Abstract
IFN1@ (interferon, type 1, cluster, also called IFNα) has been extensively studied as a treatment for patients with chronic myeloid leukemia (CML). The mechanism of anticancer activity of IFN1@ is complex and not well understood. Here, we demonstrate that autophagy, a mechanism of cellular homeostasis for the removal of dysfunctional organelles and proteins, regulates IFN1@-mediated cell death. IFN1@ activated the cellular autophagic machinery in immortalized or primary CML cells. Activation of JAK1-STAT1 and RELA signaling were required for IFN1@-induced expression of BECN1, a key regulator of autophagy. Moreover, pharmacological and genetic inhibition of autophagy enhanced IFN1@-induced apoptosis by activation of the CASP8-BID pathway. Taken together, these findings provide evidence for an important mechanism that links autophagy to immunotherapy in leukemia.
Keywords: IFN1@, autophagy, apoptosis, immunotherapy, chronic myeloid leukemia
Introduction
IFN1@ (interferon, type 1, cluster, also called IFNα) has been extensively studied as a treatment for patients with chronic myeloid leukemia (CML) since 1981, with up to 80% of early chronic phase patients achieving hematologic remission.1 Following the discovery of the leukemic oncogene BCR-ABL1 and its causal association with CML, potent BCR-ABL1 tyrosine kinase inhibitors such as imatinib have been developed.2 Nevertheless, IFN1@ remains important and its use in combination therapy with tyrosine kinase inhibitors has attracted considerable interest.3 Although IFN1@ therapy is widely accepted for selected human malignant diseases such as metastatic renal cell carcinoma and hepatocellular carcinoma, the treatment regimen is often complicated by the emergence of IFN1@ resistance, such as in CML.4,5 A more detailed understanding of how IFN1@ resistance develops will allow for improved therapeutic strategies to enhance overall patient survival.
Autophagy is a catabolic process involving the degradation of a cell’s own components such as aggregated/misfolded proteins and damaged organelles, through the lysosomal machinery.6 It is an important cellular response to stress or starvation,7 and it is implicated in certain human diseases.8 Autophagy is now an exciting field in translational cancer research. During tumor development and in cancer therapy, autophagy has paradoxically been reported to have roles in promoting both cell survival and cell death depending on the context.9 Others have noted that autophagy promotes the growth of BCR-ABL1-associated CML,10 and imatinib induces autophagy in several mammalian cells.11 Moreover, we and others have demonstrated that inhibition of autophagy enhances imatinib-induced cytotoxicity in CML cells, including stem cells.12,13 These findings suggest a unique role for autophagy in CML development and therapy.14,15 However, whether autophagy regulates the sensitivity of CML cells to IFN1@ remains unknown. In this study, we demonstrated that treatment with IFN1@ led to a dose-dependent activation of autophagy in CML cells. Inhibition of autophagy enhanced the anticancer activity of IFN1@. This process involved the activation of Janus kinase 1 (JAK1), signal transducer and activator of transcription 1, 91 kDa (STAT1), nuclear factor of kappa light polypeptide gene enhancer in B-cells (NFKB) signals, and autophagy-mediated cell survival. Therefore, these findings provide the first evidence for the development of novel therapeutic strategies based on IFN1@ and autophagy inhibition in CML.
Results
IFN1@ induces autophagy flux in CML cells
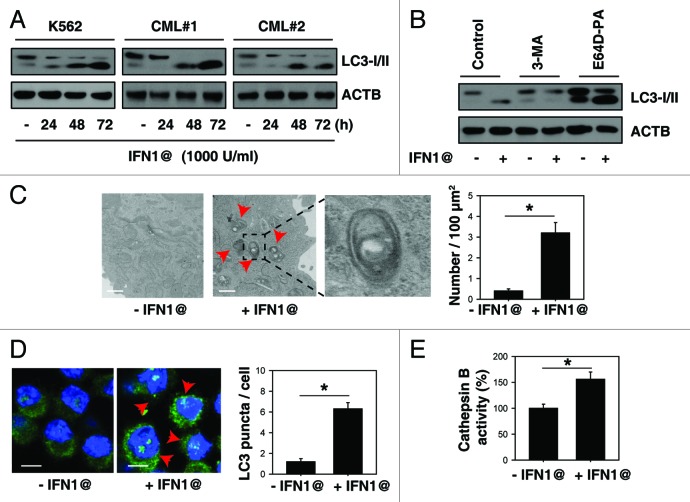
To investigate whether IFN1@ is a direct activator of autophagic flux, we detected microtubule-associated protein 1 light chain 3 α (LC3) conversion (LC3-I to LC3-II) in the absence and presence of lysosomal protease inhibitors. LC3-II is the processed form of LC3 located on the autophagosomal membrane. Treatment with IFN1@ induced a time-dependent increase in the expression of LC3-II in immortalized K562 and primary CML cells (Fig. 1A). Cells treated with IFN1@ in combination with E64D/pepstatin A (“E64D-PA”) exhibited an increase in LC3-II (Fig. 1B). In contrast, autophagic sequestration blocker 3-methyladenine (“3-MA”) decreased IFN1@-induced LC3-II accumulation (Fig. 1B). Morphological hallmarks of autophagy can be observed by transmission electron microscopy (TEM) and confocal microscopy.17 TEM revealed that autophagic vacuoles (autophagosomes/autolysosomes) were dramatically increased in K562 cells after IFN1@ treatment, whereas minimal vacuole formation was evident in the control group (Fig. 1C). Additionally, LC3 puncta and lysosomal activity were significantly increased after IFN1@ treatment, with immunofluorescent staining (Fig. 1D) and cathepsin B enzymatic activity analysis (Fig. 1E), respectively. Taken together, these findings suggest that IFN1@ is an inducer of autophagy in CML cells.
