Abstract
A chemical screen designed to identify novel inducers of autophagy led to the discovery that signal transducer and activator of transcription 3 (STAT3) inhibitors can potently stimulate the autophagic flux. Although STAT3 is best known as a pro-inflammatory and oncogenic transcription factor, mechanistic analyses revealed that autophagy is regulated by the cytoplasmic, not nuclear, pool of STAT3. Cytoplasmic STAT3 normally interacts with the eukaryotic translation initiation factor 2, subunit 1α, 35kDa (EIF2S1/eIF2α) kinase 2/protein kinase, RNA-activated (EIF2AK2/PKR), a sensor of double-stranded RNA. This interaction, which could be recapitulated using recombinant proteins in pull-down experiments, involves the catalytic domain of EIF2AK2 as well as the SH2 domain of STAT3, which can adopt a fold similar to that of EIF2S1. Thus, STAT3 may act as a competitive inhibitor of EIF2AK2. Indeed, pharmacological or genetic inhibition of STAT3 stimulates EIF2AK2-dependent EIF2S1 phosphorylation and autophagy. Conversely, the overexpression of wild-type STAT3 as well as of STAT3 mutants that cannot be phosphorylated by JAK2 or are excluded from the nucleus inhibits autophagy. However, STAT3 mutants that fail to interact with EIF2AK2 are unable to suppress autophagy. Both STAT3-targeting agents (i.e., Stattic, JSI-124 and WP1066) and EIF2AK2 activators (such as the double-strand RNA mimetic polyinosinic:polycytidylic acid) are capable of disrupting the inhibitory interaction between STAT3 and EIF2AK2 in cellula, yet only the latter does so in cell-free systems in vitro. A further screen designed to identify EIF2AK2-dependent autophagy inducers revealed that several fatty acids including palmitate trigger autophagy via a pathway that involves the disruption of the STAT3-EIF2AK2 complex as well as the phosphorylation of mitogen-activated protein kinase 8/c-Jun N-terminal kinase 1 (MAPK8/JNK1) and EIF2S1. These results reveal an unsuspected crosstalk between cellular metabolism (fatty acids), pro-inflammatory signaling (STAT3), innate immunity (EIF2AK2), and translational control (EIF2S1) that regulates autophagy.
Keywords: EIF2S1S51A, endoplasmic reticulum, IRS1, palmitate, polyI:C, STAT3Y705F
STAT3 can be phosphorylated by receptor-associated kinases (like JAK2 and MET) on Y705 to form homo- or heterodimers that, upon translocation to the nucleus, operate as transcription factors. Since STAT3 is involved in oncogenesis, inflammation and immunosuppression, clinical trials are underway to evaluate STAT3 inhibitors for their antineoplastic and immunostimulatory effects (source http://clinicaltrials.gov).
EIF2AK2 was originally characterized as a pathogen recognition receptor. The N-terminus of EIF2AK2 binds viral double-stranded RNA (dsRNA), leading to activation of the C-terminal catalytic domain. Among several substrates, active EIF2AK2 phosphorylates EIF2S1, hence shutting down the translation of viral proteins. EIF2AK2 also responds to multiple nonviral stimuli including fatty acids, extracellular ATP, monosodium urate, the adjuvant aluminum, the mitochondrial inhibitor rotenone, and the lethal anthrax toxin. EIF2AK2 interacts not only with multiple components of the inflammasome including NOD-like receptor (NLR) family pyrin domain-containing 1 and 3 (NLRP1 and NLRP3), NLR family CARD domain-containing protein 4 (NLRC4), and absent in melanoma 2 (AIM2), but also with the stress-related MAPK8. Thus, EIF2AK2 operates at the interface between inflammation, stress signaling and metabolism.
