Abstract
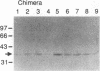
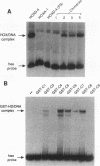
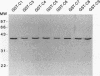
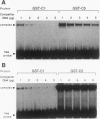
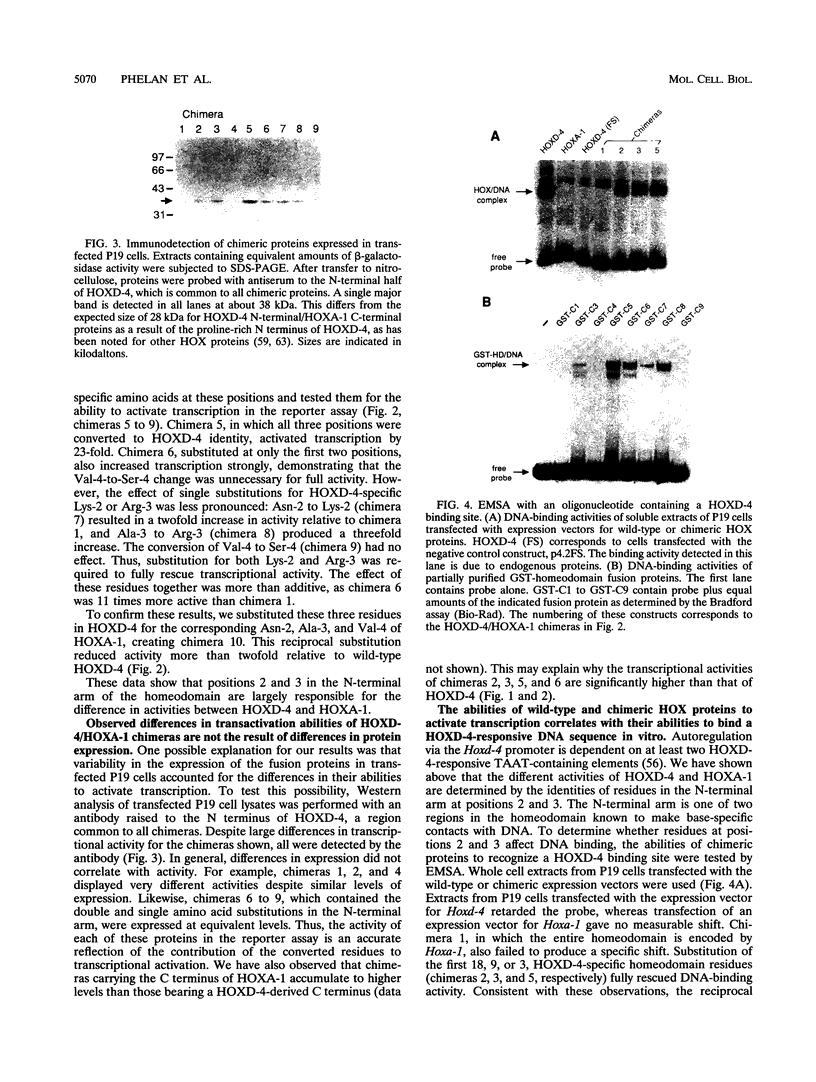
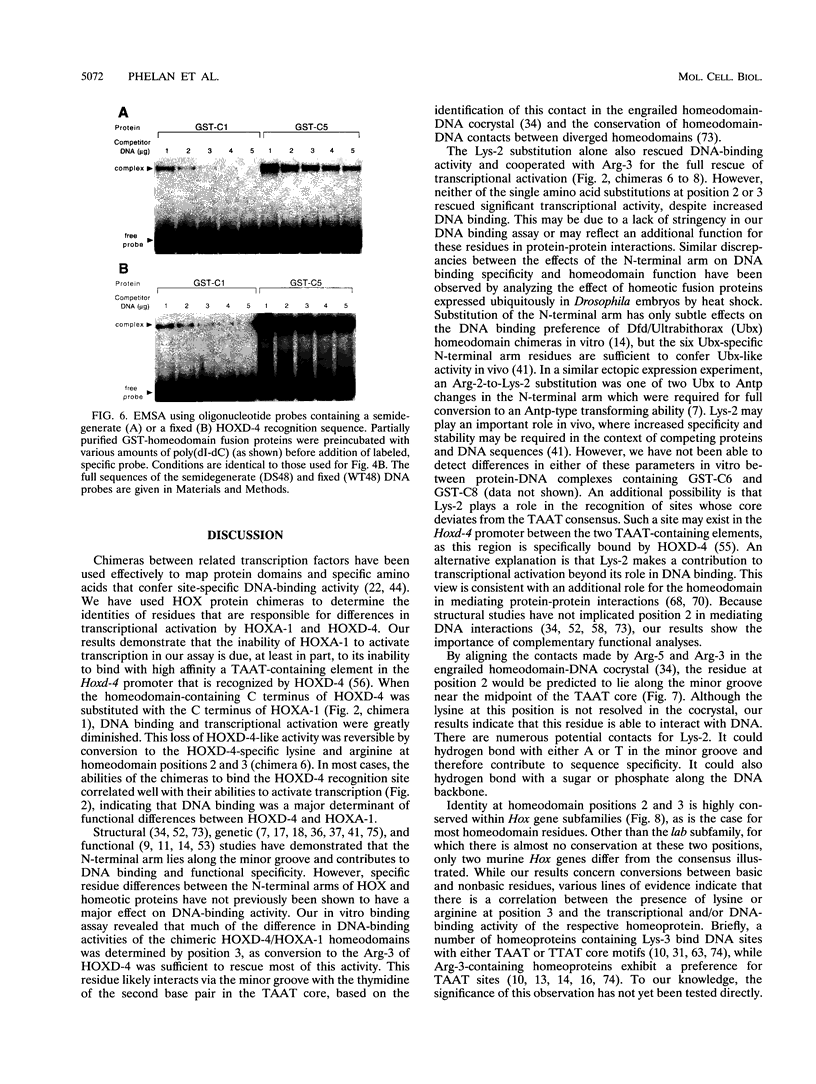
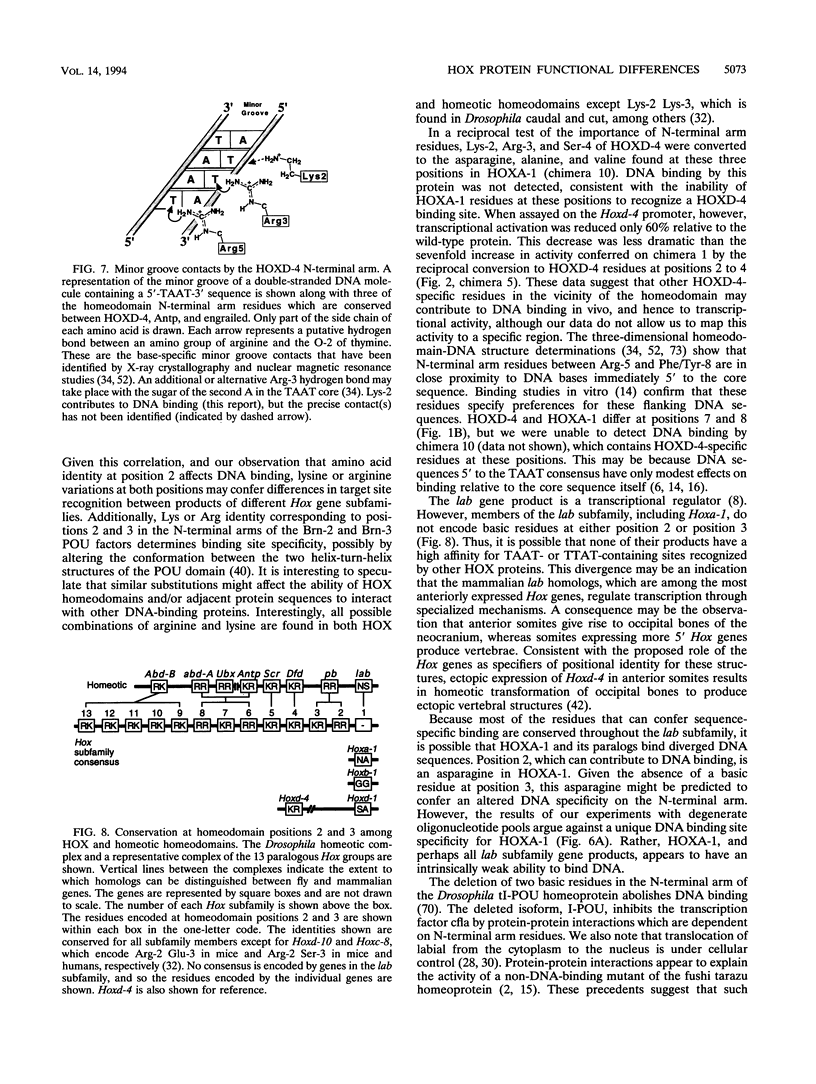
Hox genes encode homeodomain-containing transcriptional regulators that function during development to specify positional identity along embryonic axes. The homeodomain is composed of a flexible N-terminal arm and three alpha helices, and it differentially binds DNA. A number of homeodomains recognize sites containing a TAAT core motif. The product of the murine Hoxd-4 (Hox-4.2) gene functions in a positive autoregulatory fashion in P19 cells that is dependent on two TAAT motifs in the Hoxd-4 promoter. This effect is specific in that murine HOXA-1 (HOX-1.6) is unable to activate transcription through the Hoxd-4 autoregulatory element. Here we show that this is due to an inability of the HOXA-1 homeodomain to bind a HOXD-4 recognition site effectively. We have produced chimeras between HOXD-4 and HOXA-1 to map specific residues responsible for this functional difference. When positions 2 and 3 in the N-terminal arm of HOXA-1 were converted to HOXD-4 identity, both strong DNA binding and transcriptional activation were rescued. This substitution appears to confer an increased DNA-binding ability on the HOXA-1 homeodomain, since we were unable to detect a high-affinity recognition sequence for HOXA-1 in a randomized pool of DNA probes. The contribution of position 3 to DNA binding has been implicated by structural studies, but this is the first report of the importance of position 2 in regulating homeodomain-DNA interactions. Additionally, specific homeodomain residues that confer major differences in DNA binding and transcriptional activation between Hox gene products have not been previously determined. Identity at these two positions is generally conserved among paralogs but varies between Hox gene subfamilies. As a result, these residues may be important for the regulation of target gene expression by specific Hox products.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adra C. N., Boer P. H., McBurney M. W. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60(1):65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- Ananthan J., Baler R., Morrissey D., Zuo J., Lan Y., Weir M., Voellmy R. Synergistic activation of transcription is mediated by the N-terminal domain of Drosophila fushi tarazu homeoprotein and can occur without DNA binding by the protein. Mol Cell Biol. 1993 Mar;13(3):1599–1609. doi: 10.1128/mcb.13.3.