Figure 1. IFN1@ induces autophagic flux in CML cells. (A) K562 cells and primary BMMCs from CML patients were treated with IFN1@ (1000 U/ml) for 24–72 h and the LC3 level was assayed by western blot (n = 3, *p < 0.05). AU, arbitrary units. (B) Analysis of LC3 processing by autophagy in the presence or absence of lysosomal protease inhibitors pepstatin A (PA, 10 μg/ml) and E64D (10 μg/ml) or 3-methyladenine (3-MA, 10 mM) after IFN1@ (1000 U/ml) treatment for 48 h (n = 3, *p < 0.05). AU, arbitrary units. (C) Ultrastructural features in K562 cells with or without IFN1@ (1000 U/ml, 48 h) treatment. By definition, autophagic vacuoles are limited by a double, or occasionally multilayered membrane.16 They contain cytoplasmic material or organelles. Sometimes the autophagic vacuole membrane does not have any contrast in thin sections. This is probably caused by extraction of lipids during sample preparation, as lipids are not optimally preserved in conventional aldehyde fixation.16 The inset in (C) shows a magnified double-membrane autophagic vacuole. The number of autophagic vacuoles (indicated by the red arrows) under TEM was calculated (*p < 0.05). (D and E) K562 cells were treated with IFN1@ (1000 U/ml) for 48 h and LC3 puncta per cell and enzymatic activity of cathepsin B were assayed by confocal microscopy (D) and ELISA (E) as described in the methods section (*p < 0.05). Representative images in K562 cells are shown in the left panel (D). Scale bar, 10 μm.
BECN1-ATG5-ATG7 autophagy pathway is required for IFN1@-induced autophagy
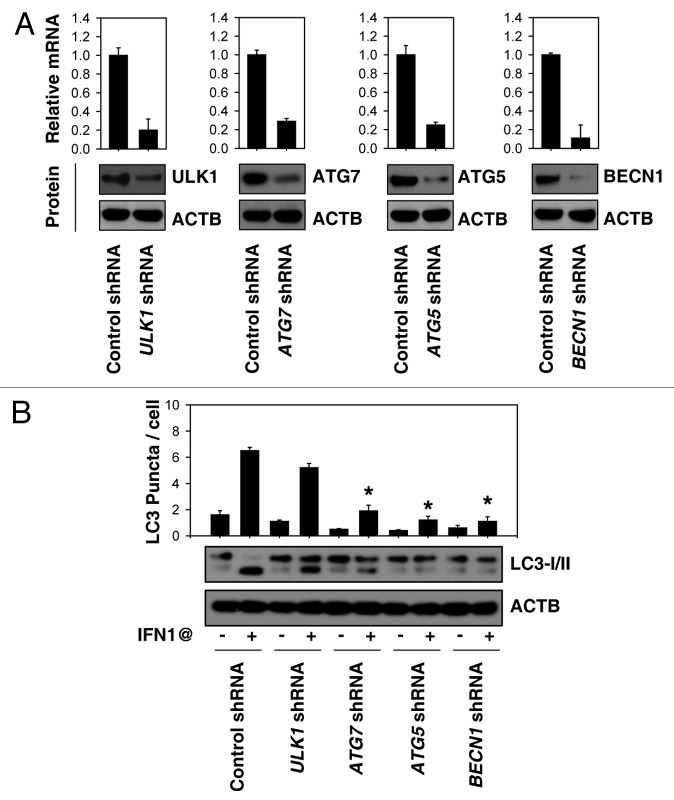
Autophagy involves a series of dynamic membrane-rearrangement reactions mediated by a core set of autophagy proteins (ATGs). Among these, ULK1 (the mammalian ortholog of yeast Atg1), BECN1 (the mammalian ortholog of yeast Vps30/Atg6), ATG5, and ATG7 are the four major regulators of the classical autophagy pathway in mammalian cells.6-8 However BECN1-independent,18 ATG5-ATG7-independent19 and ULK1-independent20 alternative autophagy pathways have been described. To further characterize the role of ATG in IFN1@-induced autophagy, target-specific shRNA against ATGs were transfected into K562 cells. Transfection of BECN1, ATG5, ULK1, and ATG7 shRNA led to a significant and persistent decrease in mRNA and protein level at 48 h post-transfection (Fig. 2A). Notably, suppression of BECN1, ATG5, and ATG7, but not ULK1 expression decreased IFN1@-induced autophagy as evaluated by LC3-II expression and LC3 puncta formation (Fig. 2B). This suggests that the BECN1-ATG5-ATG7 autophagy pathway is required for IFN1@-induced autophagy in CML cells.
Figure 2. The classical autophagy pathway is required for IFN1@-induced autophagy. (A) K562 cells were transfected with indicated shRNA for 48 h, and then the mRNA and protein expression of these shRNA targeted genes were analyzed by real-time PCR and western blot, respectively. (B) K562 cells were transfected with indicated shRNA for 48 h and then treated with IFN1@ (1000 U/ml) for 48 h. Autophagy was assayed by western blot analysis of LC3 expression or quantitation of average number of LC3 puncta per cell (B). *p < 0.05 vs. control shRNA group.
JAK-STAT1 activation promotes IFN1@-induced autophagy
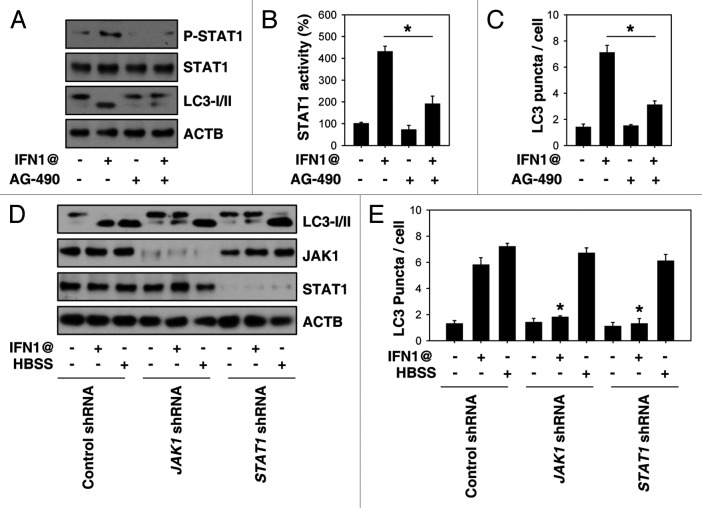
Cells respond rapidly following stimulation with IFNs via the JAK-STAT signal transduction pathway. We explored whether JAK-STAT activation is required for IFN1@-induced autophagy. Potential JAK inhibitors (e.g., AG-490) decreased IFN1@-induced phosphorylation of STAT1 (Fig. 3A), STAT1 transcriptional activity (Fig. 3B), and LC3 puncta formation (Fig. 3C). To further explore whether JAK-STAT is required for IFN1@-induced autophagy, we knocked down JAK1 and STAT1 expression by shRNA. Suppression of these proteins decreased LC3-II levels (Fig. 3D) and accumulation of LC3 puncta (Fig. 3E) after IFN1@ treatment. In contrast, knockdown of JAK1 and STAT1 did not influence starvation/HBSS-induced LC3 puncta formation in K562 cells (Fig. 3D and E), suggesting that JAK1-STAT1 signaling is specifically required for IFN1@-induced autophagy.
