Abstract
In recent years, substantial research interests have arisen to decipher miRNA function in cell death modules such as apoptosis and autophagy. Despite enormous success, it is increasingly evident that systems biology approaches that analyze the intra- and inter-modular connectivity are essential to uncover the generic organizing principles of the miRNA-mediated cell death network. We therefore constructed the miRDeathDB database to archive experimentally confirmed programmed cell death (PCD)-associated miRNAs together with their target proteins. Based on global analysis of the miRNA-mediated cell death network, we find there are two classes of miRNAs important to the cell death output. One class tends to have more regulatory links to proteins within death modules, whereas the other one lies on the boundary between modules, and tends to connect with more protein pairs across the modules. These findings indicate that miRNAs provide a novel communication mechanism between PCD-related proteins involved in both intra- and inter-cell death modules. Furthermore, miRDeathDB can also facilitate researcher access to a variety of resources centered on PCD-associated miRNAs.
Keywords: microRNAs, programmed cell death, network analysis
Mammalian cells have evolved several elegant mechanisms to die, including apoptosis, autophagy and programmed necrosis. After decades of classic cell biology and genetics studies, the morphological distinctions and molecular players regulating each module of cell death have become widely appreciated, especially for apoptosis. A notable conceptual advancement is that the three apparently different cell death phenotypes are actually intimately connected within a global network. In the PCD network model, the proteins can be simplified as functional nodes and the interactions among them should be considered as edges, which connect the nodes. Recently, a class of small (~22 nucleotides in length) noncoding RNAs, miRNA has been identified that participate in this network. miRNAs usually negatively regulate protein coding genes through base pairing with the targeted sequences in the 3′ untranslated region. Functional defects in miRNAs have been implicated in the etiology of many forms of human diseases.
miRNAs can modulate PCD-related proteins and eventually affect the cell death output, adding an additional layer to the complexity of the PCD network. Regarding the roles of miRNAs in the PCD network, many questions have been raised. For example, a single miRNA can modulate multiple PCD-related proteins, and conversely one PCD-related protein can be targeted by multiple miRNAs, so how are these regulations balanced to achieve appropriate network output? Recent evidence indicates that some miRNAs can participate in multiple death modules, or execute opposing regulations on cell death under certain circumstances; so are these miRNAs involved in the inter-modular switch, and what are the underlying mechanisms? Answers to the above questions cannot be merely based on the functional delimitation of a single miRNA, but rather through analyzing the intra- and inter-modular connectivity in the miRNA-mediated cell death network. Therefore, systems biology approaches, which ‘connect the dots’ into a global network, are warranted.
miRDeathDB archives PCD knowledge centered on miRNAs
For the first step toward understanding the network architecture, we have collected experimentally-confirmed PCD-associated miRNAs from literature mining. Core functional data such as the target proteins, the affected cell death module, organism and tissue specificity were also integrated into the miRDeathDB database (http://rna-world.org/mirdeathdb/). Furthermore, the PCD-associated miRNAs were annotated to established miRNA databases including miRBase (www.mirbase.org) and miR2Disease (www.mir2disease.org). Correspondingly, each PCD-associated protein was linked to commonly used resources such as NCBI gene (www.ncbi.nlm.nih.gov/gene), Human Protein Reference Database (HPRD, http://www.hprd.org) and Online Mendelian Inheritance in Man (OMIM, www.ncbi.nlm.nih.gov/omim) (Fig. 1). Users can efficiently retrieve a large amount of genomic, proteomic and disease-associated data via links to these external resources. To the best of our knowledge, miRDeathDB provides the first all-in-one information resource centered on PCD-associated miRNAs.
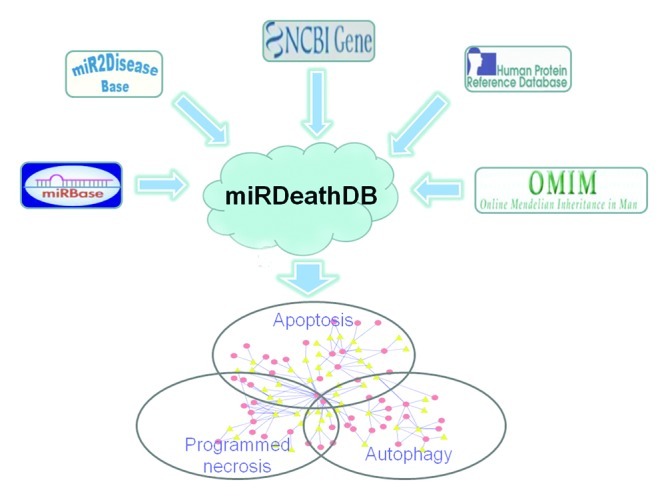
Figure 1. miRDeathDB archives PCD knowledge centered on miRNAs. Based on hundreds of experimental reports, miRDeathDB stores functional information for miRNAs and their targets in the PCD network. miRDeathDB also provides a wealth of annotation data via links to external resources such as miRBase, miR2Disease, NCBI gene, Human Protein Reference Database and Online Mendelian Inheritance in Man.