EIF2S1 phosphorylation is required for the optimal induction of autophagy, both in yeast and human cells, although the underlying mechanisms remain elusive. As a possibility, the arrest of protein translation at the endoplasmic reticulum may favor the emergence of cup-shaped protrusions, named omegasomes, which serve as platforms for autophagosome biogenesis. The phosphorylation of EIF2S1 by EIF2AK2 mediates potent pro-autophagic effects, both in yeast cells (in which the sole yeast EIF2S1 kinase can be functionally replaced by human EIF2AK2) and in the mammalian system (in which viral infection can stimulate EIF2AK2-dependent autophagy).
Taking advantage of a human osteosarcoma cell line (U2OS) that stably expresses a GFP-LC3 chimera, we performed a screen to identify novel autophagy inducers, among which we found the STAT3 inhibitor JSI-124. Subsequent validation experiments demonstrated that several other chemically unrelated STAT3 inhibitors (i.e., Stattic and WP1066), the transient knockdown of STAT3 with specific small interfering RNAs, as well as the knockout of the Stat3 gene by homologous recombination stimulate the autophagic flux, both in vitro (in human and murine cell lines) and in vivo (in the liver of mice bearing a hepatocyte-specific Stat3 knockout). Conversely, the transfection-enforced overexpression of STAT3 suppressed autophagy. Such an inhibitory effect was observed not only with wild-type STAT3 but also when a STAT3 variant that exclusively localizes to the cytoplasm, and a nonphosphorylatable STAT3 mutant (STAT3Y705F) were employed. On the contrary, an exclusively nuclear variant of STAT3 failed to repress autophagy. Thus, STAT3 inhibits autophagy via a cytoplasmic mechanism that does not involve the phosphorylation of Y705 and the consequent relocalization of STAT3 to the nuclear compartment.
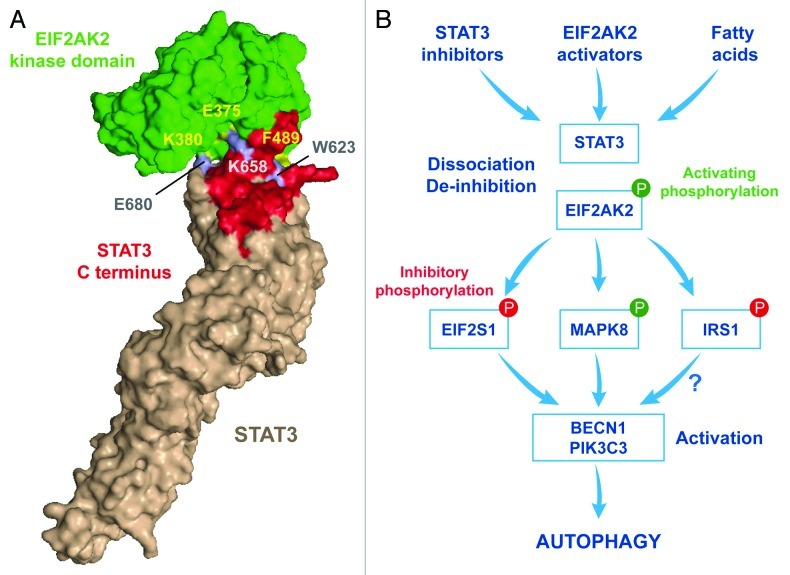
Next, we performed an in silico screen to identify autophagy-relevant proteins that interact with STAT3. This approach led us to recognize EIF2AK2 as a candidate link between STAT3 and autophagy regulation. We found that in control conditions, when autophagy is off, STAT3 and EIF2AK2 interact with each other in the cytoplasm. This interaction is specific (because EIF2AK2 failed to co-immunoprecipitate with STAT family members other than STAT3, and STAT3 failed to interact with EIF2S1 kinases other than EIF2AK2) and direct (because it could be recapitulated with recombinant proteins in pull-down experiments). Molecular modeling was then used to get further insights into the interaction between STAT3 and EIF2AK2 (Fig. 1A). The SH2 domain of STAT3 exhibits a conformational fold that can be superimposed with that of EIF2S1, suggesting that STAT3 might competitively inhibit EIF2AK2 by binding to its catalytic domain. We obtained three lines of evidence in support of this model. First, site-directed mutagenesis followed by co-immunoprecipitation experiments or pull-down assays confirmed a prominent role for the STAT3 residues that were predicted to be important for the STAT3-EIF2AK2 interaction, namely W623, K658A and E680A. Second, STAT3 mutants with a reduced affinity for EIF2AK2 (like those bearing the W623A K658A and W623 E680A double substitutions) fail to inhibit autophagy when they are overexpressed in human cancer cells. Third, inhibition or depletion of STAT3 leads to the activation of EIF2AK2, as indicated by its autophosphorylation as well as by the phosphorylation of EIF2S1. Autophagy induced by STAT3 inhibitors is abolished or attenuated when EIF2AK2 is depleted with specific siRNAs or when EIF2S1 is replaced by a nonphosphorylatable mutant (EIF2S1S51A), respectively. Taken together, our data suggest that STAT3 inhibitors promote the dissociation of STAT3 from EIF2AK2 in an indirect manner.