Figure 3. JAK1-STAT1 signaling is required for IFN1@-induced autophagy. (A–C) K562 cells were treated with IFN1@ (1000 U/ml) for 48 h in the presence or absence of AG-490 (10 μM). Then P-STAT1 was assayed by western blot (A). STAT1 transcriptional activity was assayed by a luciferase reporter system. Control group was set as 100%. (B) Autophagy was assayed by quantitation of average number of LC3 puncta per cell (C). n = 3, *p < 0.05. (D and E) K562 cells were transfected with JAK1 and STAT1 shRNA for 48 h, and then treated with IFN1@ (1000 U/ml, 48 h) or starvation (HBSS, 2 h). Autophagy was assayed by western blot analysis of LC3 expression (D) or quantitation of average number of LC3 puncta per cell (E). n = 3, *p < 0.05 vs. control shRNA group.
STAT1 and NFKB are required for IFN1@-induced BECN1 expression
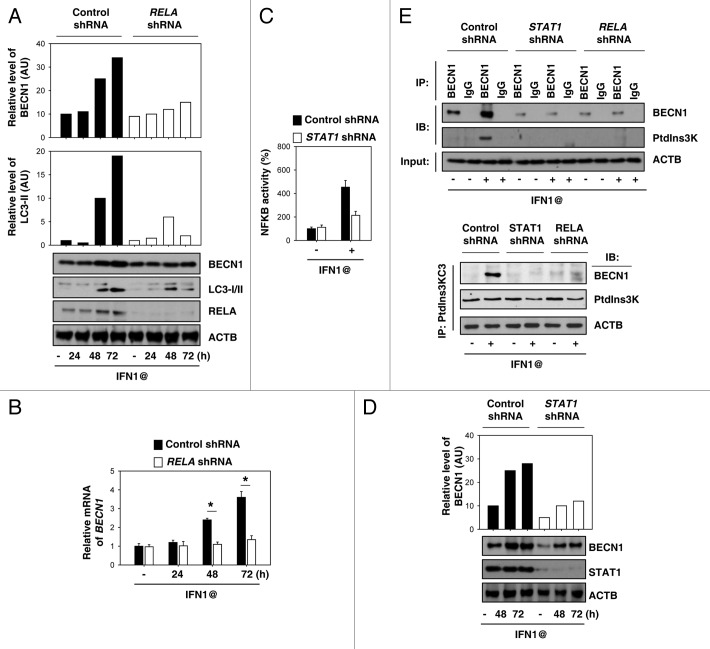
Increasing evidence suggests that NFKB is involved in the regulation of BECN1 expression in autophagy.21 Similarly, knockdown of RELA (also called NFKB P65) significantly decreased IFN1@-induced BECN1 mRNA and protein expression (Fig. 4A and B). A previous study demonstrated that STAT1 regulates NFKB activity after IFN1@ treatment in human melanoma cells.22 Consistently, knockdown of STAT1 impaired IFN1@-induced NFKB activation (Fig. 4C) and subsequently BECN1 expression (Fig. 4D). BECN1 has a critical role in inducing autophagy by promoting formation of BECN1-class III type phosphatidylinositol 3-kinase (PtdIns3K) core complexes.23 Notably, knockdown of STAT1 or RELA decreased the interaction between BECN1 and PtdIns3K (Fig. 4E). These findings suggest that STAT1-NFKB crosstalk is required for IFN1@-induced BECN1 expression, and subsequently BECN1-PtdIns3K complex formation.
Figure 4. STAT1 and NFKB are required for IFN1@-induced BECN1 expression. (A and B) K562 cells were transfected with RELA shRNA for 48 h, and then treated with IFN1@ (1000 U/ml) for 24–72 h. Protein (A) and mRNA (B) levels of BECN1 were analyzed by western blot and real-time PCR, respectively. (C and D) K562 cells were transfected with STAT1 shRNA for 48 h, and then treated with IFN1@ (1000 U/ml) for 48–72 h. NFKB transcriptional activity was assayed by a luciferase reporter system (C). The indicated proteins were analyzed by western blot (D). (E) K562 cells were treated with IFN1@ (1000 U/ml) for 48 h, then cell lysates were prepared for immunoprecipitation (IP) as indicated. The resulting immune complexes and inputs were analyzed by western blotting (IB) as shown.