Systemwide analyses of miRNAs that mediate the cell death network
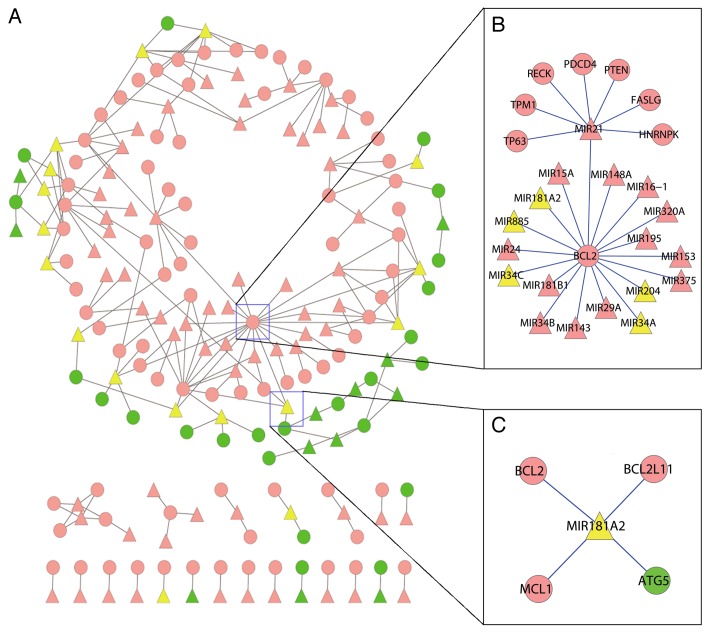
miRDeathDB is not only a knowledge depository but also the basis for globally analyzing the miRNAs in the PCD network. As shown in Figure 2A, we have constructed a simplified miRNA-mediated cell death network based on PCD-associated miRNAs together with the targeted proteins. In this network, miRNAs and proteins are represented as nodes and the miRNA targeting protein regulation is denoted as an edge. Since the molecular analysis of miRNAs in programmed necrosis is still in its infancy, this model focuses on the apoptosis and autophagy death module. Analyzing even such a simplified model has provided insights at the systems level. First, among the total 92 PCD-associated miRNAs and their 106 targets in the miRDeathDB, most of them (74% and 75% for miRNAs and targets, respectively) can be integrated into a big interlocking network. Due to the intrinsic property of miRNAs having multiple targets, proteins with shared miRNA binding sites can be efficiently organized by miRNA mediators. This result also implies that, in addition to the canonical protein-protein interaction between cell death proteins, miRNAs provide another dimension of communication for proteins in both the intra- and intercellular death module. Second, our follow-up characterization by standard network analysis techniques identified several miRNA ‘hubs’ (nodes that are highly connected with other nodes) within the apoptosis or autophagy module. Furthermore these miRNA ‘hubs’ usually connect with key cell death-related protein ‘hubs’. For example, the anti-apoptotic protein BCL2 is intensively regulated by 18 miRNAs including the MIR21 ‘hub’, which again targets eight PCD-related proteins (Fig. 2B).
Figure 2. miRNA-mediated cell death network based on literature mining. (A) Most of PCD-associated miRNAs and their targets can be integrated into a big interlocking network. Trianglar and circular nodes represent cell death-associated miRNAs and proteins, respectively. Edges represent the miRNA targeting protein regulation. Apoptotic miRNAs and proteins are in pink, while autophagy-associated ones are in green. Note that yellow-colored miRNAs play dual roles in both apoptosis and autophagy. (B) An example of miRNA hubs in the PCD network. The MIR21 hub targets 8 PCD-related proteins including the BCL2 hub, which is again intensively regulated by 18 miRNAs. (C) An example of miRNAs which potentially mediate the crosstalk between autophagic and apoptotic death modules. The MIR181A2 connects 3 apoptotic proteins (BCL2, BCL2L11 and MCL1) with 1 autophagic protein, ATG5.
Network analysis techniques were also applied to analyze the crosstalk between the autophagy and apoptosis modules. We used the betweenness centrality (BC) to assess the capability of each miRNA node to link two distant nodes. BC equals the number of shortest paths from all nodes to all others that pass through a given node. Nodes with higher BC are usually connected to two relatively isolated sub-modules in the global network. In the miRNA-mediated PCD network, we found there is a group of miRNAs that have statistically higher BC than others (Wilcoxon rank-sum test). As shown in Figure 2C, MIR181A, the one with the highest BC, connects 3 apoptotic proteins (BCL2, BCL2L11 and MCL1) with 1 autophagic protein, ATG5. Since all these proteins possess a MIR181A binding site, they can modulate each other by competing for miRNA binding. More importantly, this miRNA-dependent mechanism may mediate the conversion between autophagic and apoptotic death modules, leading to cellular homeostasis upon external stimuli.
Potential use of miRNAs in anticancer therapy
Understanding the miRNA-mediated cell death network has clinical implications. Currently, many antitumor agents have been developed to target the core proteins of the cell death machinery. However, intrinsic mechanisms, such as evasion of apoptotic cell death and induction of autophagy to tolerate chemotherapy, are often utilized by tumor cells to resist cell death. As we discussed above, it is now clear that this is only half of the story, since miRNAs provide alternative signal transduction routes in cell death modules. Furthermore, systemwide analyses of PCD-associated miRNAs indicate that some miRNAs may be involved in inter-modular communication in the cell death network. Therefore miRNAs may be exploited therapeutically to globally manipulate the cell death network and provide a promising target for anticancer therapy.
Acknowledgments
We are grateful to Dr. Daniel J. Klionsky for help in editing the manuscript. We would like to thank Dr. Dapeng Hao for helpful discussion. This work was supported by grants from Henan University of Technology (2009BS040 and 11JCYJ11 to J.X.), National Natural Science Foundation of China (31100901 to D.W.) and Natural Science Foundation of Heilongjiang Province of China (QC2010012 to D.W.).
Glossary
Abbreviations:
- PCD
programmed cell death
- HPRD
Human Protein Reference Database
- OMIM
Online Mendelian Inheritance in Man
- BC
betweenness centrality
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23096