Figure 1. Regulation of fatty acid-induced autophagy by STAT3 and EIF2AK2. (A) In normal conditions, when autophagy is inhibited, cytoplasmic STAT3 and EIF2AK2, engage in a direct inhibitory interaction that appears to involve the following residues: W623, K658 and E680 in STAT3 and E375, K380 and F489 in EIF2AK2. (B) Upon treatment with STAT3 inhibitors, EIF2AK2 activators or fatty acids, the STAT3-EIF2AK2 complex dissociates and EIF2AK2 becomes available to phosphorylate EIF2S1, hence inhibiting translation. EIF2AK2 is also required for the activating phosphorylation of MAPK8, the inhibitory phosphorylation of IRS1, the phosphatidylinositol-3-kinase activity of the BECN1-PIK3C3 complex, and the induction of macroautophagy.
The aforementioned results established that STAT3 represses the pro-autophagic activity of EIF2AK2, yet failed to provide physiologically relevant information. Therefore, we screened a library of autophagy triggers for their dependence on EIF2AK2. This screen led to the identification of palmitate (and other fatty acids) as EIF2AK2-dependent inducers of autophagy. In line with this notion, the palmitate-induced phosphorylation of MAPK8, insulin receptor substrate 1 (IRS1) and EIF2S1 could be suppressed by the siRNA-mediated knockdown of EIF2AK2 (but not of other EIF2S1 kinases) as well as by the overexpression of STAT3. Altogether, these results indicate the existence of a molecular circuitry whereby fatty acids can trigger the dissociation of STAT3 and EIF2AK2, hence de-inhibiting EIF2AK2, allowing it to phosphorylate EIF2S1 (together with other substrates) and to set off the autophagic cascade (Fig. 1B).
STAT3 is not the only transcription factor that represses autophagy when present in the cytoplasm; the cytoplasmic pool of oncosuppressor protein TP53 has a similar autophagy-inhibitory function. Although the mechanisms through which STAT3 and TP53 suppress autophagy are entirely different, they have one common denominator: as they are activated and translocate to the nucleus, both STAT3 and TP53 lose their capacity to suppress autophagy.
Acknowledgments
G.K. is supported by the Ligue Nationale Contre le Cancer (Equipes Labelisée), Agence Nationale pour la Recherche (ANR), Fondation Axa (Chair For Longevity Research), European Commission (ArtForce), Fondation pour la Recherche Médicale (FRM), Institut National du Cancer (INCa), Cancéropôle Ile-de-France, Fondation Bettencourt-Schueller and the LabEx Immuno-Oncology. M.N.S., S.A., S.A.M., L.G. and M.C.M. are supported by Junta de Extremadura-Fondo Social Europeo, Ligue Nationale contre le Cancer, the Higher Education Commission (HEC) of Pakistan, the LabEx Immuno-Oncology and the Association pour la Recherche sur le Cancer (ARC), respectively.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/22910