Abstract
CCN family proteins 2 and 3 (CCN2 and CCN3) belong to the CCN family of proteins, all having a high level of structural similarity. It is widely known that CCN2 is a profibrotic molecule that mediates the development of fibrotic disorders in many different tissues and organs. In contrast, CCN3 has been recently suggested to act as an anti-fibrotic factor in several tissues. This CCN3 action was shown earlier to be exerted by the repression of the CCN2 gene expression in kidney tissue, whereas different findings were obtained for liver cells. Thus, the molecular action of CCN3 yielding its anti-fibrotic effect is still controversial. Here, using a general model of fibrosis, we evaluated the effect of CCN3 overexpression on the gene expression of all of the CCN family members, as well as on that of fibrotic marker genes. As a result, repression of CCN2 gene expression was modest, while type I collagen and α-smooth muscle actin gene expression was prominently repressed. Interestingly, not only CCN2, but also CCN4 gene expression showed a decrease upon CCN3 overexpression. These findings indicate that fibrotic gene induction is under the control of a complex molecular network conducted by CCN family members functioning together.
Keywords: CCN2, CCN3, CCN4, NOV, Fibrosis, CCN family
Introduction
The mammalian CCN family of proteins consists of 6 members having a high degree of structural similarity (Brigstock et al. 2003; Perbal and Takigawa 2005). These members display multiple functionalities under different biological conditions. Indeed, the same CCN family member can either promote or repress the progression of malignant tumors, depending upon the property of the tumors and the surrounding microenvironment (Leask and Abraham 2006; Chen and Lau 2009). Such apparently contradictory effects of CCN family proteins may be ascribed to their unique mode of molecular action. Except for CCN5, all of the other CCN family members are composed of 4 distinct modules connected in tandem. The modules, designated as insulin-like growth factor binding protein-like (IGFBP), von Willebrand factor type C repeat (VWC), thrombospondin type 1 repeat (TSP1) and carboxyl terminal cystine knot (CT) modules, are highly interactive with a number of different types of molecules including growth factors, extracellular matrix (ECM) components, and cell-surface receptors (Kubota and Takigawa 2007; Chen and Lau 2009). Using these modules, CCN family proteins manipulate the multiple molecules present in their microenvironments, thus resulting in highly context-dependent and diverse biological outcomes.
One of the classical family members, CCN2, is known to be a key factor that promotes harmonized development and regeneration of mesenchymal tissues (Kikuchi et al. 2008; Ono et al. 2008; Kubota and Takigawa 2011). Nevertheless, the distinct profibrotic function of this same molecule has been widely recognized as well, which function is firmly supported by scientific evidence presented by a number of investigators. In fact, overexpression of the CCN2 gene is observed in fibrotic disorders found in a variety of tissues and organs including kidney, lung, liver, pancreas, skin, and periodontal tissues (Leask et al. 2009; Kubota 2012). Moreover, the induction or overproduction of CCN2 is known to provoke fibrotic changes in several cell cultures of different origins. As such, concerning fibrosis, no negative effect of CCN2 on it has been presented. In this context, it should be noted that a few recent reports have described the anti-fibrotic effect of CCN3, another classical member of the CCN family, in renal and hepatic systems (Riser et al. 2009; Borkham-Kamphorst et al. 2012).
Despite its high degree of structural homology with other CCN family molecules, CCN3 has been shown to be the CCN family member that functionally counteracts CCN2. According to previous reports, the application of recombinant CCN2 or CCN3 protein promotes or inhibits the proliferation and differentiation of growth-plate chondrocytes (Kawaki et al. 2008). Of note, in Ccn2-null mice, the CCN3 gene expression is elevated (Ivkovic et al. 2003; Kawaki et al. 2008). Therefore, CCN2 and CCN3 appear to act in a “yin-yang” manner both functionally and genetically (Leask 2009). Indeed, one previous study described that overexpression of the CCN3 gene in renal fibroblasts causes a remarkable reduction in CCN2 production and fibrotic response (Riser et al. 2009), whereas knocking-down the CCN3 gene yields the opposite effects. However, another recent study showed contradictory results regarding this yin-yang relationship of these 2 CCN family members (Borkham-Kamphorst et al. 2012). Additionally, since most CCN family members are functionally, as well as structurally related, the behavior of the other members should be taken into account for comprehensive interpretation of the biological outcome (Kawaki et al. 2011).
