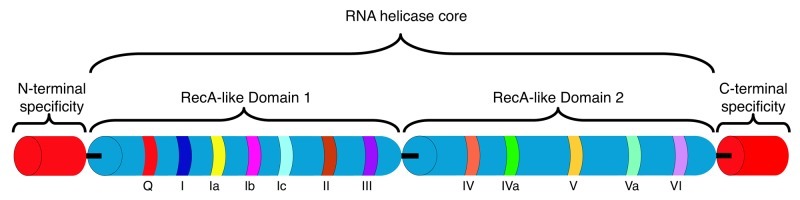
Figure 1. DEAD-box RNA helicase structure. The DEAD-box helicase core consists of two RecA-related domains containing a minimum of 12 conserved domains that characterize the family.4,5,15,126 The sequence of the conserved domains in the E. coli DeaD protein are: Q, PSPIQ; I, GSGKTAAF; Ia, LAPTRELAVQV; Ib, GG; Ic, VGTPGRLLD; II, VLDEADEM; III, FSATM; IV, IIFVRTK; IVa, NGDMNQALR; V, LIATDVA; Va, ARGLDVERISLVVNYD; VI, YVHRIGRTGRAG. The relative order for the domains with RNA-binding or unwinding and ATP binding and/or hydrolysis functions are shown. For example, domain II contains the DEAD motif (Asp-Glu-Ala-Asp). In addition to the helicase core, DEAD-box RNA helicases frequently contain N- and C-terminal extensions that provide RNA and/or protein interaction specificity. The figure is not drawn to scale and has been adapted from Linder and Jankowsky.4
Figure 2. RNA helicases and RNA structure remodeling. The majority of RNA helicases are believed to rearrange RNA structure by unwinding dsRNA into ssRNA. Relatively few also catalyze annealing of complementary ssRNA into dsRNA, the annealing and unwinding activities combining to promote RNA strand exchange. These processes require ATP hydrolysis, not for the structure rearrangement but for helicase association with the RNA substrate.2,127,128 Unwinding generally occurs over short distances, 1–2 helical turns,3 however protein cofactors can increase the processivity and also contribute to RNA substrate specificity.
Similar to eukaryotic systems, molecular, structure/function and genetic analysis has recently provided unique insights into the roles RNA helicases perform in prokaryotes. Prokaryotes generally encode significantly fewer RNA helicases than eukaryotes, implying a multi-tasking role for individual prokaryotic RNA helicases. Multiple roles are also indicated by protein network alignments indicating that RNA helicases function in conserved pathways between bacteria and yeast.27 For example the bacterial helicase, DeaD (also called CsdA), is structurally related to three helicases in yeast, suggesting that DeaD functions in multiple pathways in Escherichia coli. These observations indicate that bacterial RNA helicases can perform numerous roles, facilitating crosstalk between different cellular pathways in response to changing environmental conditions. This review discusses recent insights associating DEAD-box RNA helicase activity with prokaryotic response to abiotic stress.
RNA Helicases in Cold Stress
Induction of RNA helicase expression in response to a reduction in growth temperature (Table 1) intimates that RNA helicase activity is required for normal cellular function at temperatures below the growth optimum. While the exact biochemical nature of this requirement is not known, it is generally assumed that helicase activity resolves RNA secondary structures that are thermodynamically stabilized at low temperature. This general function would potentially explain the pleiotropic nature of cellular processes associated with cold-induced RNA helicase inactivation.28-33 Resolving of secondary structure is not expected to occur over long distances as the majority of DEAD-box RNA helicases are not processive, exhibiting RNA unwinding activity over short artificial RNA duplexes containing less than two helical turns in vitro although it is possible that protein cofactors and/or specific RNA substrates may enhance processivity in vivo (Fig. 2).4
Table 1. Overview of the roles performed by prokaryotic RNA helicases in response to abiotic stress.
| RNA helicase |
Bacterium |
Role in RNA metabolism |
Reference |
|---|---|---|---|
| Cold Stress | |||
| DeaD (CsdA) |
Various |
Pleiotropic effects at all temperatures; formation of a cold-adapted ribosome; replaces RhlB in degradosome at low temperature |
29, 54, 57, 58, 74, 75, 76 |
| SrmB |
E. coli |
Ribosome assembly |
28, 54, 77, 130 |
| RhlE |
Various |
Degradosome composition; suppression of deaD mutation phenotype |
54, 77, 99 |
| CrhR |
Synechocystis PCC 6803 |
DeaD-related homolog, unique C-terminal extension; pleiotropic effects at all temperatures |
32, 41, 44, 45, 46 |
| CrhC |
Anabaena PCC 7120 |
RhlE homolog; Cold stress specific; membrane localized at cell poles |
37–40 |
| DeaD |
Psychrobacter arcticus 273-4 |
DeaD exchanges with RhlB in degradosome during cold stress |
101 |
| DeaD |
Methanococcoides burtonii |
Psychrotolerant increases dead expression from 4 to 23°C |
50 |
| DeaD |
Methanococcus jannaschii |
Hyperthermophilic methanarchaeon isolated from a deep-ocean hydrothermal vent, grows optimally at 85°C; DEAD homolog is the most significantly upregulated gene (15-fold) in response to cold stress at 65°C |
51 |
| TK0306 |
Thermococcus kodakaraensis |
DeaD homolog; expression decreases as temperature increased from 60 to 93°C |
52 |
| CshA CshB CshC |
Bacillus cereus |
Five DEAD box helicase mutants; cshA, cshB and cshC have different effects at different temperatures (12, 30 and 37°C) |
30, 134, 135 |
| CshA CshB YxiN |
Bacillus subtilis |
DeaD homologs; transcriptome, CshA or CshB required for viability; YxiN, the DbpA homolog, interacts with 23S rRNA |
56, 87, 105, 108, 136 |
| DeaD |
Listeria monocytogenes EGD-e |
All four DEAD box genes increase; 3 affected growth and motility Δlmo0866, Δlmo1450 and Δlmo1722, one affected growth at all temperatures (lmo1450) |
47, 48, 137 |
| SCD_00930 |
Sulfuricella denitrificans skB26 |
RhlE homolog; one of three DEAD box helicases increases at 4°C vs. 22°C |
49 |
| DeaD |
Yersinia pseudotuberculosis |
Inactivation reduces growth and deaD expression increased at 3°C vs. 28°C |
65 |
| DeaD |
Shewanella piezotolerans |
Psychrotolerant bacterium isolated from deep-sea sediment; deaD expression increases 23-fold at 0°C vs. optimal at 20°C |
55 |
| CC_0835 | Caulobacter crescentus | RhlE homolog; upregulated and severely reduces growth of mutant at 15°C vs. 30°C | 138 |
| Other Stresses | |||
|---|---|---|---|
| DVU3310 |
Desulfovibrio vulgaris Hildenborough |
Salt; DeaD homolog, proteome |
60 |
| CrhR |
Synechocystis 6803 |
Salt; subtractive RNA hybridization |
42 |
| DeaD |
Clostridium perfringens |
Oxidative stress; DeaD homolog mutation enhances cell resistance |
62 |
| DeaD |
E. coli |
Isobutanol tolerance |
63 |
| DR 1624 (DeaD) |
Deinococcus radiodurans |
Cadmium stress; DeaD homolog transcript levels rapidly reduced |
64 |
| DR B0136 (HepA) |
Deinococcus radiodurans | Cadmium stress; HepA homolog transcript levels progressively reduced | 64 |
| Ribosome Biogenesis | |||
|---|---|---|---|
| DeaD |
E. coli |
50S subunit assembly |
29, 54, 76, 130 |
| SrmB |
E. coli |
50S subunit assembly |
28, 80, 81, 139 |
| RhlE |
E. coli |
50S subunit assembly |
77 |
| DbpA | E. coli | 70S subunit assembly; 23S rRNA conformational change | 53, 76, 86 |
| RNA Turnover: Degradosome | |||
|---|---|---|---|
| RhlB |
E. coli and other |
Subunit 37°C; forms membrane associated helical structures |
90, 112, 113 |
| RhlE |
|
Cold shock degradosome in vitro |
92 |
| DeaD |
|
Cold shock degradosome in vivo |
93–97 |
| CshA |
Bacillus subtilis |
RhlB equivalent |
104–107 |
| CshB DeaD |
B. subtilis |
Evidence for transient association under specific conditions but not cold stress; nucleoid associated |
107 |
| CshA |
Staphylococcus aureus |
RhlB equivalent |
103 |
| RhlE |
Pseudomonas syringae |
RhlE-homologue interacts with RNase R and RNase E |
102 |
| RhlE | Caulobacter crescentus | Nucleoid localized degradosome containing a RhlE-homolog | 140, 141 |
Diversity of cold induced RNA helicases
Cold induction of RNA helicase expression has been observed in a variety of bacteria including psychrotolerant and hyperthermophilic Gram-negative, Gram-positive and archaeal species (Table 1). While many of the examples originate from transcriptomic or proteomic analysis, some cases have been examined in more detail. Eukaryotic genomes generally encode a number of genes coding for DEAD-box RNA helicases, 26 in the Saccharomyces cerevisiae and 37 in the human genome,4 at least two of which are temperature regulated in yeast, DED1 and DBP2.34 Indeed, the smallest genome of a free-living eukaryote from the fungus Ashbyz gosypii contains a DEAD-box RNA helicase repertoire essentially identical to that observed in S. cerevisiae.35 Functionally, eukaryotic RNA helicases do not genetically complement one another. Thus the genetic repertoire and functional analysis suggests that each RNA helicase generally performs a specific function in eukaryotes. This is not the situation in prokaryotes whose genomes encode a variable number of RNA helicase genes with evidence that individual proteins participate in divergent pathways. The phylogeny of bacterial RNA helicases has been extensively analyzed in a recent review.36 Although not a general characteristic of these organisms, only a limited number of obligate endo-mutualist genomes do not encode an RNA helicase gene, presumably since they do not encounter rapid changes in growth conditions, especially temperature, and thus do not require RNA helicase activity. A substantial number (116) of sequenced genomes only contain a single DEAD-box helicase while gene expansion has resulted in 12 genes in some γ-Proteobacteria, for example members of the Shewanella and Vibrio genera.36 The clade to which these RNA helicase genes group is essentially random, with no clear association between the number or repertoire of RNA helicases encoded in a given bacterial genome. The number of helicase genes is also not related to ORF number or GC content although there is a correlation between the number of RNA helicase genes and 23S rRNA copy number, most probably a reflection of RNA helicase activity association with ribosome biogenesis.36 Thus, predictions relating growth habitat with RNA helicase repertoire is not currently possible.