Inhibition of autophagy enhances anticancer activity of IFN1@
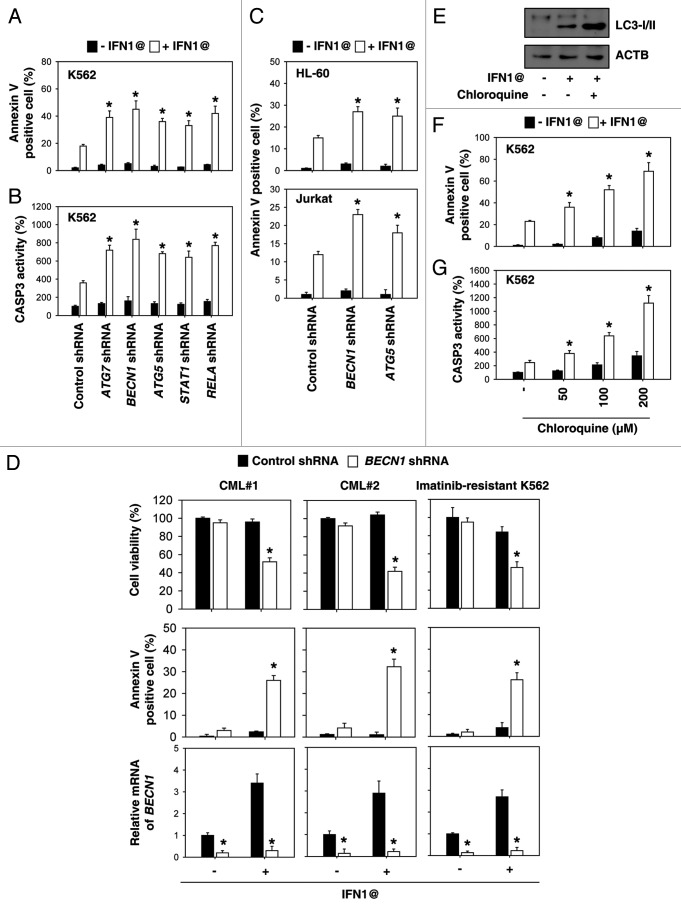
To examine the effects of autophagy on the anticancer activity of IFN1@, we analyzed apoptosis after knockdown of autophagy regulators by shRNA. Compared with cells transfected with control shRNA, knockdown of ATG7, BECN1 and ATG5 in K562 cells increased IFN1@-induced apoptosis by flow cytometric analysis (Fig. 5A). Moreover, knockdown of STAT1 and RELA increased IFN-induced cell apoptosis (Fig. 5A). CASP3 as a final effector is activated in apoptotic cells by both extrinsic and intrinsic pathways. Knockdown of the autophagy genes STAT1 and RELA promoted IFN1@- induced CASP3 activation compared with control shRNA cells (Fig. 5B). IFN1@ has been reported to induce apoptosis in other leukemia cells, such as HL-60 human acute myeloid leukemia cells24 and Jurkat human T-cell acute lymphoblastic leukemia cells.25 Knockdown of BECN1 and ATG5 in these cells also increased IFN1@-induced apoptosis (Fig. 5C), suggesting a wide range of anti-apoptosis effects of autophagy in leukemia cells after IFN1@ treatment. Next, we determined whether IFN1@-mediated autophagy is a potential survival mechanism for CML stem cells and imatinib-resistant cells. We observed that knockdown of BECN1 by shRNA restored the sensitivity of CD34+ cells and imatinib-resistant K562 cells to IFN1@ (Fig. 5D). Recent studies have shown that inhibition of therapy-induced autophagy with chloroquine derivatives can enhance cell death in established tumors, leading to enhanced tumor regression and delayed tumor regrowth.9,12,13 To determine the effects of chloroquine on autophagic flux, the processing of LC3 was assessed via western blot.26 We confirmed that chloroquine elicited an increase in LC3-II in K562 cells after treatment with IFN1@ (Fig. 5E). Moreover, we found that chloroquine resulted in a dose-dependent increase in IFN1@-induced apoptosis (Fig. 5F) as well as CASP3 activity (Fig. 5G) in K562 cells. Together, these results demonstrate that autophagy plays an important role in modulating apoptosis in response to IFN1@.
Figure 5. Inhibition of autophagy enhances anticancer activity of IFN1@. (A and B) K562 cells were treated with IFN1@ (1000 U/ml) for 48 h. Apoptosis was analyzed by measuring annexin V-positive cells by flow cytometry (A). In parallel, CASP3 activation was assayed (B). n = 3, *p < 0.05 vs. control shRNA group. (C) Indicated HL-60 and Jurkat cells were treated with IFN1@ (1000 U/ml) for 48 h. Apoptosis was analyzed by measuring annexin V-positive cells by flow cytometry. n = 3, *p < 0.05 vs. control shRNA group. (D) CD34+ and CD34- cells isolated from bone marrow mononuclear cells of CML patients by CD34 MicroBead Kit (Miltenyi Biotec) or imatinib-resistant K562 cells were transfected with BECN1 shRNA or control shRNA for 48 h, and then treated with IFN1@ (1000 U/ml) for 48 h. Cell viability was analyzed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay, and apoptosis was analyzed by measuring annexin V-positive cells by flow cytometry. In parallel, mRNA levels of BECN1 were analyzed by real-time PCR. n = 3, *p < 0.05 vs. control shRNA group. (E) Western blot analysis of LC3 processing by autophagy in the presence or absence of chloroquine (200 μM) after IFN1@ (1000 U/ml) treatment for 48 h in K562 cells. (F and G) K562 cells were treated with IFN1@ (1000 U/ml) for 48 h in the presence or absence of chloroquine (50–200 μM). Apoptosis was analyzed by measuring annexin V-positive cells by flow cytometry (F). In parallel, CASP3 activation was assayed (G). n = 3, *p < 0.05 vs. the group without chloroquine treatment.