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcioni L., Simeone A., Guazzi S., Zappavigna V., Boncinelli E., Mavilio F. The upstream region of the human homeobox gene HOX3D is a target for regulation by retinoic acid and HOX homeoproteins. EMBO J. 1992 Jan;11(1):265–277. doi: 10.1002/j.1460-2075.1992.tb05049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron A., Featherstone M. S., Hill R. E., Hall A., Galliot B., Duboule D. Hox-1.6: a mouse homeo-box-containing gene member of the Hox-1 complex. EMBO J. 1987 Oct;6(10):2977–2986. doi: 10.1002/j.1460-2075.1987.tb02603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Catron K. M., Iler N., Abate C. Nucleotides flanking a conserved TAAT core dictate the DNA binding specificity of three murine homeodomain proteins. Mol Cell Biol. 1993 Apr;13(4):2354–2365. doi: 10.1128/mcb.13.4.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. K., Mann R. S. The segment identity functions of Ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev. 1993 May;7(5):796–811. doi: 10.1101/gad.7.5.796. [DOI] [PubMed] [Google Scholar]
- Chouinard S., Kaufman T. C. Control of expression of the homeotic labial (lab) locus of Drosophila melanogaster: evidence for both positive and negative autogenous regulation. Development. 1991 Dec;113(4):1267–1280. doi: 10.1242/dev.113.4.1267. [DOI] [PubMed] [Google Scholar]
- Damante G., Di Lauro R. Several regions of Antennapedia and thyroid transcription factor 1 homeodomains contribute to DNA binding specificity. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5388–5392. doi: 10.1073/pnas.88.12.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearolf C. R., Topol J., Parker C. S. The caudal gene product is a direct activator of fushi tarazu transcription during Drosophila embryogenesis. Nature. 1989 Sep 28;341(6240):340–343. doi: 10.1038/341340a0. [DOI] [PubMed] [Google Scholar]
- Dessain S., Gross C. T., Kuziora M. A., McGinnis W. Antp-type homeodomains have distinct DNA binding specificities that correlate with their different regulatory functions in embryos. EMBO J. 1992 Mar;11(3):991–1002. doi: 10.1002/j.1460-2075.1992.tb05138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D., Dollé P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989 May;8(5):1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker S. C., Young K. E., von Kessler D. P., Beachy P. A. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 1991 May;10(5):1179–1186. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker S. C., von Kessler D. P., Beachy P. A. Differential DNA sequence recognition is a determinant of specificity in homeotic gene action. EMBO J. 1992 Nov;11(11):4059–4072. doi: 10.1002/j.1460-2075.1992.tb05499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick V. D., Percival-Smith A., Ingles C. J., Krause H. M. Homeodomain-independent activity of the fushi tarazu polypeptide in Drosophila embryos. Nature. 1992 Apr 16;356(6370):610–612. doi: 10.1038/356610a0. [DOI] [PubMed] [Google Scholar]