RNA helicase gene complement
The RNA helicase gene complement of an organism is also not associated with a free-living growth style. For example, low temperature induction of RNA helicase expression has been examined in the free-living obligate photoautotrophic cyanobacteria. In Anabaena PCC 7120, one of the two DEAD-box RNA helicase genes, crhC, is specifically expressed only in response to temperature stress induced by a temperature downshift from 30 to 20°C.37,38 CrhC exhibits limited RNA helicase activity in vitro39 and is localized to the cell poles in this filamentous organism.40 This cellular localization has led to the speculation that CrhC is associated with the co-translational-translocation of proteins into or across the inner membrane. Expression of the second Anabaena PCC 7120 helicase, crhB, resembles that of crhR in Synechocystis PCC 6803.37 A single DEAD-box RNA helicase, crhR, is encoded in the Synechocystis genome whose expression is regulated by the redox status of the electron transport chain41 and is enhanced by stresses that increase reduction of the chain, including salt42 and temperature.43 crhR inactivation has a pleiotropic effect with mutant cells exhibiting a severe growth phenotype at 20°C and physiological and morphological effects derived from a decrease in photosynthetic carbon fixation.32 An important observation is that crhR inactivation produces effects at all temperatures, indicating that RNA helicase activity is constitutively required although the effects of mutation are exacerbated at low temperature.32 CrhR is most closely related to DeaD, however this homology only encompasses the RNA helicase core, as CrhR possesses a unique C-terminal extension that is specific to related genes only found in cyanobacteria (Owttrim, unpublished). This domain is presumably related to the ability of CrhR to catalyze dsRNA unwinding, annealing and strand exchange, providing the potential to perform RNA structural rearrangements in addition to those normally performed by canonical RNA helicases.24 Functionally, CrhR activity has been associated with regulation of protein chaperone expression,44 photosystem stoichiometry45 and various pathways in response to cold stress.46 These genetic, structural and biochemical attributes indicate that CrhR may perform a variety of roles in RNA metabolism in Synechocystis.
Species containing more than one DEAD box RNA helicase family member frequently differentially express each gene. For example, the effect of mutation of the five DEAD box helicases in Bacillus cereus in response to temperature, pH and oxidative stress indicated that cshD and cshE deletion had no effect under any condition tested while ∆cshA, B and C did not grow at 12°C.30 ∆cshA, B and C produce differential effects in response to temperature fluctuation, deletion of cshA reduced growth at all temperatures up to 45°C, deletion of cshB had an impact below 30°C and over 37°C, while deletion of cshC led mainly to a cold-sensitive phenotype. Similar differentially effects were observed in response to oxidative stress while the response to pH stress was only affected in ∆cshA cells. The results indicate that CshA, CshB and CshC participate in general response pathways to stress conditions with CshA activity playing more important roles in cellular response to low temperature and pH stress. This appears to be a common phenomena as similar effects are observed in psychrotolerant organisms, the Gram-positive pathogen Listeria monocytogenes EGD-e47,48 and the sulfur oxidizing Gram-negative bacterium Sulfuricella denitrificans skB26.49
RNA helicase “cold induction” temperature
RNA helicase association with cellular response to low temperature is not based on the absolute temperature but related to growth at temperatures below the growth optimum. Expression of DeaD homologs is temperature regulated in Archaebacteria in response to a temperature decrease from 23 to 4°C in the psychrotolerant Methanococcoides burtonii50 and for TK0306, from the hyperthermophilic archaeons Methanococcus jannaschii and Thermococcus kodakaraensis grown at 60 or 65°C compared with the optimal growth temperature of 85°C.51,52 It is therefore tempting to speculate that the hyperthermophilic data suggest that RNA helicase activity is performing alternative roles unrelated to problems associated with the stabilization of RNA structure at the non-permissive temperature.
An important observation is that RNA helicase activity of the temperature-regulated gene is required at all temperatures, similar to the effect of the R331A mutation in DbpA53 and ∆deaD29,54 in E. coli and ∆crhR in Synechocystis.32 In instances where this is not the case, another RNA helicase gene presumably complements the RNA helicase mutation, as exemplified by Shewanella piezotolerans, a psychrotolerant Gram-negative bacterium isolated from deep-sea sediment that expresses 11 DEAD-box RNA helicases.55 Inactivation of the S. piezotolerans gene most significantly induced by low temperature, the deaD homolog, has no effect on growth at any temperature, suggesting that another RNA helicase complements its role. Complementation of helicase inactivation in organisms expressing multiple RNA helicases appears to be common, as also observed in B. subtilis in which at least one of the cold-induced helicases, cshA or cshB, is required for viability.56
Expression of a non-conserved number of RNA helicases in bacterial species deserves further investigation. One potential explanation involves the switching of RNA helicases in multi-subunit complexes in response to changes in growth temperature, as discussed below. RNA helicase isozyme switching may also apply to other abiotic stress responses as well. These observations indicate that bacterial RNA helicase involvement in cellular response to abiotic stress is flexible, with species-specific approaches to the problem. Correlation between the type of helicase and abiotic stress is not always observed although cold stress is associated with alteration of ribosome and RNA degradosome composition (see below).
RNA helicase inactivation differentially affects growth in E. coli
Inactivation of RhlE, SrmB, CsdA and DbpA differentially affect growth in response to temperature change. deaD mutation is primarily responsible for the growth defects observed in rich media at 37°C, as the doubling time is increased by 20% in ∆deaD strains while individual deletion of the other four helicases has no effect on growth rate.54 The ∆deaD and ∆srmB strains both exhibit a cold sensitive phenotype that is progressively enhanced as the temperature decreases below 30°C.28,29,54 In the ∆deaD strain the doubling time increases 2 to 4.5-fold29,54,57 and results in a filamentous growth phenotype at 15°C.57 In conjunction, deaD expression is induced in response to a reduction in growth temperature, increasing from basal levels at 37°C to significant accumulation at 15°C, with the DeaD protein associating with ribosomes.57 At reduced temperature, DeaD overexpression suppresses the cold sensitive phenotype of rpsB, encoding ribosomal protein S258 and deaD mutation influences expression of specific genes, including heat- and cold-shock proteins57 and degradation of specific mRNA transcripts (Table 1).59 These results are reflective of DeaD performing roles in translation initiation and RNA degradation (see below). Deletion of multiple DEAD box genes in E. coli has provided further insights into the roles performed by RNA helicases, at temperatures below 25°C additive effects of the deletion of multiple helicases is observed with the most severe effect created by deletion of all five genes (∆5 strain), however the primarily effect results from the absence of DeaD activity.54
RNA Helicases in Other Abiotic Stresses
Regulation of RNA helicase expression is associated with a variety of environmental stress conditions other than low temperature, as summarized in Table 1. The examples include situations in which helicase inactivation either enhances or decreases cellular sensitivity to the stress. Similar to cold stress, cellular requirement for increased RNA helicase abundance is generally associated with alteration of the stability of RNA secondary structure in response to the stress. An example is salt stress, which, like low temperature, also causes water stress, is thought to reflect an increased requirement for RNA unwinding caused by the stabilizing effect of increased salt concentrations on RNA stability.42,60 This conclusion is supported by the observation that osmolytes decrease RNA stability, suggesting that cells subjected to osmotic stress utilize a combination of osmolyte synthesis and RNA helicase activity to overcome the restrictions imposed on RNA metabolism.61 Examination of how multiple stresses influence cell growth in organisms that express multiple RNA helicases is just beginning. An extensive analysis of the roles performed by four putative DEAD-box RNA helicase genes on growth of L. monocytogenes EGD-e under heat, pH, osmotic, ethanol, and oxidative stresses has recently been presented.48 Again mutational studies indicate that each helicase functions in divergent cellular processes in response to different conditions, although changes in expression level are not correlated with changes in stress tolerance for two of the helicases.48
RNA helicase inactivation enhancement of stress tolerance
Instances in which helicase mutation enhances cellular tolerance to stress are accumulating. Mutation of a deaD homolog in the strictly anaerobic Gram-positive pathogen Clostridium perfringens, that exhibits a strong oxidative stress response, generates a mutant that is more resistant to oxidative stress.62 Enhancement of stress tolerance by RNA helicase mutation has also proven useful in a biotechnological application, contributing to the reallocation of carbon flow and enhancement of the production of second-generation biofuels. E. coli induced to evolve isobutanol tolerance are mutated in deaD and hrpA.63 Interestingly, the deaD mutation “evolved” during the selection process subsequently to mutations in rpsB, coding for the 30S ribosomal subunit S2, and the RNA chaperone, hfq. The physiological reasons for the enhanced tolerance are not known but possibly involve abnormal production of a protein that is detrimental during the stress condition. These observations indicate that alteration of RNA metabolic pathways contributes to cellular tolerance of butanol stress and suggest that targeting RNA helicases will provide a fruitful approach toward enhancement of additional biotechnological applications.