Autophagy regulates IFN1@-induced CASP8 dependent apoptosis
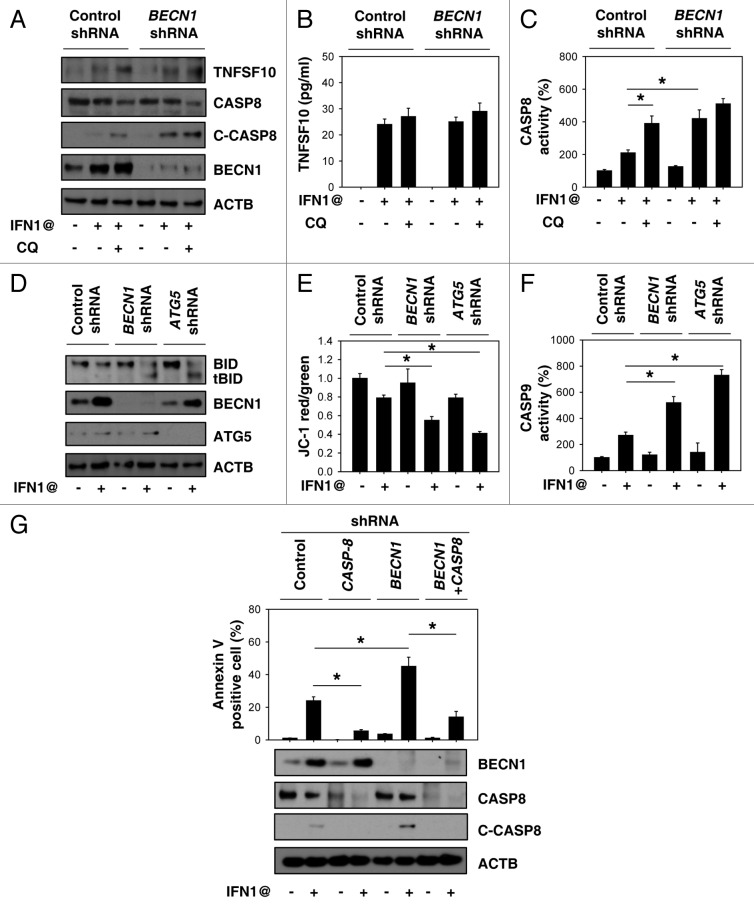
IFN1@-induced apoptosis is associated with induction of tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10, also called TRAIL) expression and release in several types of cancer cells.27 However, inhibition of autophagy by chloroquine or BECN1 shRNA did not influence IFN1@-induced expression and release of TNFSF10 (Fig. 6A and B). In contrast, the activity of CASP8 (Fig. 6C) and cleaved CASP8 (Fig. 6A) was increased after inhibition of autophagy, suggesting that autophagy regulates CASP8 during apoptosis. Cleavage of BID, a BH3 domain-containing BCL2 protein, by CASP8 mediates the crosstalk between the extrinsic (membrane receptor) and intrinsic (mitochondria) pathways.28 Inhibition of autophagy by BECN1 or ATG5 shRNA increased cleavage of BID (tBID), decreased mitochondrial membrane potential (Δψm) and increased CASP9 activation (Fig. 6D–F). Moreover, knockdown of CASP8 reversed BECN1 shRNA-induced apoptosis in K562 cells (Fig. 6G). These results demonstrate that autophagy plays an important role in modulating IFN1@-induced CASP8 dependent apoptosis.
Figure 6. Autophagy regulates IFN1@-induced CASP8 dependent apoptosis. (A–C) K562 cells were treated with IFN1@ (1000 U/ml) for 48 h in the presence or absence of chloroquine (CQ, 100 μM). The expression and release of TNFSF10 was analyzed by western blot (A) and ELISA (B) respectively. In parallel, CASP8 activation (C) was assayed as described in Materials and Methods (n = 3, *p < 0.05). (D–F) Indicated K562 cells were treated with IFN1@ (1000 U/ml) for 48 h. Cleavage of BID (tBID) was analyzed by western blot (D). In parallel, mitochondrial membrane potential (Δψm) (E) and CASP9 activity (F) was assayed as described in Materials and Methods (n = 3, *p < 0.05). (G) K562 cells were transfected with indicated shRNA and then treated with IFN1@ (1000 U/ml) for 48 h. Apoptosis was analyzed by measuring annexin V-positive cells by flow cytometry (n = 3, *p < 0.05).
Discussion
Autophagy, as a major survival mechanism, is induced by cellular stress to regulate cellular homeostasis.6,7 Our current study indicated that induction of autophagy conferred resistance to IFN1@ in leukemia cells. This signal transduction event is involved in activation of JAK-STAT1 and NFKB, which contributes to IFN1@-induced autophagy. The molecular basis of resistance due to autophagy stems partially from regulation of CASP8-mediated apoptosis. Thus, inhibition of autophagy may be an attractive strategy for enhancing the clinical activity of IFN1@-mediated immunotherapy in hematologic malignancies, as well as other tumors.
IFNs, as pleiotropic cytokines, affect the activities of macrophages, natural killer cells, dendritic cells, T cells, and tumor cells by enhancing antigen presentation and cell differentiation, inducing an antiviral state, inhibiting cell growth and promoting apoptosis.29 IFNs also have a role in regulating autophagy with cell type-specific differences. For example, IFNG/IFNγ has been reported to induce autophagy in macrophages30 and fibroblasts.31 However, IFN1@ does not induce autophagy in macrophages.30 In contrast, IFN1@ induces autophagy as a protective mechanism in bladder cancer and normal urothelial cells.32 IFN1@ inhibits cell-mediated autophagy in some cell types.33 In this study, we found that IFN1@ is a strong inducer of autophagy in CML cells. The classic BECN1-ATG5-ATG7 pathway is required for IFN1@-induced autophagy. Others have demonstrated that autophagy can itself regulate the production and secretion of IFN1@ in virally-infected cells.34 However, research into the crosstalk between autophagy and IFN1@ is still not well characterized.
Recently, several studies have linked autophagy to leukemia. (1) Autophagy promotes leukemogenesis and development. ATG3 functions as an E2-like enzyme during the initial stages of autophagosome formation.10 ATG3 deletion prevents BCR-ABL1-mediated leukemogenesis in a cell transfer model.10 Autophagy is a hallmark of T-lineage lymphoblastic lymphoma (T-LBL) in a transgenic myc-driven zebrafish model of T-LBL.35 Chloroquine significantly delays the onset of spontaneous tumors in a transgenic mouse model of Myc-induced lymphoma.36 (2) Autophagy inhibition enhances the efficacy of chemotherapy. Autophagy upregulation occurs in response to chemotherapeutic agents used in the treatment of hemopoietic malignancies including CML.15,37 Pharmacological or RNAi-mediated inhibition of autophagy potentiates the effects of imatinib, bafetinib, suberoylanilide hydroxamic acid, and OSI-027-induced death of CML cells13,38-40 and stem cells.12,13 Our previous studies demonstrate that high mobility group box 1 (HMGB1) is a BECN1 binding protein,41 and HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells.42 In this study, we found that knockdown of BECN1 and ATG5 or chloroquine treatment enhances the anticancer activity of IFN1@ in CML, including in stem cells and other leukemia cells. It is unknown whether HMGB1-mediated autophagy is involved in this process. (3) Under some conditions, autophagy inhibition decreases the efficacy of chemotherapy. For example, induction of autophagy is essential for steroid sensitization in childhood acute lymphoblastic leukemia cells, which requires necroptosis,43 a form of programmed necrosis. Inhibition of autophagy also decreases resveratrol-induced cell death in CML cells.44 These studies suggest that inhibition or induction of autophagy may improve the therapeutic efficacy of treatments for leukemia depending on the types of anticancer agent and pathway of stress response.