In this way, by employing a general in vitro model of fibrosis using TGF-β-stimulated NIH3T3 cells, as described herein we investigated the effect of CCN3 on the fibrogenic phenotype and gene expression profile of CCN family members in order to gain a comprehensive insight into this issue.
Materials and methods
Cell culture
Murine embryonic skin fibroblast-derived NIH3T3 cells were seeded into a 24-well plate at a density of 104 cells/cm2 and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal bovine serum (FBS) purchased from Nissui Pharmaceutical Co. Ltd. (Tokyo, Japan) and Cancera International (Rexcalale, Ontario, Canada), respectively. The cells were cultured at 37 °C with 5 % CO2 in air.
For the production of pseudovirions, Phoenix packaging cells (http://www.stanford.edu/group/nolan/retroviral_systems/phx.html) were seeded at a density of 8 × 105 cells per 100-mm cell culture plate in regular DMEM and allowed to reach an early sub-confluent state. The medium was then replaced with fresh medium 3 h before the transfection, for which we used ProFection® Mammalian Transfection System-Calcium phosphate purchased from Promega Corporation (Madison, WI, USA). The transfection reagents were prepared according to manufacturer’s instructions and then added to the proviral plasmid construct, after which the combined solutions were incubated at room temperature for 30 min. The solution was then added together with chloroquine (Takara, Ohtsu, Japan) at 25 μM to the cells, which were incubated at 37 °C with 5 % CO2 in the air for up to 24 h. Afterwards, the cells were checked with a fluorescence microscope to ensure the success of transfection; then the supernatant was discarded and replaced with fresh medium. After another 24 h, the supernatant with the pseudovirions was collected. The last step was repeated for 6 more days to obtain a sufficient amount of viral stocks.
Generation of a retroviral vector for CCN3 overexpression
Prior to the insertion of a CCN3 cDNA, the parental vector, pLEGFP-N1 (Clontech, Mountain View, CA, USA), was modified to contain an internal ribosomal entry site (IRES) sequence immediately upstream of the EGFP cDNA at the unique Bam HI site. Afterwards, human CCN3 cDNA with the signal peptide-encoding region was built in the modified parental vector between unique Bgl II and Sal I sites. The human CCN3 cDNA was obtained from a pGEX-4 T-1 (GE Healthcare, Waukesha, WI, USA)—derived prokaryotic CCN3 expression vector that was kindly provided by Prof. B. Perbal.
CCN3 overexpression and induction of fibrotic response in NIH3T3 cells
The previously collected virus-containing supernatant was suspended in fresh medium and used to infect NIH3T3 cells for 24 h in the presence of hexadimethrine bromide (Sigma-Aldrich, St Louis, MO). Then the medium was changed to regular DMEM, and incubation continued for 8 h to regain the integrity of the cells. One group of cells was infected with pseudovirions for the overexpression of CCN3 and GFP; and the other group with control ones expressing GFP alone. After multiple rounds of infection, the cells were then checked by fluorescence microscope to ensure the successful transfection and subsequently allowed to reach confluence. Thereafter, the cells were collected by using 0.05 % trypsin with ethylenediamine tetraacetate (EDTA) and reseeded in fresh medium in a 6-well plate and allowed to reach confluence again, after which the RNA or protein extract was obtained. To induce a fibrotic response in NIH3T3 cells, we added transforming growth factor-β (TGF-) at a final concentration of 5 ng/ml to the cells and then incubated them for 12 h before RNA collection. This factor was purchased from Biomedical Technologies (Stoughton, MA, USA) or R & D Systems (Minneapolis, MN, USA). Before obtaining protein extract, MEK1/2 inhibitor (PD98059: Calbiochem, San Diego, CA, USA), p38 MAPK inhibitor (SB203580: Calbiochem, San Diego, CA, USA), or JNK inhibitor (SP600125: BIOMOL, Plymouth Meeting, PA, USA) were occasionally added at the same time with TGF-β.
RNA extraction and real-time RT-PCR analysis
Total RNA was extracted and purified from the cells with an RNeasy kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. Total RNA (500 ng) was reverse-transcribed by avian myeloblastosis virus (AMV) Reverse Transcriptase (Takara) at 42 °C for 30 min, according to the manufacturer’s protocol. Real-time PCR was performed by using TOYOBO SYBR Green PCR Master Mix (TOYOBO, Osaka, Japan) in a LightCycler™ system (Roche, Basel, Switzerland). The nucleotide sequences of the primers used are listed in Table 1.