RNA helicase downregulation in response to abiotic stress
It is interesting that the downregulation of RNA helicase expression in response to stress has rarely been reported, possibly an indication that RNA helicases perform essential functions during optimal growth. An example occurs in Deinococcus radiodurans in which transcript accumulation of the DEAH-box homolog of hepA (DR_B0136) progressively decreases (12-fold within 1 h) and the deaD homolog, DR_1624, is rapidly reduced (15-fold within 15 min) in response to cadmium stress.64 The reason(s) for the decrease are not understood, however the mechanism is potentially related to the situations in which deletion of RNA helicases enhance cellular tolerance to stress.
RNA helicases in pathogenesis
RNA helicases performing diverse roles in cellular stress responses suggests a potential involvement in pathogenesis, such as the example given above in C. perfringens.62 Recently, deaD inactivation in the food-borne pathogen, Yersinia pseudotuberculosis, indicates that DeaD is dispensable at 28°C and growth is impaired at 3°C and thus may play a role in food-borne pathogenesis.65 This appears to be an underexploited area of investigation and more examples relating RNA helicases to pathogenicity are expected.
RNA helicase response to abiotic stress is not universal
Cellular requirement for enhanced RNA helicase expression in response to abiotic stress is not universal, being extremely variable and frequently not observed even in response to extreme conditions.66-68 For example, RNA helicase gene expression was not altered in B. subtilis spores exposed to outer space conditions or a simulated Martian environment for 559 d on a space shuttle mission.69 The mechanism(s) why the alteration of RNA helicase expression varies in stress responses in different organisms is not known, presumably other RNA chaperones are fulfilling the role performed by RNA helicases in these systems. For example, cyanobacteria and E. coli express members of the RNA binding protein families, Rbp and Csp, in response to abiotic stresses including low temperature59,70,71 and osmotic and nitrogen stress.72 This suggests Rbp and Csp proteins perform roles required at low temperature with CspA being inferred to destabilize RNA secondary structure.73
RNA Helicases in Ribosome Biogenesis
RNA helicases associated with E. coli ribosome biogenesis
Since more than 20 RNA helicases are associated with eukaryotic ribosome biogenesis,74 it is not unexpected that four of the five DEAD-box RNA helicases encoded in the E. coli genome,75 RhlE, SrmB, CsdA and DbpA, are associated with this process. In E. coli, these genes are non-essential under normal growth conditions with mutant’s only manifesting problems at low temperature. This implies that the defect in ribosome assembly is created by a thermal induced event involving stabilization of RNA secondary structure, trapping it in a misfolded state, which is alleviated by RNA helicase catalyzed unwinding. Ribosome assembly is a tightly controlled process involving the correct folding of rRNAs coordinated with ribosomal protein (r-protein) loading, in which the E. coli helicases function in a sequential manner, a perfect scenario for pathway regulation by RNA helicase activity.9 The primary objective is formation of active ribosomes, potentially via pathways that differ at low and high temperature, producing temperature-adapted ribosomes.
Individual roles for RNA helicases in E. coli ribosome biogenesis
The progressive growth defects observed in response to RNA helicase mutation correlate with accumulation of aberrant forms of the 50S ribosomal subunit, sedimenting at 40S, that accumulate in ∆deaD and ∆srmB mutants at all temperatures, but more pronounced at low temperature.28,29,54 In the ∆deaD mutant, 40S particle accumulation is growth stage specific, accumulating to a lesser extent at 37°C during early log- but not mid log-phase growth.76 The 40S particles are deficient in a differing subset of 50S proteins in the ∆deaD and ∆srmB mutants.29 This observation, combined with the non-additive nature of the defects in the ∆deaD∆srmB double mutant, indicates that the two helicases perform different roles in 50S biogenesis. There is some functional overlap, however, as DeaD overexpression partially complements SrmB mutation, but not the reverse (Fig. 3).28,29
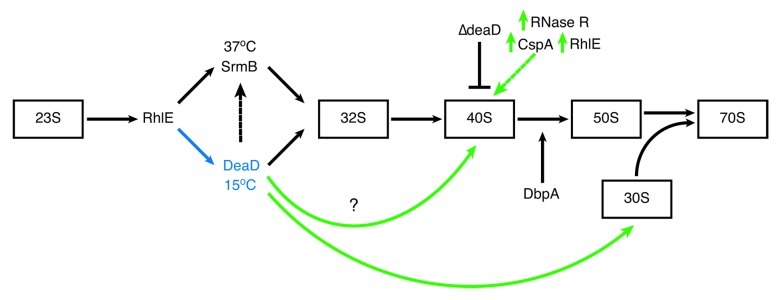
Figure 3. RNA helicases in E. coli ribosome assembly. The relative order of RNA helicase function in ribosome assembly and the sizes of intermediate 50S subunit particles that accumulate in RNA helicase mutants are indicated. RhlE functions early in the assembly process and is proposed to dictate whether assembly will follow an SrmB- or DeaD-specific pathway.77 Whether the ribosome biosynthetic pathway involves both RNA helicases or if separate SrmB- and DeaD-specific pathways function at high and low temperature, respectively, is not known. SrmB interaction with L4, L24 and the 23S rRNA81 indicates that it functions before DeaD however there is crosstalk between the two helicases as DeaD overexpression partially rescues the assembly defect in ΔsrmB cells (black dotted arrow).29 SrmB is required to form the L1024 G-ribo wrench pseudoknot, providing the nucleation site for L13 and subsequent events early in ribosome assembly.81 The role of DbpA in assembly is controversial76 but is reported to act late in the process, performing a function that occurs in the absence of DbpA, but not in cells expressing both wild type and mutant R331A proteins.53,86 DeaD has also been shown to be required for a very late step in 30S assembly involving 16S rRNA.117deaD inactivation is partially alleviated by overexpression of cold-induced proteins, RNase R or CspA,98,100 and also RhlE (green dotted arrow),98 which catalyze RNA unwinding.73,99,129 These observations suggest that DeaD functions to remove inhibitory RNA secondary structures whose stability is enhanced at low temperature. The overall objective is to produce stress-condition-specific ribosomes differing in their subunit composition e.g., a cold adapted ribosome. The figure is adapted from Shajani et al.130
Association of the deficient r-proteins at different points in the assembly process indicates that SrmB functions before DeaD in the assembly process (Fig. 3).29 SrmB associates with the aberrant 40S particle but not mature 30S, 50S or 70S subunits, suggesting that SrmB completes its functional role and normally leaves the assembling 50S subunit before assembly is complete, a step that cannot be completed in the absence of SrmB. Identical ribosome profiles are observed in wild type and ∆deaD cells grown at 37°C while 40S particles lacking a specific subset of L-proteins and containing improperly processed pre-23rRNA accumulate progressively as the temperature is decreased.29,76 Thus, DeaD appears to be only crucial for ribosome assembly at low temperature29,76 in conjunction with its expression pattern.57 Again, the function performed by the RNA helicases in ribosome assembly appears to be related to the removal of inhibitory RNA secondary structures and/or RNP complexes that form in the absence of RNA helicase activity and whose stability is enhanced in response to a reduction in growth temperature as ribosomal defects in the ∆5 strain are progressively alleviated by increasing the growth temperature to 41 and 45°C.54
rRNA defects in deaD and srmB mutants
The ∆deaD and ∆srmB strains also accumulate improperly processed precursors of the 23S and 16S rRNA but not 5S rRNA.28,29,77 These observations are not considered to be a direct effect of RNA helicase mutation but are ascribed to a defect in final 70S ribosome assembly as final processing of rRNA precursors occurs in the mature ribosome.78
Roles of DeaD and SrmB in E. coli ribosome biogenesis
What are the site(s) and mode of DeaD and SrmB action? Are the defects observed in ΔdeaD helicase mutants resulting from lack of translation initiation of r-proteins or rRNA maturation or actual assembly? Peil et al.76 proposed that DeaD could affect r-protein translation leading to the lack of specific r-proteins in deaD mutants, as observed by Charollais et al.29 While DeaD has been associated with translaton, Peil et al.76 also show that the final activation step, as indicated by reduced peptide bond formation by the 40S particles, is affected in deaD mutants, suggesting a direct involvement of DeaD in ribosome assembly. DeaD interaction with a variety of nucleic acid binding proteins including large and small subunit ribosomal proteins, in addition to RNA and DNA methylases and RNase R in a non-RNA-dependent manner suggests that DeaD may perform additional roles in ribosome assembly and/or cellular metabolism in addition to alteration of rRNA secondary structure,79 roles which are required at low temperature.
SrmB interacts with L4, L24 and domain 1 located near the 5′ end of the 23S rRNA suggesting that the protein cofactors L4 and L24 directs SrmB to its correct RNA target in vivo.80 The L4-L24–23S rRNA RNP complex is stable in vitro, leading Trubetskoy et al.80 to propose that SrmB functions to clamp the 23S rRNA in a stable conformation until this stage of assembly can be completed. Insights into the basis of SrmB-rRNA interaction were revealed by an elegant genetic screen to identify rRNA mutations compensating for the lack of SrmB. Two heptanucleotide repeats were identified in domain II of the 23S rRNA (nucleotides 1022–1028 and 1034–1040) and a complementary hexanucleotide repeat in the 5S rRNA (nucleotides 25–30).81 These mutations potentially disrupt complementary base pairing between the 23S and 5S rRNAs. These sites do not Watson-Crick base pair in the mature ribosome suggesting that SrmB may function to resolve improper folding of the 23S and 5S rRNAs that would result in the inhibition of 50S subunit assembly. These interactions indicate that SrmB is required to form the L1024 G-ribo wrench pseudoknot, providing the nucleation site for L13 and subsequent events early in ribosome assembly.81 Low temperature would be predicted to enhance stability of these misfolded conformations, necessitating alleviation catalyzed by SrmB unwinding.