Our study suggests an interaction between STAT1 and NFKB in regulating BECN1 expression. Mammalian BECN1 was originally isolated in a yeast two-hybrid screen for BCL2 interacting proteins. BECN1 interacts with several cofactors to regulate the lipid kinase PtdIns3K and promote the formation of BECN1-PtdIns3K core complexes, thereby inducing autophagy.23 RELA directly binds the BECN1 promoter and upregulates its mRNA and protein levels, leading to autophagy in T cells.21 We found that knockdown of RELA impaired IFN1@-induced BECN1 expression. Activation of the JAK-STAT pathway is the crucial event during IFN1@ treatment.29,45 STAT proteins are a family of cytoplasmic proteins that function as secondary messengers and transcription factors. The antitumor effects of IFN1@ are abrogated in the Stat1-deficient mouse.46 Moreover, STAT1 is able to directly interact with RELA to regulate NFKB activity.22 We found that knockdown of STAT1 inhibits NFKB activity and subsequent IFN1@-induced BECN1 expression. Another possibility is that STAT1 promotes BECN1 transcription directly. Furthermore, the interaction between BECN1 and PtdIns3K was decreased after inhibition of STAT1 and the NFKB pathway. These findings provide a mechanism by which IFN1@ enhances autophagy in CML cells.
The role of autophagy in the regulation of apoptosis remains unclear. Our findings suggest that inhibition of autophagy increases IFN1@-induced apoptosis by regulation of CASP8 activity. Others have demonstrated that active CASP8 is directly degraded by the autophagic pathway in human colon cancer cells.47 Thus, inhibition of autophagy reduces the degradation of active CASP8 and increases levels of CASP8 activity. A link exists between the death receptor pathway and the mitochondria apoptosis pathway through CASP8-mediated cleaved BID (tBID). Inhibition of autophagy increases IFN1@-mediated tBID formation, which accompanies mitochondrial apoptosis events such as decreased Δψm. These findings provide a crosstalk mechanism between autophagy and apoptosis.
In summary, we demonstrate here that inhibition of autophagy by RNAi or chloroquine increased the anticancer activity of IFN1@ in leukemia cells by the CASP8-dependent apoptosis pathway. Therapy of selected human malignancies with IFN1@ is widely accepted but is often complicated by the emergence of IFN1@ resistance. In addition, IFN1@ has both acute and chronic toxicities associated with its administration which have resulted in poor patient compliance during clinical trials.2 Chloroquine and hydroxychloroquine (HCQ) are used as antimalarial drugs and several clinical trials have now been approved for cancer treatment with the use of chloroquine and HCQ as an autophagy inhibitor.9 Thus, use of the combination of chloroquine or HCQ with IFN1@ in the clinic might decrease IFN1@ resistance and result in a lower incidence of side effects, which will improve outcomes obtained with IFN1@ alone.
Materials and Methods
Reagents
The antibodies to LC3 (NB100-2220), BECN1 (NB500-249), ATG5 (NB110-53818) and ULK1 (NBP1-03502) were obtained from Novus. The antibodies to CASP8 (9746), cleaved CASP8 (9496), RELA (4764), PtdIns3K (3811) and ACTB/β-ACTIN (3700) were obtained from Cell Signaling Technology. The antibodies to TNFSF10 (sc-7877), STAT1 (sc-98783), P-STAT1 (sc-8394), and JAK1 (sc-1677) were obtained from Santa Cruz Technology. Recombinant human IFN1@ (11200) was obtained from R&D Systems. The NE-PER Nuclear and Cytoplasmic Extraction Kit (78833) were obtained from Thermo Fisher Scientific. The cathepsin B ELISA Kit (ab119584) was obtained from Abcam. All other reagents not listed came from Sigma.
Cell culture and patients
The human leukemia cell line, K562 (chronic myeloid leukemia cells), HL-60 (acute myeloid leukemia cells), and Jurkat (T-cell acute lymphoblastic leukemia cells) from the American Type Culture Collection were cultured in Iscove’s Modified Dulbecco’s medium or RPMI-1640 medium with 10% heat-inactivated FBS and 2 mM glutamine in a humidified incubator with 5% CO2 and 95% air. Bone marrow mononuclear cells (BMMCs) from CML patients were isolated by Ficoll density gradient centrifugation. The diagnosis of CML was made on the basis of clinical features, hematologic characteristics, and the presence of the Philadelphia chromosome, which has been described in detail previously.48 Described briefly, a study on newly diagnosed childhood CML patients who had adequate hepatic and renal function and no serious medical conditions was conducted. The trial was approved by the institution’s review boards and ethics committees and all patients gave written informed consent in accordance with the Declaration of Helsinki. Samples were enriched for CD34+ cells using CliniMACS (130–046–702) according to the manufacturer’s instructions. The purity of CD34+ was routinely above 95% as assessed by flow cytometric analysis. CD34+ cells were cultured in X-VIVO 15 (BioWhittaker, 04380-Q) containing 1% BSA, 2 mM L-glutamine, and a cytokine cocktail (StemCell Technologies, 02697).
RNAi
Short hairpin RNA (shRNA) against human ULK1 (SHCLNV-NM_003565), BECN1 (SHCLNV-NM_003766), ATG5 (SHCLNV-NM_004849), ATG7 (SHCLNV-NM_006395), CASP8 (SHCLNV-NM_001228), JAK1 (SHCLNV-NM_002227), STAT1 (SHCLNV-NM_007315) and RELA (SHCLNV-NM_021975) were obtained from Sigma and were transfected into cells by lentiviral delivery systems according to the manufacturer’s instructions.