- Florence B., Handrow R., Laughon A. DNA-binding specificity of the fushi tarazu homeodomain. Mol Cell Biol. 1991 Jul;11(7):3613–3623. doi: 10.1128/mcb.11.7.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukubo-Tokunaga K., Flister S., Gehring W. J. Functional specificity of the Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6360–6364. doi: 10.1073/pnas.90.13.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukubo-Tokunaga K., Müller M., Affolter M., Pick L., Kloter U., Gehring W. J. In vivo analysis of the helix-turn-helix motif of the fushi tarazu homeo domain of Drosophila melanogaster. Genes Dev. 1992 Jun;6(6):1082–1096. doi: 10.1101/gad.6.6.1082. [DOI] [PubMed] [Google Scholar]
- Gould A. P., White R. A. Connectin, a target of homeotic gene control in Drosophila. Development. 1992 Dec;116(4):1163–1174. doi: 10.1242/dev.116.4.1163. [DOI] [PubMed] [Google Scholar]
- Graba Y., Aragnol D., Laurenti P., Garzino V., Charmot D., Berenger H., Pradel J. Homeotic control in Drosophila; the scabrous gene is an in vivo target of Ultrabithorax proteins. EMBO J. 1992 Sep;11(9):3375–3384. doi: 10.1002/j.1460-2075.1992.tb05416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A., Papalopulu N., Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989 May 5;57(3):367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Green S., Chambon P. Oestradiol induction of a glucocorticoid-responsive gene by a chimaeric receptor. Nature. 1987 Jan 1;325(6099):75–78. doi: 10.1038/325075a0. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. A genetic model for interaction of the homeodomain recognition helix with DNA. Science. 1991 Jan 25;251(4992):426–430. doi: 10.1126/science.1671176. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989 Jun 30;57(7):1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Scott M. P. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990 Nov 30;63(5):883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- Hoey T., Warrior R., Manak J., Levine M. DNA-binding activities of the Drosophila melanogaster even-skipped protein are mediated by its homeo domain and influenced by protein context. Mol Cell Biol. 1988 Nov;8(11):4598–4607. doi: 10.1128/mcb.8.11.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. W., Hogan B. L. Expression of homeo box genes during mouse development: a review. Genes Dev. 1988 Jul;2(7):773–782. doi: 10.1101/gad.2.7.773. [DOI] [PubMed] [Google Scholar]
- Hoppler S., Bienz M. Specification of a single cell type by a Drosophila homeotic gene. Cell. 1994 Feb 25;76(4):689–702. doi: 10.1016/0092-8674(94)90508-8. [DOI] [PubMed] [Google Scholar]
- Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989 Apr 15;77(1):61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Immerglück K., Lawrence P. A., Bienz M. Induction across germ layers in Drosophila mediated by a genetic cascade. Cell. 1990 Jul 27;62(2):261–268. doi: 10.1016/0092-8674(90)90364-k. [DOI] [PubMed] [Google Scholar]