SrmB and DeaD therefore appear to have differing yet overlapping effects on 50S ribosome maturation, a proposal supported by the observation that the effects are not additive in a ∆srmB∆deaD mutant.29 Involvement of another factor, possibly another RNA helicase or RNA chaperone, that compensates for the absence of DeaD is provided by the observation that essentially all of the 40S particles in ∆deaD cells can be converted into functional 50S subunits although the rate is reduced 4-fold compared with wild type.76
RhlE and E. coli ribosome biogenesis
RhlE is proposed to function in 50S subunit assembly before DeaD and SrmB, aiding DeaD and SrmB activity in ribosome biogenesis (Fig. 3). rhlE is a multicopy suppressor that partially alleviates the ∆deaD mutation but exacerbates the ∆srmB cold sensitive phenotype.77 Similar differential effects of RhlE on ∆deaD and ∆srmB mutants were observed on 40S particle accumulation, 23S rRNA maturation and the effect of rhlE deletion in ∆deaD∆rhlE and ∆srmB∆rhlE double mutants. Jain77 proposed that the concentration of RhlE regulates pre-50S ribosome maturation through two separate pathways in which high and low RhlE levels favor separate SrmB- or DeaD-dependent maturation pathways, respectively. Since deaD expression is cold induced, RhlE selection of DeaD for 50S assembly could contribute to a cold-specific ribosome assembly pathway, resulting in formation of a cold-adapted ribosome (Fig. 3). Evidence for further roles in ribosome biogenesis or activity is potentially provided by the observation that RhlE associates with 50S but also 30S and 70S ribosomes,77 a situation similar to that observed for DeaD29 but not SrmB which primarily associates with the 50S subunit.28 The mechanism regulating RhlE expression levels or how RhlE interacts with or directs activity of the other helicases is not known. These observations indicate that the functional interactions between these helicases are complex and require further investigation.
DbpA and E. coli ribosome biogenesis
DbpA is a non-essential gene and mutant strains do not display a growth defect in rich media at either 37°C (82) or 25°C.76 Any role for DbpA in ribosome biogenesis is therefore controversial83 and, although lower temperatures were not investigated, Peil et al.76 were unable to provide conclusive evidence for DbpA involvement in ribosome biogenesis at 25°C in rich media. A link with ribosome biogenesis has only been observed in response to mutation of the terminal Arg residue in the DbpA HRIGR box (R331A in conserved motif VI).53 R331A creates a dominate-negative mutant, generating a small colony phenotype only in the presence of wild type DbpA, an effect observed at 37°C and more severely at 22°C. The marginal reduction in growth rate, 1.6- and 2.2-fold at 37°C and 22°C, reinforces the conclusion that temperature has a minor effect on DbpA function.
What is the role performed by DbpA? DbpA is one of the few RNA helicases for which a specific RNA sequence is known to activate RNA-dependent unwinding, helix 92 of the 23S rRNA peptidyl transferase center, suggesting a role altering the structure of this rRNA domain during ribosome assembly.84,85 Aberrant 45S particles accumulate in R331A cells, lacking L-proteins that bind late in assembly and contain incompletely modified 23S rRNA, effects that are enhanced at 22°C.53,86 In R331A, the 45S particles stimulate DbpA ATPase activity86 indicating that hairpin 92 is not sequestered and this assembly step has not been completed. These results suggest that DbpA functions at a late stage of ribosome assembly, possibly required to catalyze a conformational change necessary for 23S rRNA processing, a step involved in peptidyl transferase center formation (Fig. 3). The lack of a phenotype in dbpA mutants suggests that DbpA activity is not crucial for ribosome biogenesis and can be catalyzed by another RNA helicase, RNA chaperone or spontaneously at a rate sufficient to produce a wild type phenotype. The mechanism by which appearance of a phenotype only when R331A is overexpressed in the presence of the wild type gene is not known.86 In these cells, the low levels of wild type DbpA will be completely sequestered by R331A.86 The wild type DbpA-R331A complex may bind to the normal structure in the 92 hairpin but cannot catalyze the required conformational change, thereby stalling assembly and creating the 45S particle. This clamping scenario could also explain the observation that overexpression of R331A in a ∆dpbA background does not produce the phenotype and why the reaction does not occur spontaneously, as observed in the ∆dpbA mutant.53,86 This proposal is consistent with the observation that the 45S particles contain reduced levels of seven L-proteins, many of which bind late in the assembly process.86 A clamping hypothesis does not immediately appear to account for the observation that either DbpA or R331A are ribosome-associated under any condition tested,86 a situation differing from SrmB, DeaD and RhlE.28,29,77 In Bacillus subtilis, the DbpA homolog, YxiN, interacts specifically with the 23 rRNA87 however further analysis of association with stress responses or ribosome biogenesis has not been reported.
RNA helicase role in ribosome biogenesis in other bacteria
RNA helicase involvement in ribosome biogenesis in a temperature dependent fashion is not restricted to E. coli. For example similar effects on growth are obtained in response to deletion of the DExD family member, Lmo1722, in L. monocytogenes EGD-e. Lmo1722 is highly conserved in Gram-positive bacteria, a homolog of which is not present in E. coli. Lmo1722 is required for optimal growth at 25°C but dispensable at 37°C. The deletion strain exhibits reduced motility, decreased 50S and 70S accumulation and has defects in 23S rRNA processing, defects associated with deletion of the C-terminal region.88
RNA Helicases in RNA Turnover
Temperature dependent RNA helicase composition of the E. coli RNA degradosome
In E. coli and other bacteria a major player in RNA turnover is a multisubunit complex termed the degradosome, consisting of the DEAD-box RNA helicase RhlB, RNase E, PNPase and enolase at 37°C.89,90 The complex catalyzes RNA degradation from an initial endonucleolytic site created by RNase E and subsequent 3′ to 5′ exonucleolytic nucleotide removal catalyzed by PNPase. Structured RNA inhibits PNPase function and thus the RNA helicase activity of RhlB removes impeding RNA secondary structure, allowing PNPase degradation of the generated ssRNA.91
RhlB exhibits limited RNA helix unwinding activity and can be functionally replaced by RhlE in vitro92 or DeaD in vitro and in vivo at low temperature (Fig. 4).93 Proteomic analysis confirmed the presence of DeaD and not RhlB in E. coli degradosomes purified from cells grown at 10°C, indicating not only that degradosome RNA helicase composition varies in response to low temperature stress but also that RhlB association is the rate-limiting factor in E. coli degradosome assembly.94 Replacement of RhlB and potentially other components is believed to create a “cold shock degradosome” that unwinds and promotes degradation of specific RNAs whose duplex structures are thermodynamically stabilized at low temperature.93 Degradosome composition therefore alters in response to various growth conditions, presumably functioning in cellular adaptation to the new environmental condition, providing the potential that the standard subunits may vary in response to other stress conditions and that the exchanged subunits may associate with other RNP complexes and thus function in alternative pathways.95,96
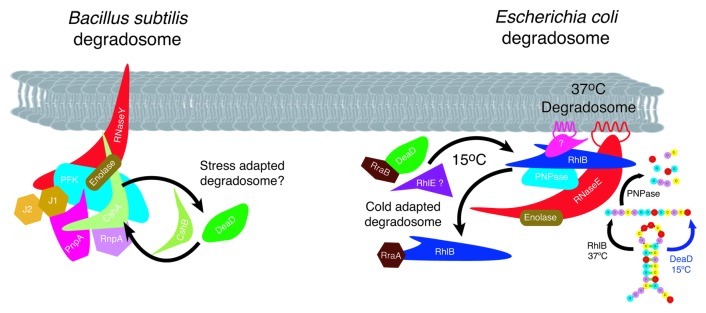
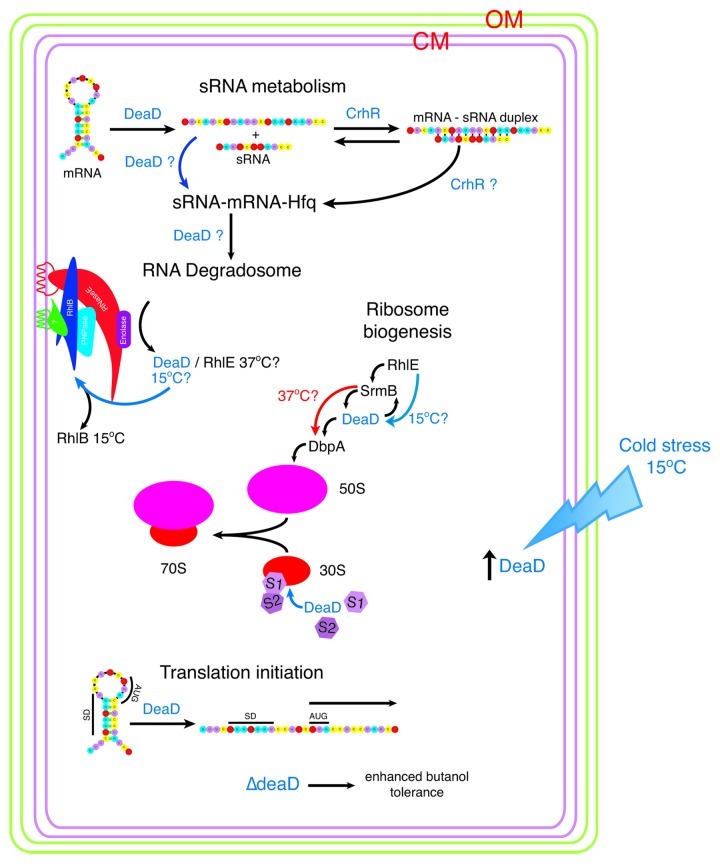
Figure 4. RNA helicases and stress adapted RNA degradosomes. Subunit composition of the B. subtilis and E. coli degradosome is diagrammed. Under normal and conditions at 37°C, the E. coli degradosome protein subunits are shown with the identified protein-protein interactions indicated by overlapping symbols. The non-catalytic C-terminal domain of RNase E acts as the scaffold for subunit assembly109 and associates with the membrane via an amphipathic α-helix.92 RNase E and RhlB independently form helical cytoskeletal structures.112,113 RhlB interacts directly with RNase E and PNPase at 37°C. In response to cold stress, RhlB can be exchanged with DeaD (in vitro and in vivo) or RhlE (in vitro) to produce a cold stress degradosome.93,94 Stoichiometry of subunits is not known, however a model in which PNPase:RNase E:enolase:RhlB associate in a ratio of 4:3:8:8 binding to the C-terminus of RNase E has been proposed.131 Similar to E. coli, the B. subtilis and Staphylococcus aureus degradosomes assemble on RNase Y (RNase E equivalent)105 and are composed of the RNA helicase CshA (RhlB equivalent), enolase and PnpA (PNPase equivalent). In addition the complex contains the RNases J1, J2 and RnpA and the glycolytic enzyme, phosphofructokinase (Pfk).103,104 RNase Y contains a single transmembrane domain, anchoring the complex in the membrane105 but it is not known if the Bacillus degradosome complex forms helical cytoskeletal structures similar to those observed in E. coli. CshA interacts directly with enolase, Pfk and PnpA however evidence for exchange with other helicases in response to low temperature, although it could not be ruled out, possibly does not occur as CshA levels remain unaltered in response to growth conditions.107 The diversity of interacting proteins provides the potential for alteration of degradosome composition in response to a range of abiotic stress conditions in B. subtilis.