Western blot analysis
Proteins in cell lysates was first resolved by SDS-PAGE, then transferred to nitrocellulose membrane and subsequently incubated with primary antibodies. After incubation with peroxidase-conjugated secondary antibodies, the signals were visualized by enhanced chemiluminescence according to the manufacturer’s instruction. The relative band intensity was quantified using Gel-pro Analyzer® software.
Immunofluorescence analysis
Cells were fixed in 4% formaldehyde for 15 min at room temperature prior to cell permeabilization with 0.1% Triton X-100 (4°C, 10 min). Cells were saturated with PBS containing 2% BSA for one h at room temperature and processed for immunofluorescence with anti-LC3 antibody, followed by Alexa Fluor 488-conjugated Ig and Hoechst 33258. Between all incubation steps, cells were washed three times for three minutes with PBS containing 0.2% BSA. Fluorescence signals were analyzed using an Olympus Fluoview 1000 confocal microscope. The average LC3 puncta per cell from at least 50 cells was determined using Image-Pro Plus 5.1 software as previously described.49
Immunoprecipitation analysis
Cells were lysed at 4°C in ice-cold RIPA lysis buffer (Cell Signaling Technology, 9806). Concentrations of proteins in the supernatant were determined by BCA assay. Prior to immunoprecipitation, samples containing equal amounts of proteins were pre-cleared with Protein A and subsequently incubated with various irrelevant IgG or specific antibodies in the presence of protein A beads for 2 h or overnight at 4°C with gentle shaking. Following incubation, beads were washed extensively with PBS and proteins eluted by boiling in 2 × SDS sample buffer before SDS-PAGE electrophoresis.
Transmission electron microscopy analysis
Cells were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.4), followed by 1% OsO4. After dehydration, thin sections were stained with uranyl acetate and lead citrate for observation under a JEM 1011CX electron microscope (JEOL). Images were acquired digitally from a randomly selected pool of 10–15 fields under each condition. The quantification of autophagosomes and autophagolysosomes was performed as previously described.50
Assessment of apoptosis
Apoptosis in cells was assessed using an annexin V/PI Apoptosis Detection Kit (BD PharMingen, 556570) by flow cytometric analysis.51 Mitochondrial membrane potential depolarization (Δψm) was measured by flow cytometry using a fluorescent cationic dye, 1,1′3,3′-tetraethylbenzamidazolocarbocyanin iodide (Molecular Probes, JC-1, M34152). CASPASE activity was assayed using the CASP3, CASP8, and CASP9 Colorimetric Assay Kit (Calbiochem, 235419, 218770, 218824) according to the manufacturer’s instructions.
STAT1 and NFKB activation assay
Cells were transiently transfected in a 12-well plate with a STAT1 or NFKB luciferase reporter plasmid (SABiosciences, CCS-013L, CLS-008L) or control empty plasmid according to the manufacturer’s instructions. After 24–48 h, the cells were exposed to various agents. The luciferase activity was determined using the luciferase assay system with the reporter lysis buffer obtained from Promega (E1500). The results are expressed as relative transcription activity after normalizing to the control empty plasmid.
TNFSF10 release measurements
Commercially available enzyme linked immunosorbant assay (ELISA) kits (RD system, DTRL00) were used to measure the concentrations of TNFSF10 in the culture medium according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction
cDNA from various cell samples were amplified by real-time quantitative PCR with specific primers from SABiosciences according to the manufacturer’s instructions. The control group was set as 100%.
Statistical analysis
Data are expressed as means ± SD of three independent experiments. Significance of differences between groups was determined by two-tailed Student’s t test or ANOVA LSD test. A p value < 0.05 was considered significant.
Acknowledgments
We thank Christine Heiner (Department of Surgery/Department of Anesthesiology, University of Pittsburgh) for editing work. This work was supported by grants from the National Natural Sciences Foundation of China (30973234 and 31171328 to L.C.) and a grant from the University of Pittsburgh (D.T.). It was supported in part by grants from the National Institutes of Health (R01CA160417 to D.T.).
Glossary
Abbreviations:
- Δψm
mitochondrial membrane potential
- 3-MA
3-methyladenine
- ATG
autophagy-related
- BMMCs
bone marrow mononuclear cells
- CML
chronic myeloid leukemia
- E64D-PA
E64D/pepstatin A
- HCQ
hydroxychloroquine
- HMGB1
high mobility group box 1
- IFN1@
interferon, type 1, cluster
- JAK1
Janus kinase 1
- LC3
microtubule-associated protein 1 light chain 3
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide
- NFKB
nuclear factor of kappa light polypeptide gene enhancer in B-cells
- PtdIns3K
class III phosphatidylinositol 3-kinase
- shRNA
short hairpin RNA
- STAT1
signal transducer and activator of transcription 1, 91kDa
- tBID
cleaved BID
- TNFSF10
tumor necrosis factor (ligand) superfamily, member 10
- T-LBL
T-lineage lymphoblastic lymphoma
- TEM
transmission electron microscopy
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22923
References
- 1.Talpaz M, McCredie KB, Mavligit GM, Gutterman JU. Leukocyte interferon-induced myeloid cytoreduction in chronic myelogenous leukemia. Blood. 1983;62:689–92. [PubMed] [Google Scholar]
- 2.Kujawski LA, Talpaz M. The role of interferon-alpha in the treatment of chronic myeloid leukemia. Cytokine Growth Factor Rev. 2007;18:459–71. doi: 10.1016/j.cytogfr.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Hehlmann R, Berger U, Pfirrmann M, Hochhaus A, Metzgeroth G, Maywald O, et al. German CML-Study Group Randomized comparison of interferon alpha and hydroxyurea with hydroxyurea monotherapy in chronic myeloid leukemia (CML-study II): prolongation of survival by the combination of interferon alpha and hydroxyurea. Leukemia. 2003;17:1529–37. doi: 10.1038/sj.leu.2403006. [DOI] [PubMed] [Google Scholar]
- 4.Wong N, Chan KY, Macgregor PF, Lai PB, Squire JA, Beheshti B, et al. Transcriptional profiling identifies gene expression changes associated with IFN-alpha tolerance in hepatitis C-related hepatocellular carcinoma cells. Clin Cancer Res. 2005;11:1319–26. [PubMed] [Google Scholar]