- Jones F. S., Prediger E. A., Bittner D. A., De Robertis E. M., Edelman G. M. Cell adhesion molecules as targets for Hox genes: neural cell adhesion molecule promoter activity is modulated by cotransfection with Hox-2.5 and -2.4. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2086–2090. doi: 10.1073/pnas.89.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappen C., Schughart K., Ruddle F. H. Early evolutionary origin of major homeodomain sequence classes. Genomics. 1993 Oct;18(1):54–70. doi: 10.1006/geno.1993.1426. [DOI] [PubMed] [Google Scholar]
- Kessel M., Balling R., Gruss P. Variations of cervical vertebrae after expression of a Hox-1.1 transgene in mice. Cell. 1990 Apr 20;61(2):301–308. doi: 10.1016/0092-8674(90)90810-2. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kuziora M. A., McGinnis W. A homeodomain substitution changes the regulatory specificity of the deformed protein in Drosophila embryos. Cell. 1989 Nov 3;59(3):563–571. doi: 10.1016/0092-8674(89)90039-1. [DOI] [PubMed] [Google Scholar]
- Kuziora M. A., McGinnis W. Altering the regulatory targets of the Deformed protein in Drosophila embryos by substituting the Abdominal-B homeodomain. Mech Dev. 1990 Dec;33(1):83–93. doi: 10.1016/0925-4773(90)90137-b. [DOI] [PubMed] [Google Scholar]
- LaRosa G. J., Gudas L. J. Early retinoic acid-induced F9 teratocarcinoma stem cell gene ERA-1: alternate splicing creates transcripts for a homeobox-containing protein and one lacking the homeobox. Mol Cell Biol. 1988 Sep;8(9):3906–3917. doi: 10.1128/mcb.8.9.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mouellic H., Lallemand Y., Brûlet P. Homeosis in the mouse induced by a null mutation in the Hox-3.1 gene. Cell. 1992 Apr 17;69(2):251–264. doi: 10.1016/0092-8674(92)90406-3. [DOI] [PubMed] [Google Scholar]
- Li P., He X., Gerrero M. R., Mok M., Aggarwal A., Rosenfeld M. G. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes Dev. 1993 Dec;7(12B):2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- Lin L., McGinnis W. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev. 1992 Jun;6(6):1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- Lufkin T., Dierich A., LeMeur M., Mark M., Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991 Sep 20;66(6):1105–1119. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- Lufkin T., Mark M., Hart C. P., Dollé P., LeMeur M., Chambon P. Homeotic transformation of the occipital bones of the skull by ectopic expression of a homeobox gene. Nature. 1992 Oct 29;359(6398):835–841. doi: 10.1038/359835a0. [DOI] [PubMed] [Google Scholar]
- Mader S., Kumar V., de Verneuil H., Chambon P. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature. 1989 Mar 16;338(6212):271–274. doi: 10.1038/338271a0. [DOI] [PubMed] [Google Scholar]
- Malicki J., Schughart K., McGinnis W. Mouse Hox-2.2 specifies thoracic segmental identity in Drosophila embryos and larvae. Cell. 1990 Nov 30;63(5):961–967. doi: 10.1016/0092-8674(90)90499-5. [DOI] [PubMed] [Google Scholar]