RNA helicase subunit switching in the degradosome is mediated by the RNase E binding proteins, RraA and RraB, with the level of RhlB being reduced by RraA and DeaD increasing in the presence of RraB (Fig. 4).97 The physiological importance of RraA- and RraB-mediated degradosome subunit switching is illustrated by the alteration of RNA decay patterns generating global transcript abundance profiles that differ from those observed in response to RNase E mutation.97
DeaD is implicated in RNA turnover by the observation that cspA mRNA is stabilized in ∆deaD cells, an effect that is partially reversed by overexpression of RhlE, CspA or RNase R (Fig. 3).98 CspA and RNase R are cold-induced proteins while RhlE and RNase R possess RNA helix destabilizing activity,73,99,100 a function also inferred for CspA.73 The results suggest that these RNA interacting proteins complement deaD deletion by promoting unwinding of DeaD RNA targets, providing ssRNA substrates for degradation. It is also possible that DeaD is required for the expression of an RNase, possibly RNase R, whose expression is cold induced.100 These interactions again exemplify the plasticity of RNA helicase interaction with different protein cofactors in response to alterations in growth conditions, interactions that alter the repertoire of cellular biochemical capabilities.
RNA helicase composition of the RNA degradosome in other bacteria
Variations in degradosome composition are observed in other bacteria where evidence for RNA helicase isozyme exchange in response to temperature change has been obtained from genomic and proteomic analysis. For example in the Gram-negative γ-proteobacterium Psychrobacter arcticus 273–4 which grows in permafrost soils, transcriptome analysis indicated that deaD transcript abundance generally decreased while psyc_0943 levels increased as the temperature increased from -6°C to 22°C.101 Psyc_0943 is an RhlB homolog, suggesting that Psychrobacter arcticus 273–4 switches RhlB for DeaD in the degradosome during cold stress. Alteration of degradosome composition in response to low temperature extends to the replacement of RhlB with RhlE and PNPase by RNase R in the cold adapted Gram-negative organism, Pseudomonas syringae Lz4W.102 It should be noted that expression of RNase R100 and PNPase95 are enhanced at low temperature in E. coli but RNase R is not detected in cold-stress degradosomes.94
Gram-positive bacteria potentially possess an even wider diversity of degradosome complexes based on the variety of proteins known to associate with the complex.103,104 Similar degradosomes assemble in B. subtilis and Staphylococcus aureus containing the RhlB-equivalent DEAD-box RNA helicase, CshA, RNases Y (functional equivalent of RNase E),105 J1, J2 and RnpA, PNPase and the glycolytic enzymes enolase and phosphofructokinase (Fig. 4).103,104 Ribonuclease interaction suggests that CshA helicase activity is crucial for degradosome activity, supported by the observation that strong secondary structure impedes RNase J1 degradation.106 Evidence for association of two of the other three DEAD-box RNA helicases, CshB and DeaD, with the degradosome could not be ruled out,107 suggesting that subunit composition can be altered under additional growth conditions in B. subtilis (Fig. 4). The mechanisms regulating this switch appear to differ from those occurring in E. coli, as while cshA and cshB are temperature regulated at the transcript level in B. subtilis108 this is not reflected at the protein level where the level of the degradosome-associated CshA remains constant in response to both temperature and growth phase.107 This implies that additional, more complex mechanisms would be required to regulate degradosome subunit switching in B. subtilis.
RNA helicase cellular compartmentalization
RNA helicases also function in cellular compartmentalization in prokaryotic systems. In E. coli, the degradosome complex assembles through association with the non-catalytic C-terminus of RNase E,109 the N-terminal domain of which localizes the complex to the inner cellular membrane.110,111 Degradosomes are components of the bacterial cytoskeleton, associating with the membrane as helical filaments, although only RNase E is required for formation of the coiled structures.112,113 RhlB also independently assembles into coiled, cytoskeleton-like structures, in the absence of RNase E.113 It is not known how RhlB associates with the inner membrane, most likely indirectly by interaction with an unidentified membrane protein or lipid. Demonstration that RhlE, DeaD and RNase R replace their respective components in the cytoskeletal degradosome structures at low temperature would further establish the diversity of roles performed by RNA helicases in RNA degradation and cellular compartmentalization in response to temperature stress.
RNA Helicases in Translation
Role of DeaD in E. coli translation
In eukaryotes, the DEAD box helicase eIF-4A is required to initiate translation of essentially all mRNAs, removing secondary structure that inhibits Kozak scanning.114,115 Although the E. coli DeaD helicase has also been proposed to aid translation of structured mRNAs in a similar fashion,116,117 the actual function of DeaD in translation is controversial. Butland et al.79 showed that DeaD overexpression removes the inhibitory effect of secondary structure artificially introduced into the 5′ UTR of the chloramphenicol acetyltransferase gene (cat) in vivo at 25°C (Figs. 2 and 5). Derepression in this system also occurs simply in response to an increase in growth temperature, implying that DeaD is removing RNA secondary structure whose stability is enhanced by low temperature. This DeaD-induced enhancement of translation initiation potentially also contributes to stabilization of mRNAs observed in cells overexpressing DeaD118,119 and the impairment of heat shock protein expression observed in ∆deaD E. coli at low temperature.57
Figure 5. DEAD box RNA helicases function in the E. coli cold stress response. Association of the five E. coli DEAD-box RNA helicases with cellular pathways associated with cold stress is indicated. DeaD is present at significantly reduced levels at 37°C with expression induced in response to cold stress at 15°C.57 DeaD helicase activity is associated with a number of cellular pathways. RhlE, SrmB, DeaD and DbpA function in ribosome biogenesis, including 50S and 30S subunit assembly.130 Whether these helicases function in a linear sequence of reactions associated with 50S subunit assembly or RhlE dictates either an SrmB- or DeaD-specific pathway, as proposed by Jain,77 remains to be elucidated. At 37°C, RhlE is proposed to direct initial assembly followed by SrmB while at 15°C, RhlE is proposed to direct a DeaD-dependent pathway,77 possibly associated with formation of a cold-adapted ribosome. Evidence for DeaD functioning at a late stage in 30S assembly has also been presented, aiding association of S1 and thus S2 onto the ribosome.117 RhlB is an integral subunit of the RNA degradosome at 37°C, catalyzing RNA turnover. The degradosome complex assembles into two interacting cytoskeletal helices, one formed by RNase E and one by RhlB. At 15°C, DeaD and potentially RhlE can functionally replace RhlB, creating a cold-specific degradosome. DeaD also enhances translation at low temperature, potentially by removing RNA secondary structure which prevents translation initiation.79 However, this effect has been ascribed to reflect DeaD loading of S1 and S2 onto the 30S subunit late in assembly, rather than DeaD unwinding of 5′ UTR secondary structure.117 Evidence for RNA helicase function in the degradosome catalyzed turnover of sRNA-mRNA complexes, directed to the degradosome by the RNA chaperone Hfq, is circumstantial132,133 although evidence has been presented indicating that RNA helicase activity could be required under specific conditions, for example cold stress. Resch et al.122 have shown that DeaD is a cofactor required for regulation of rpoS translation by the sRNA, DsrA. At low growth temperature, rpoS translation depends upon DsrA binding which relieves the iss blocking ribosome entry to the rpoS ribosome-binding site. DeaD is proposed to unwind the iss secondary structure prior to Hfq annealing of DsrA to rpoS.122 It is also possible that DeaD RNA helicase activity extends to a requirement to unfold DsrA and/or Hfq removal once the rpoS-DsrA duplex is formed and/or the requirement for an annealing RNA helicase, such as CrhR, to anneal the mRNA-sRNA duplex before interaction with Hfq at low temperature.