Table 1.
Nucleotide sequences of the primers used in this study
| Gene name | Nucleotide sequence | Direction |
|---|---|---|
| mouse CCN 1 | 5’-ATGAAGACAGCATTAAGGACTC-3’ | sense |
| 5’-TGCAGAGGGTTGAAAAGAAC-3’ | anti-sense | |
| mouse CCN 2 | 5’-CCACCCGAGTTACCAATGAC-3’ | sense |
| 5’-GTGCAGCCAGAAAGCTCA-3’ | anti-sense | |
| mouse CCN 3 | 5’-TGAAGTCTCTGACTCCAGCATT-3’ | sense |
| 5’-TGGCTTTCAGGGATTTCTTG-3’ | anti-sense | |
| mouse CCN 4 | 5’-TGAGAACTGCATAGCCTACAC-3’ | sense |
| 5’-TACACAGCCAGGCATTTC-3’ | anti-sense | |
| mouse CCN 5 | 5’-GCTGTGATGACGGTGGTT-3’ | sense |
| 5’-GACAAGGGCAGAAAGTTGG-3’ | anti-sense | |
| mouse/human | 5’-GCGAATTCCTGCCAGTAGCATATGCTG-3’ | sense |
| ribosomal S18 | 5’-GGAAGCTTAGAGGAGCAGCGACCAAAGC-3’ | anti-sense |
| human CCN 3 | 5’-CCAGAGCAGCCAACAGATAA-3’ | sense |
| 5’-TGGTTTTGGTATTGTGGGGA-3’ | anti-sense | |
| mouse type 1 collagen | 5’-CTGGTCCTGCTGGTCCTGCTG-3’ | sense |
| 5’-TCTGTCACCTTGTTCGCCTGTCTC-3’ | anti-sense | |
| mouse α-smooth | 5’-CCCGCTCTGTCTCTAGCAC-3’ | sense |
| muscle actin | 5’-CACACGAGTAACAAATCAAAGC-3’ | anti-sense |
Western blotting analysis
Total proteins were extracted from the cells with RIPA buffer (50 mM Tris–HCl, 0.15 M NaCl, 4 mM EDTA, 1 % Nonidet P-40, 0.1 % sodium deoxycholate, 10 mM Na4P2O7, 10 mM NaF, 2 mM Na3VO3, 1 mM phenylmethylsulfonyl fluoride, pH 7.5). Extracted total proteins (10 μg) were heated at 95 °C for 5 min in a lithium dodecyl sulfate (LDS) sample buffer with 2-mercaptoethanol, separated by NuPAGE® Novex 4–12 % Bis-Tris Gel (Invitrogen, Carlsbad, CA, USA) electrophoresis and then were transferred onto a polyvinylidenedifluoride (PVDF) membrane (GE Healthcare, Waukesha, WI). After having been blocked with 5 % skim milk, the membrane was incubated for 24 h at 4 °C with 1000-fold-diluted goat antibody against CCN2 (R & D Systems), 5000-fold-diluted rabbit antibody against CCN3 (AbD Serotec, Raleigh, NC, USA) or 5000-fold-diluted mouse monoclonal anti-actin antibody (SIGMA, St. Louis, MO, USA). An anti-rabbit (GE Healthcare) or anti-mouse (GE Healthcare) or anti-Goat (Santa Cruz Biotechnology, Santa Cruz, CA, USA) IgG horseradish peroxidase (HRP) conjugate was then applied as a secondary antibody at a 2000-fold dilution. The blot was visualized by use of an enhanced chemiluminescence (ECL) system.
Statistical analysis
Unless otherwise specified, all of the experiments were performed 3 times with different samples. Data were analyzed for statistical significance by use of the unpaired Student’s t-test.