- McGinnis N., Kuziora M. A., McGinnis W. Human Hox-4.2 and Drosophila deformed encode similar regulatory specificities in Drosophila embryos and larvae. Cell. 1990 Nov 30;63(5):969–976. doi: 10.1016/0092-8674(90)90500-e. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Garber R. L., Wirz J., Kuroiwa A., Gehring W. J. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984 Jun;37(2):403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. Homeobox genes and axial patterning. Cell. 1992 Jan 24;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Mihara H., Kaiser E. T. A chemically synthesized Antennapedia homeo domain binds to a specific DNA sequence. Science. 1988 Nov 11;242(4880):925–927. doi: 10.1126/science.2903553. [DOI] [PubMed] [Google Scholar]
- Murphy P., Hill R. E. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991 Jan;111(1):61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- Müller M., Affolter M., Leupin W., Otting G., Wüthrich K., Gehring W. J. Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J. 1988 Dec 20;7(13):4299–4304. doi: 10.1002/j.1460-2075.1988.tb03328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Müller M., Affolter M., Gehring W. J., Wüthrich K. Protein--DNA contacts in the structure of a homeodomain--DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990 Oct;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith A., Müller M., Affolter M., Gehring W. J. The interaction with DNA of wild-type and mutant fushi tarazu homeodomains. EMBO J. 1990 Dec;9(12):3967–3974. doi: 10.1002/j.1460-2075.1990.tb07617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri K. G., Gervais F. G., Weil R., Davidson D., Gish G. D., Veillette A. Interactions of the SH2 domain of lymphocyte-specific tyrosine protein kinase p56lck with phosphotyrosine-containing proteins. Oncogene. 1993 Oct;8(10):2765–2772. [PubMed] [Google Scholar]
- Pöpperl H., Featherstone M. S. An autoregulatory element of the murine Hox-4.2 gene. EMBO J. 1992 Oct;11(10):3673–3680. doi: 10.1002/j.1460-2075.1992.tb05452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöpperl H., Featherstone M. S. Identification of a retinoic acid response element upstream of the murine Hox-4.2 gene. Mol Cell Biol. 1993 Jan;13(1):257–265. doi: 10.1128/mcb.13.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y. Q., Otting G., Furukubo-Tokunaga K., Affolter M., Gehring W. J., Wüthrich K. NMR structure determination reveals that the homeodomain is connected through a flexible linker to the main body in the Drosophila Antennapedia protein. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10738–10742. doi: 10.1073/pnas.89.22.10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaldi I., Kovàcs E. N., Featherstone M. S. A proline-rich transcriptional activation domain in murine HOXD-4 (HOX-4.2). Nucleic Acids Res. 1994 Feb 11;22(3):376–382. doi: 10.1093/nar/22.3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Solis R., Zheng H., Whiting J., Krumlauf R., Bradley A. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell. 1993 Apr 23;73(2):279–294. doi: 10.1016/0092-8674(93)90229-j. [DOI] [PubMed] [Google Scholar]