The observed enhancement of DeaD on translation initiation may not be a direct effect. DeaD does not influence translation via unwinding of mRNA secondary structure in leaderless and canonical mRNAs but instead is involved in ribosome biogenesis, associated with loading of ribosomal proteins S1 and thus S2 onto the 30S subunit late in assembly.117 The effect on translation initiation may therefore not directly involve mRNA unwinding but is a consequence of the requirement for S1 and S2 loading to form an active ribosome. S1 and S2 loading on the ribosome possibly involve DeaD rearrangement of 16S rRNA secondary structure (Fig. 3).117 Thus, the exact role DeaD performs in the enhancement of translation remains unknown, as it is potentially possible that DeaD performs two independent roles involving RNA structure rearrangement of both mRNA and the 16S rRNA during translation initiation at low temperature.
Association of DeaD with the E. coli transcription-translation machinery
Butland et al.79 also showed that DeaD interacts with a number of other nucleic acid binding proteins, including SrmB, 30 and 50S ribosomal proteins, RNase R, and DNA and RNA methylases, at 25°C. Although not shown exhaustively, formation of this complex appears to be RNA dependent. These results again indicate that DeaD associates with ribosomes and they contribute to the concept that DeaD performs multiple roles in E. coli. DeaD interaction with two DNA methylases, YcbY and YhiR, potentially expand the roles to include DeaD association with the regulation of transcription or with a transcription-translation complex. Further evidence for RNA helicase association with transcription in prokaryotic systems comes from the observation that the Rhodobacter capsulatus degradosome contains two RhlB-related DEAD-box RNA helicases that associate with Rho.120 Rho functions as the transcription termination factor ring-shaped RNA-DNA helicase belonging to helicase SF5, thereby linking physical interaction between various RNA helicases and the transcriptional machinery.
DeaD is therefore associated with numerous cellular functions, however a common theme appears to involve removal of inhibitory RNA secondary structure. Further elucidation of the abiotic and biotic factors regulating DeaD association with a variety of protein cofactors and complexes and how DeaD is specifically targeted to the correct RNA substrates under different growth conditions are crucial outstanding questions.
RNA helicases in sRNA function
RNA helicases are well known players in sRNA metabolism in eukaryotic systems, having implications in cell development, viral infection and cancer.10,11,121 It is therefore likely that RNA helicases perform a role in sRNA metabolism in prokaryotes. Evidence of this activity is starting to appear.
DeaD and sRNA stimulation of translation
Resch et al.122 show that DeaD is a cofactor required for positive regulation of rpoS translation by the sRNA, DsrA. Binding of bacterial sRNAs can enhance translation by opening intramolecular inhibitory stem-loop structures (iss) that inhibit ribosome binding. At low growth temperature, rpoS translation depends on DsrA binding which relieves the iss blocking ribosome entry to the rpoS ribosome-binding site, with rpoS-DsrA duplex formation requiring the RNA chaperone, Hfq.123 The lack of rpoS-DsrA duplex formation at low growth temperature in a deaD mutant combined with evidence for Hfq-DeaD interaction suggests that DeaD is required to unwind the iss secondary structure prior to Hfq annealing of DsrA to rpoS (Fig. 5).122 It is also possible that DeaD RNA helicase activity extends to a role in DsrA unfolding and/or Hfq removal once the rpoS-DsrA duplex is formed. The ability of RNA helicases to rearrange RNA or RNP structure by performing both unwinding and annealing of sRNA-mRNA complexes indicates that additional and possibly more complex examples of a central role for RNA helicase regulation of sRNA metabolism await discovery.
RhlB requirement for sRNA-mRNA duplex degradation
In contrast to the example given above, the majority of sRNA-mRNA interactions decrease gene expression through RNA degradosome mediated degradation of the RNA duplex. In E. coli at 37°C, the sRNA-mRNA duplex is targeted to the degradosome by Hfq interaction with RNase E.124 Overexpression of RhlB prevents Hfq binding to RNase E suggesting that RNA helicase activity is not required for degradosome activity on sRNA-mRNA duplexes.125 This indicates the potential for different pathways by which specific RNAs are degraded by a degradosome containing different subunits, highlighting the versatility of the degradosome to be modified in order to respond to different conditions and/or RNA substrates. It is possible that a DeaD-associated degradosome is required for sRNA-mRNA degradation at low temperature.
Concluding Remarks
RNA helicases function in a variety of prokaryotic RNA pathways associated with abiotic stress. A common theme involves resolution of complications associated with stabilization of RNA secondary structure. The most studied stress is low temperature although responses to other stresses have also been reported. Whether this common theme limits RNA helicase association with other abiotic stress responses remains to be answered. Regulated expression and/or protein exchange of RNA helicases is involved in formation of cold-adapted ribosomes and RNA degradosomes. Similar functions for RNA helicases in transcription-translation complexes and roles in RNA processing and maturation whose efficiency is also affected by RNA secondary structure stabilization at low temperature are expected.
RNA helicases therefore contribute to functional crosstalk between helicase and protein cofactors thereby altering the composition of functional protein complexes in response to changing environmental conditions. Bacterial helicases may also function in different cellular pathways contributing to the compartmentalization of cellular function. Common themes include cold stress, 50S ribosome biogenesis, RNA turnover and membrane association.
While significant advances have been made in deciphering RNA helicase biochemistry and cellular structure/location, a model describing the mechanisms by which RNA helicase expression is regulated and the factors that target RNA helicases to different RNA substrates and protein complexes in response to changing environmental conditions is incomplete. Furthermore, while E. coli continues to be the dominant source for investigation, divergent prokaryotic species will no doubt exhibit species-specific RNA helicase function, yielding unique insights into additional roles alteration of RNA secondary structure performs in the regulation of prokaryotic cellular metabolism. Evidence that RNA helicases are prime targets for improving biotechnological applications, indicates that alteration of RNA helicase activity has implications in biotechnological, food safety and potentially other medical applications.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The work is supported by grant 171319 from the Natural Sciences and Engineering Research Council of Canada (NSERC) to G.W.O.
Glossary
Abbreviations:
- 30S
small ribosomal subunit
- 50S
large ribosomal subunit
- DEAD-box
Asp-Glu-Ala-Asp
- DEAH-box
Asp-Glu-Ala-His
- DExD-box
Asp-Glu-X-Asp
- dsRNA
double stranded RNA
- iss
intramolecular inhibitory stem-loop structures
- PNPase
polynucleotide phosphorylase
- r-protein
ribosomal protein
- RecA
recombinase A
- RNP
ribonucleoprotein
- rRNA
ribosomal RNA
- ssRNA
single stranded RNA
- S1
ribosomal protein S1
- S2
ribosomal protein S2
- sRNA
small RNA
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22638
References
- 1.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–5. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E, Fairman ME. Duplex unwinding and RNP remodeling with RNA helicases. Methods Mol Biol. 2008;488:343–55. doi: 10.1007/978-1-60327-475-3_22. [DOI] [PubMed] [Google Scholar]
- 3.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA. 2011;2:135–52. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 5.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–41. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 6.Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–45. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 7.Chen HC, Cheng SC. Functional roles of protein splicing factors. Biosci Rep. 2012;32:345–59. doi: 10.1042/BSR20120007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin R, Straub AU, Doebele C, Bohnsack MT. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 2012;9 doi: 10.4161/rna.21879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owttrim GW. RNA helicases and abiotic stress. Nucleic Acids Res. 2006;34:3220–30. doi: 10.1093/nar/gkl408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambrus AM, Frolov MV. The diverse roles of RNA helicases in RNAi. Cell Cycle. 2009;8:3500–5. doi: 10.4161/cc.8.21.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linder P, Owttrim GW. Plant RNA helicases: linking aberrant and silencing RNA. Trends Plant Sci. 2009;14:344–52. doi: 10.1016/j.tplants.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Fuller-Pace FV, Moore HC. RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol. 2011;7:239–51. doi: 10.2217/fon.11.1. [DOI] [PubMed] [Google Scholar]
- 13.Steimer L, Klostermeier D. RNA helicases in infection and disease. RNA Biol. 2012;9:751–71. doi: 10.4161/rna.20090. [DOI] [PubMed] [Google Scholar]
- 14.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol. 2007;17:316–24. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–9. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 17.Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, et al. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19:5363–72. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Westermarck J, Weiss C, Saffrich R, Kast J, Musti AM, Wessely M, et al. The DEXD/H-box RNA helicase RHII/Gu is a co-factor for c-Jun-activated transcription. EMBO J. 2002;21:451–60. doi: 10.1093/emboj/21.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walstrom KM, Schmidt D, Bean CJ, Kelly WG. RNA helicase A is important for germline transcriptional control, proliferation, and meiosis in C. elegans. Mech Dev. 2005;122:707–20. doi: 10.1016/j.mod.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–15. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem. 2010;148:381–92. doi: 10.1093/jb/mvq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rössler OG, Straka A, Stahl H. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 2001;29:2088–96. doi: 10.1093/nar/29.10.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chamot D, Colvin KR, Kujat-Choy SL, Owttrim GW. RNA structural rearrangement via unwinding and annealing by the cyanobacterial RNA helicase, CrhR. J Biol Chem. 2005;280:2036–44. doi: 10.1074/jbc.M409700200. [DOI] [PubMed] [Google Scholar]