- 5.Sakai I, Takeuchi K, Yamauchi H, Narumi H, Fujita S. Constitutive expression of SOCS3 confers resistance to IFN-alpha in chronic myelogenous leukemia cells. Blood. 2002;100:2926–31. doi: 10.1182/blood-2002-01-0073. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman BJ, Jacobs SR, Mason EF, Michalek RD, MacIntyre AN, Coloff JL, et al. Autophagy is essential to suppress cell stress and to allow BCR-Abl-mediated leukemogenesis. Oncogene. 2011;30:1855–67. doi: 10.1038/onc.2010.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J, et al. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21:936–42. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- 12.Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–23. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, Yang L, Zhao M, Zhu S, Kang R, Vernon P, et al. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia. 2012;26:1752–60. doi: 10.1038/leu.2012.65. [DOI] [PubMed] [Google Scholar]
- 14.Helgason GV, Karvela M, Holyoake TL. Kill one bird with two stones: potential efficacy of BCR-ABL and autophagy inhibition in CML. Blood. 2011;118:2035–43. doi: 10.1182/blood-2011-01-330621. [DOI] [PubMed] [Google Scholar]
- 15.Calabretta B, Salomoni P. Inhibition of autophagy: a new strategy to enhance sensitivity of chronic myeloid leukemia stem cells to tyrosine kinase inhibitors. Leuk Lymphoma. 2011;52(Suppl 1):54–9. doi: 10.3109/10428194.2010.546913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol. 2009;452:143–64. doi: 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith DM, Patel S, Raffoul F, Haller E, Mills GB, Nanjundan M. Arsenic trioxide induces a beclin-1-independent autophagic pathway via modulation of SnoN/SkiL expression in ovarian carcinoma cells. Cell Death Differ. 2010;17:1867–81. doi: 10.1038/cdd.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 20.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A. 2011;108:11121–6. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krämer OH, Baus D, Knauer SK, Stein S, Jäger E, Stauber RH, et al. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–85. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakashima A, Kumakura S, Mishima S, Ishikura H, Kobayashi S. IFN-alpha enhances TNF-alpha-induced apoptosis through down-regulation of c-Myc protein expression in HL-60 cells. J Exp Clin Cancer Res. 2005;24:447–56. [PubMed] [Google Scholar]
- 25.Romero-Weaver AL, Wang HW, Steen HC, Scarzello AJ, Hall VL, Sheikh F, et al. Resistance to IFN-alpha-induced apoptosis is linked to a loss of STAT2. Mol Cancer Res. 2010;8:80–92. doi: 10.1158/1541-7786.MCR-08-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papageorgiou A, Lashinger L, Millikan R, Grossman HB, Benedict W, Dinney CP, et al. Role of tumor necrosis factor-related apoptosis-inducing ligand in interferon-induced apoptosis in human bladder cancer cells. Cancer Res. 2004;64:8973–9. doi: 10.1158/0008-5472.CAN-04-1909. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/S0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 29.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 31.Chang YP, Tsai CC, Huang WC, Wang CY, Chen CL, Lin YS, et al. Autophagy facilitates IFN-gamma-induced Jak2-STAT1 activation and cellular inflammation. J Biol Chem. 2010;285:28715–22. doi: 10.1074/jbc.M110.133355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang XQ, Dunner K, Jr., Benedict WF. Autophagy is induced by adenoviral-mediated interferon alpha treatment in interferon resistant bladder cancer and normal urothelial cells as a cell death protective mechanism but not by the bystander factors produced. Cancer Gene Ther. 2010;17:579–84. doi: 10.1038/cgt.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchser WJ, Laskow TC, Pavlik PJ, Lin HM, Lotze MT. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970–9. doi: 10.1158/0008-5472.CAN-11-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 35.Feng H, Stachura DL, White RM, Gutierrez A, Zhang L, Sanda T, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010;18:353–66. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helgason GV, Karvela M, Holyoake TL. Kill one bird with two stones: potential efficacy of BCR-ABL and autophagy inhibition in CML. Blood. 2011;118:2035–43. doi: 10.1182/blood-2011-01-330621. [DOI] [PubMed] [Google Scholar]
- 38.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–22. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamitsuji Y, Kuroda J, Kimura S, Toyokuni S, Watanabe K, Ashihara E, et al. The Bcr-Abl kinase inhibitor INNO-406 induces autophagy and different modes of cell death execution in Bcr-Abl-positive leukemias. Cell Death Differ. 2008;15:1712–22. doi: 10.1038/cdd.2008.107. [DOI] [PubMed] [Google Scholar]
- 40.Yogalingam G, Pendergast AM. Abl kinases regulate autophagy by promoting the trafficking and function of lysosomal components. J Biol Chem. 2008;283:35941–53. doi: 10.1074/jbc.M804543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25:23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- 43.Bonapace L, Bornhauser BC, Schmitz M, Cario G, Ziegler U, Niggli FK, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest. 2010;120:1310–23. doi: 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–52. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- 45.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–63. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 46.Lesinski GB, Anghelina M, Zimmerer J, Bakalakos T, Badgwell B, Parihar R, et al. The antitumor effects of IFN-alpha are abrogated in a STAT1-deficient mouse. J Clin Invest. 2003;112:170–80. doi: 10.1172/JCI16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao M, Yang M, Yang L, Yu Y, Xie M, Zhu S, et al. HMGB1 regulates autophagy through increasing transcriptional activities of JNK and ERK in human myeloid leukemia cells. BMB Rep. 2011;44:601–6. doi: 10.5483/BMBRep.2011.44.9.601. [DOI] [PubMed] [Google Scholar]
- 49.Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Cao L, Kang R, Yang M, Liu L, Zhao Y, et al. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARα oncoprotein. Autophagy. 2011;7:401–11. doi: 10.4161/auto.7.4.14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang R, Tang D, Yu Y, Wang Z, Hu T, Wang H, et al. WAVE1 regulates Bcl-2 localization and phosphorylation in leukemia cells. Leukemia. 2010;24:177–86. doi: 10.1038/leu.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]