- Reuter R., Panganiban G. E., Hoffmann F. M., Scott M. P. Homeotic genes regulate the spatial expression of putative growth factors in the visceral mesoderm of Drosophila embryos. Development. 1990 Dec;110(4):1031–1040. doi: 10.1242/dev.110.4.1031. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Yamamoto M., Kuroiwa A. Cell type dependent transcription regulation by chick homeodomain proteins. Mech Dev. 1992 Mar;37(1-2):25–36. doi: 10.1016/0925-4773(92)90012-9. [DOI] [PubMed] [Google Scholar]
- Schier A. F., Gehring W. J. Functional specificity of the homeodomain protein fushi tarazu: the role of DNA-binding specificity in vivo. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1450–1454. doi: 10.1073/pnas.90.4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Scott M. P. Vertebrate homeobox gene nomenclature. Cell. 1992 Nov 13;71(4):551–553. doi: 10.1016/0092-8674(92)90588-4. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Stern S., Tanaka M., Herr W. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989 Oct 19;341(6243):624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- Tomotsune D., Shoji H., Wakamatsu Y., Kondoh H., Takahashi N. A mouse homologue of the Drosophila tumour-suppressor gene l(2)gl controlled by Hox-C8 in vivo. Nature. 1993 Sep 2;365(6441):69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- Treacy M. N., Neilson L. I., Turner E. E., He X., Rosenfeld M. G. Twin of I-POU: a two amino acid difference in the I-POU homeodomain distinguishes an activator from an inhibitor of transcription. Cell. 1992 Feb 7;68(3):491–505. doi: 10.1016/0092-8674(92)90186-g. [DOI] [PubMed] [Google Scholar]
- Vachon G., Cohen B., Pfeifle C., McGuffin M. E., Botas J., Cohen S. M. Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell. 1992 Oct 30;71(3):437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
- Wagner-Bernholz J. T., Wilson C., Gibson G., Schuh R., Gehring W. J. Identification of target genes of the homeotic gene Antennapedia by enhancer detection. Genes Dev. 1991 Dec;5(12B):2467–2480. doi: 10.1101/gad.5.12b.2467. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Zappavigna V., Renucci A., Izpisúa-Belmonte J. C., Urier G., Peschle C., Duboule D. HOX4 genes encode transcription factors with potential auto- and cross-regulatory capacities. EMBO J. 1991 Dec;10(13):4177–4187. doi: 10.1002/j.1460-2075.1991.tb04996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Andrew D. J., Mathies L. D., Horner M. A., Scott M. P. Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functional specificity. Development. 1993 Jun;118(2):339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]
- Zhao J. J., Lazzarini R. A., Pick L. The mouse Hox-1.3 gene is functionally equivalent to the Drosophila Sex combs reduced gene. Genes Dev. 1993 Mar;7(3):343–354. doi: 10.1101/gad.7.3.343. [DOI] [PubMed] [Google Scholar]