- 25.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 26.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–6. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 27.Kelley BP, Sharan R, Karp RM, Sittler T, Root DE, Stockwell BR, et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc Natl Acad Sci U S A. 2003;100:11394–9. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol. 2003;48:1253–65. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 29.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–9. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandiani F, Chamot S, Brillard J, Carlin F, Nguyen-the C, Broussolle V. Role of the five RNA helicases in the adaptive response of Bacillus cereus ATCC 14579 cells to temperature, pH, and oxidative stresses. Appl Environ Microbiol. 2011;77:5604–9. doi: 10.1128/AEM.02974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce A, Gillette D, Jones PG. Escherichia coli cold shock protein CsdA effects an increase in septation and the resultant formation of coccobacilli at low temperature. Arch Microbiol. 2011;193:373–84. doi: 10.1007/s00203-011-0682-0. [DOI] [PubMed] [Google Scholar]
- 32.Rosana AR, Ventakesh M, Chamot D, Patterson-Fortin LM, Tarassova O, Espie GS, et al. Inactivation of a low temperature-induced RNA helicase in Synechocystis sp. PCC 6803: physiological and morphological consequences. Plant Cell Physiol. 2012;53:646–58. doi: 10.1093/pcp/pcs020. [DOI] [PubMed] [Google Scholar]
- 33.Tamura M, Kers JA, Cohen SN. Second-site suppression of RNase E essentiality by mutation of the deaD RNA helicase in Escherichia coli. J Bacteriol. 2012;194:1919–26. doi: 10.1128/JB.06652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schade B, Jansen G, Whiteway M, Entian KD, Thomas DY. Cold adaptation in budding yeast. Mol Biol Cell. 2004;15:5492–502. doi: 10.1091/mbc.E04-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–80. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.López-Ramírez V, Alcaraz LD, Moreno-Hagelsieb G, Olmedo-Álvarez G. Phylogenetic distribution and evolutionary history of bacterial DEAD-Box proteins. J Mol Evol. 2011;72:413–31. doi: 10.1007/s00239-011-9441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chamot D, Magee WC, Yu E, Owttrim GW. A cold shock-induced cyanobacterial RNA helicase. J Bacteriol. 1999;181:1728–32. doi: 10.1128/jb.181.6.1728-1732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chamot D, Owttrim GW. Regulation of cold shock-induced RNA helicase gene expression in the Cyanobacterium anabaena sp. strain PCC 7120. J Bacteriol. 2000;182:1251–6. doi: 10.1128/JB.182.5.1251-1256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu E, Owttrim GW. Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res. 2000;28:3926–34. doi: 10.1093/nar/28.20.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Fahmawi B, Owttrim GW. Polar-biased localization of the cold stress-induced RNA helicase, CrhC, in the Cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol. 2003;50:1439–48. doi: 10.1046/j.1365-2958.2003.03783.x. [DOI] [PubMed] [Google Scholar]
- 41.Kujat SL, Owttrim GW. Redox-regulated RNA helicase expression. Plant Physiol. 2000;124:703–14. doi: 10.1104/pp.124.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinnemeier J, Hagemann M. Identification of salt-regulated genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 by subtractive RNA hybridization. Arch Microbiol. 1999;172:377–86. doi: 10.1007/s002030050774. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki I, Los DA, Kanesaki Y, Mikami K, Murata N. The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 2000;19:1327–34. doi: 10.1093/emboj/19.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prakash JS, Krishna PS, Sirisha K, Kanesaki Y, Suzuki I, Shivaji S, et al. An RNA helicase, CrhR, regulates the low-temperature-inducible expression of heat-shock genes groES, groEL1 and groEL2 in Synechocystis sp. PCC 6803. Microbiology. 2010;156:442–51. doi: 10.1099/mic.0.031823-0. [DOI] [PubMed] [Google Scholar]
- 45.Sireesha K, Radharani B, Krishna PS, Sreedhar N, Subramanyam R, Mohanty P, et al. RNA helicase, CrhR is indispensable for the energy redistribution and the regulation of photosystem stoichiometry at low temperature in Synechocystis sp. PCC6803. Biochim Biophys Acta. 2012;1817:1525–36. doi: 10.1016/j.bbabio.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 46.Rowland JG, Simon WJ, Prakash JS, Slabas AR. Proteomics reveals a role for the RNA helicase crhR in the modulation of multiple metabolic pathways during cold acclimation of Synechocystis sp. PCC6803. J Proteome Res. 2011;10:3674–89. doi: 10.1021/pr200299t. [DOI] [PubMed] [Google Scholar]
- 47.Markkula A, Mattila M, Lindström M, Korkeala H. Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3°C. Environ Microbiol. 2012;14:2223–32. doi: 10.1111/j.1462-2920.2012.02761.x. A. [DOI] [PubMed] [Google Scholar]
- 48.Markkula A, Lindström M, Johansson P, Björkroth J, Korkeala H. Role of four putative DEAD-box RNA helicase genes in growth of Listeria monocytogenes EGD-e under heat, pH, osmotic, ethanol, and oxidative stresses. Appl Environ Microbiol. 2012;78:6875–82. doi: 10.1128/AEM.01526-12. B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe T, Kojima H, Fukui M. Draft genome sequence of a psychrotolerant sulfur-oxidizing bacterium, Sulfuricella denitrificans skB26, and proteomic insights into cold adaptation. Appl Environ Microbiol. 2012;78:6545–9. doi: 10.1128/AEM.01349-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim J, Thomas T, Cavicchioli R. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon, Methanococcoides burtonii. J Mol Biol. 2000;297:553–67. doi: 10.1006/jmbi.2000.3585. [DOI] [PubMed] [Google Scholar]
- 51.Boonyaratanakornkit BB, Simpson AJ, Whitehead TA, Fraser CM, El-Sayed NM, Clark DS. Transcriptional profiling of the hyperthermophilic methanarchaeon Methanococcus jannaschii in response to lethal heat and non-lethal cold shock. Environ Microbiol. 2005;7:789–97. doi: 10.1111/j.1462-2920.2005.00751.x. [DOI] [PubMed] [Google Scholar]
- 52.Shimada Y, Fukuda W, Akada Y, Ishida M, Nakayama J, Imanaka T, et al. Property of cold inducible DEAD-box RNA helicase in hyperthermophilic archaea. Biochem Biophys Res Commun. 2009;389:622–7. doi: 10.1016/j.bbrc.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 53.Elles LM, Uhlenbeck OC. Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucleic Acids Res. 2008;36:41–50. doi: 10.1093/nar/gkm926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jagessar KL, Jain C. Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA. 2010;16:1386–92. doi: 10.1261/rna.2015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Xiao X, Sun P, Wang F. Screening of genes regulated by cold shock in Shewanella piezotolerans WP3 and time course expression of cold-regulated genes. Arch Microbiol. 2008;189:549–56. doi: 10.1007/s00203-007-0347-1. [DOI] [PubMed] [Google Scholar]
- 56.Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J Bacteriol. 2006;188:240–8. doi: 10.1128/JB.188.1.240-248.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones PG, Mitta M, Kim Y, Jiang W, Inouye M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toone WM, Rudd KE, Friesen JD. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173:3291–302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamanaka K, Inouye M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J Bacteriol. 2001;183:2808–16. doi: 10.1128/JB.183.9.2808-2816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukhopadhyay A, He Z, Alm EJ, Arkin AP, Baidoo EE, Borglin SC, et al. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J Bacteriol. 2006;188:4068–78. doi: 10.1128/JB.01921-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lambert D, Draper DE. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J Mol Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Briolat V, Reysset G. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J Bacteriol. 2002;184:2333–43. doi: 10.1128/JB.184.9.2333-2343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minty JJ, Lesnefsky AA, Lin F, Chen Y, Zaroff TA, Veloso AB, et al. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb Cell Fact. 2011;10:18. doi: 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joe MH, Jung SW, Im SH, Lim SY, Song HP, Kwon O, et al. Genome-wide response of Deinococcus radiodurans on cadmium toxicity. J Microbiol Biotechnol. 2011;21:438–47. [PubMed] [Google Scholar]
- 65.Palonen E, Lindström M, Somervuo P, Johansson P, Björkroth J, Korkeala H. Requirement for RNA helicase CsdA for growth of Yersinia pseudotuberculosis IP32953 at low temperatures. Appl Environ Microbiol. 2012;78:1298–301. doi: 10.1128/AEM.07278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babu MM, Sridhar J, Gunasekaran P. Global transcriptome analysis of Bacillus cereus ATCC 14579 in response to silver nitrate stress. J Nanobiotechnology. 2011;9:49. doi: 10.1186/1477-3155-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wen J, Deng X, Li Z, Dudley EG, Anantheswaran RC, Knabel SJ, et al. Transcriptomic response of Listeria monocytogenes during the transition to the long-term-survival phase. Appl Environ Microbiol. 2011;77:5966–72. doi: 10.1128/AEM.00596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bae D, Liu C, Zhang T, Jones M, Peterson SN, Wang C. Global gene expression of Listeria monocytogenes to salt stress. J Food Prot. 2012;75:906–12. doi: 10.4315/0362-028X.JFP-11-282. [DOI] [PubMed] [Google Scholar]
- 69.Nicholson WL, Moeller R, Horneck G, PROTECT Team Transcriptomic responses of germinating Bacillus subtilis spores exposed to 1.5 years of space and simulated martian conditions on the EXPOSE-E experiment PROTECT. Astrobiology. 2012;12:469–86. doi: 10.1089/ast.2011.0748. [DOI] [PubMed] [Google Scholar]
- 70.Sato N. A family of cold-regulated RNA-binding protein genes in the cyanobacterium Anabaena variabilis M3. Nucleic Acids Res. 1995;23:2161–7. doi: 10.1093/nar/23.12.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.El-Sharoud WM, Graumann PL. Cold shock proteins aid coupling of transcription and translation in bacteria. Sci Prog. 2007;90:15–27. doi: 10.3184/003685007780440549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mori S, Castoreno A, Mulligan ME, Lammers PJ. Nitrogen status modulates the expression of RNA-binding proteins in cyanobacteria. FEMS Microbiol Lett. 2003;227:203–10. doi: 10.1016/S0378-1097(03)00682-7. [DOI] [PubMed] [Google Scholar]
- 73.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 74.Rawling DC, Baserga SJ. In vivo approaches to dissecting the function of RNA helicases in eukaryotic ribosome assembly. Methods Enzymol. 2012;511:289–321. doi: 10.1016/B978-0-12-396546-2.00014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kalman M, Murphy H, Cashel M. rhlB, a new Escherichia coli K-12 gene with an RNA helicase-like protein sequence motif, one of at least five such possible genes in a prokaryote. New Biol. 1991;3:886–95. [PubMed] [Google Scholar]
- 76.Peil L, Virumäe K, Remme J. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 2008;275:3772–82. doi: 10.1111/j.1742-4658.2008.06523.x. [DOI] [PubMed] [Google Scholar]
- 77.Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA. 2008;14:381–9. doi: 10.1261/rna.800308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Srivastava AK, Schlessinger D. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc Natl Acad Sci U S A. 1988;85:7144–8. doi: 10.1073/pnas.85.19.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Butland G, Krogan NJ, Xu J, Yang WH, Aoki H, Li JS, et al. Investigating the in vivo activity of the DeaD protein using protein-protein interactions and the translational activity of structured chloramphenicol acetyltransferase mRNAs. J Cell Biochem. 2007;100:642–52. doi: 10.1002/jcb.21016. [DOI] [PubMed] [Google Scholar]
- 80.Trubetskoy D, Proux F, Allemand F, Dreyfus M, Iost I. SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo. Nucleic Acids Res. 2009;37:6540–9. doi: 10.1093/nar/gkp685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Proux F, Dreyfus M, Iost I. Identification of the sites of action of SrmB, a DEAD-box RNA helicase involved in Escherichia coli ribosome assembly. Mol Microbiol. 2011;82:300–11. doi: 10.1111/j.1365-2958.2011.07779.x. [DOI] [PubMed] [Google Scholar]
- 82.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–, 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iost I, Dreyfus M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006;34:4189–97. doi: 10.1093/nar/gkl500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–26. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nicol SM, Fuller-Pace FV. The “DEAD box” protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc Natl Acad Sci U S A. 1995;92:11681–5. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 2009;37:6503–14. doi: 10.1093/nar/gkp711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kossen K, Uhlenbeck OC. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 1999;27:3811–20. doi: 10.1093/nar/27.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Netterling S, Vaitkevicius K, Nord S, Johansson J. A Listeria monocytogenes RNA helicase essential for growth and ribosomal maturation at low temperatures uses its C terminus for appropriate interaction with the ribosome. J Bacteriol. 2012;194:4377–85. doi: 10.1128/JB.00348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci U S A. 1996;93:3865–9. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–72. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 91.Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999;13:2594–603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prud’homme-Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol Microbiol. 2004;54:1409–21. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 94.Regonesi ME, Del Favero M, Basilico F, Briani F, Benazzi L, Tortora P, et al. Analysis of the Escherichia coli RNA degradosome composition by a proteomic approach. Biochimie. 2006;88:151–61. doi: 10.1016/j.biochi.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 95.Beran RK, Simons RW. Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol Microbiol. 2001;39:112–25. doi: 10.1046/j.1365-2958.2001.02216.x. [DOI] [PubMed] [Google Scholar]
- 96.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–66. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao J, Lee K, Zhao M, Qiu J, Zhan X, Saxena A, et al. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol Microbiol. 2006;61:394–406. doi: 10.1111/j.1365-2958.2006.05246.x. [DOI] [PubMed] [Google Scholar]
- 98.Awano N, Xu C, Ke H, Inoue K, Inouye M, Phadtare S. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J Bacteriol. 2007;189:5808–15. doi: 10.1128/JB.00655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–66. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 100.Cairrão F, Cruz A, Mori H, Arraiano CM. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol Microbiol. 2003;50:1349–60. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- 101.Bergholz PW, Bakermans C, Tiedje JM. Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J Bacteriol. 2009;191:2340–52. doi: 10.1128/JB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Purusharth RI, Klein F, Sulthana S, Jäger S, Jagannadham MV, Evguenieva-Hackenberg E, et al. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J Biol Chem. 2005;280:14572–8. doi: 10.1074/jbc.M413507200. [DOI] [PubMed] [Google Scholar]
- 103.Roux CM, DeMuth JP, Dunman PM. Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J Bacteriol. 2011;193:5520–6. doi: 10.1128/JB.05485-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Newman JA, Hewitt L, Rodrigues C, Solovyova AS, Harwood CR, Lewis RJ. Dissection of the network of interactions that links RNA processing with glycolysis in the Bacillus subtilis degradosome. J Mol Biol. 2012;416:121–36. doi: 10.1016/j.jmb.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 105.Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, et al. RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol. 2011;193:5431–41. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deikus G, Condon C, Bechhofer DH. Role of Bacillus subtilis RNase J1 endonuclease and 5′-exonuclease activities in trp leader RNA turnover. J Biol Chem. 2008;283:17158–67. doi: 10.1074/jbc.M801461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lehnik-Habrink M, Pförtner H, Rempeters L, Pietack N, Herzberg C, Stülke J. The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol Microbiol. 2010;77:958–71. doi: 10.1111/j.1365-2958.2010.07264.x. [DOI] [PubMed] [Google Scholar]
- 108.Beckering CL, Steil L, Weber MH, Völker U, Marahiel MA. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J Bacteriol. 2002;184:6395–402. doi: 10.1128/JB.184.22.6395-6402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, et al. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–81. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miczak A, Srivastava RA, Apirion D. Location of the RNA-processing enzymes RNase III, RNase E and RNase P in the Escherichia coli cell. Mol Microbiol. 1991;5:1801–10. doi: 10.1111/j.1365-2958.1991.tb01929.x. [DOI] [PubMed] [Google Scholar]
- 111.Liou GG, Jane WN, Cohen SN, Lin NS, Lin-Chao S. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc Natl Acad Sci U S A. 2001;98:63–8. doi: 10.1073/pnas.98.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Taghbalout A, Rothfield L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc Natl Acad Sci U S A. 2007;104:1667–72. doi: 10.1073/pnas.0610491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taghbalout A, Rothfield L. RNaseE and RNA helicase B play central roles in the cytoskeletal organization of the RNA degradosome. J Biol Chem. 2008;283:13850–5. doi: 10.1074/jbc.M709118200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 115.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–94. doi: 10.1017/S135583820100108X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu J, Aoki H, Ganoza MC. Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. Int J Biochem Cell Biol. 1999;31:215–29. doi: 10.1016/S1357-2725(98)00142-3. [DOI] [PubMed] [Google Scholar]
- 117.Moll I, Grill S, Gründling A, Bläsi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol Microbiol. 2002;44:1387–96. doi: 10.1046/j.1365-2958.2002.02971.x. [DOI] [PubMed] [Google Scholar]
- 118.Iost I, Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994;372:193–6. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- 119.Brandi A, Spurio R, Gualerzi CO, Pon CL. Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under non-stress conditions. EMBO J. 1999;18:1653–9. doi: 10.1093/emboj/18.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jäger S, Fuhrmann O, Heck C, Hebermehl M, Schiltz E, Rauhut R, et al. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 2001;29:4581–8. doi: 10.1093/nar/29.22.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, et al. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–41. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Resch A, Većerek B, Palavra K, Bläsi U. Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol. 2010;7:796–802. doi: 10.4161/rna.7.6.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–86. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol. 2011;79:419–32. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 126.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–14. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Henn A, Bradley MJ, De La Cruz EM. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu Rev Biophys. 2012;41:247–67. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Awano N, Rajagopal V, Arbing M, Patel S, Hunt J, Inouye M, et al. Escherichia coli RNase R has dual activities, helicase and RNase. J Bacteriol. 2010;192:1344–52. doi: 10.1128/JB.01368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–26. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 131.Burger A, Whiteley C, Boshoff A. Current perspectives of the Escherichia coli RNA degradosome. Biotechnol Lett. 2011;33:2337–50. doi: 10.1007/s10529-011-0713-6. [DOI] [PubMed] [Google Scholar]
- 132.Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005;19:328–38. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–96. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Broussolle V, Pandiani F, Haddad N, Michaud C, Carlin F, Nguyen-the C, et al. Insertional mutagenesis reveals genes involved in Bacillus cereus ATCC 14579 growth at low temperature. FEMS Microbiol Lett. 2010;306:177–83. doi: 10.1111/j.1574-6968.2010.01953.x. [DOI] [PubMed] [Google Scholar]
- 135.Pandiani F, Brillard J, Bornard I, Michaud C, Chamot S, Nguyen-the C, et al. Differential involvement of the five RNA helicases in adaptation of Bacillus cereus ATCC 14579 to low growth temperatures. Appl Environ Microbiol. 2010;76:6692–7. doi: 10.1128/AEM.00782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hunt A, Rawlins JP, Thomaides HB, Errington J. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology. 2006;152:2895–907. doi: 10.1099/mic.0.29152-0. [DOI] [PubMed] [Google Scholar]
- 137.Chan YC, Raengpradub S, Boor KJ, Wiedmann M. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl Environ Microbiol. 2007;73:6484–98. doi: 10.1128/AEM.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mazzon RR, Lang EA, Braz VS, Marques MV. Characterization of Caulobacter crescentus response to low temperature and identification of genes involved in freezing resistance. FEMS Microbiol Lett. 2008;288:178–85. doi: 10.1111/j.1574-6968.2008.01337.x. [DOI] [PubMed] [Google Scholar]
- 139.Remme J. How to avoid undesirable interactions during ribosome assembly? Mol Microbiol. 2011;82:269–71. doi: 10.1111/j.1365-2958.2011.07838.x. [DOI] [PubMed] [Google Scholar]
- 140.Hardwick SW, Chan VS, Broadhurst RW, Luisi BF. An RNA degradosome assembly in Caulobacter crescentus. Nucleic Acids Res. 2011;39:1449–59. doi: 10.1093/nar/gkq928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]