Results
Overexpression of human CCN3 and its effect on endogenous Ccn3 expression
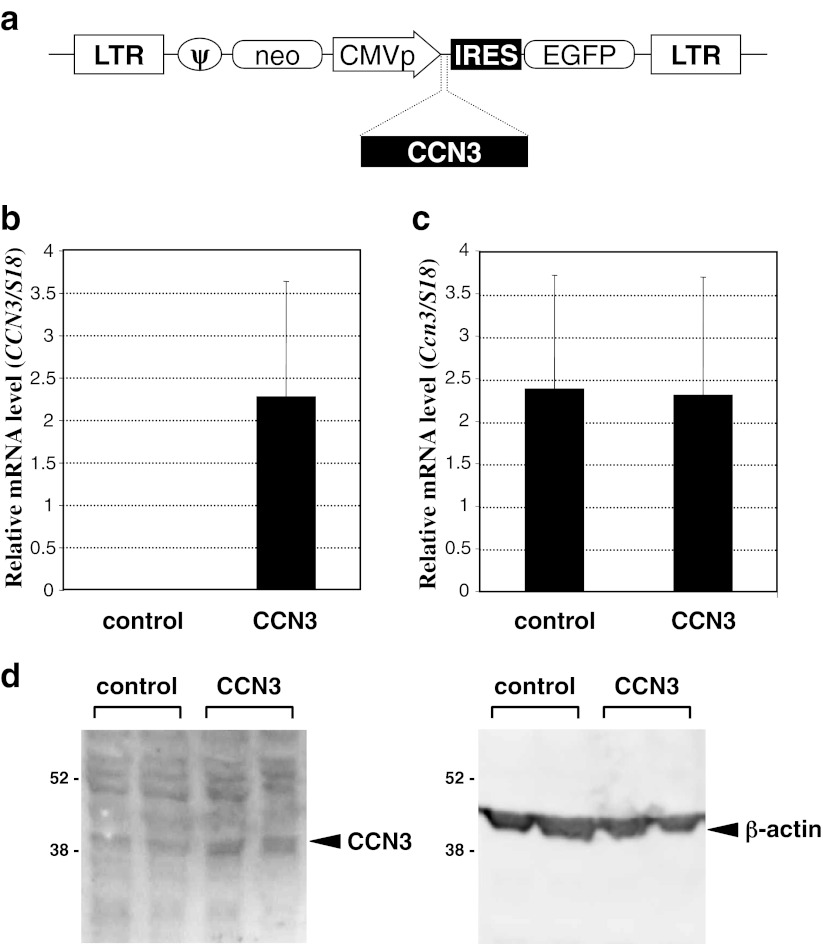
In order to overexpress the human CCN3 gene in murine NIH3T3 cells, we designed and constructed the ecotropic retroviral vector as detailed in Materials and methods and shown in Fig. 1a. Both CCN3-expressing and control pseudovirions contain neomycin-resistant and EGFP genes as selection markers. Utilizing the EGFP marker gene, we confirmed efficient transfection with the gene (data not shown). Under the same condition, successful overexpression of human CCN3 mRNA was confirmed by real-time RT-PCR analysis, whereas no expression was observed in the cells infected with the control pseudovirions (Fig. 1b). TGF-β-stimulated NIH3T3 cells also express endogenous Ccn3. Quantitative real-time RT-PCR analysis of murine CCN3 mRNA revealed no substantial change in the expression level between the human CCN3 gene-overexpressed and control cells (Fig. 1c), indicating that neither a positive nor negative feedback system was involved in the regulation of Ccn3, at least in NIH3T3 cells. Moreover, overexpression of the CCN3 gene in total was solidly confirmed by the results of Western blotting analysis, in which much stronger signals representing CCN3 protein were detected in the CCN3 gene-overexpressed cells (Fig. 1d).
Fig. 1.
Overexpression of human CCN3 in NIH3T3 embryonic skin fibroblastic cells by a retroviral vector. a Structure of the proviral construct used for the production of pseudovirions. Abbreviations: LTR, long terminal repeat of Moloney murine leukemia virus (MLV); ψ, psi sequence of MLV for packaging; neo, neomycin resistant gene; CMVp, cytomegalovirus promoter; CCN3, human CCN3 cDNA, IRES, EMCV internal ribosomal entry site sequence; EGFP, enhanced green fluorescent protein cDNA. b Human CCN3 mRNA level in TGF-β-stimulated NIH3T3 cells infected with parental (control) or CCN3-expressing (CCN3) retroviral vector. Data were standardized against those of 18 s ribosomal RNA. c Endogenous murine Ccn3 mRNA levels in the same cells as those used in panel “b” Experiments were performed 3 times, and mean values are shown with error bars denoting standard deviations. d CCN3 proteins in the lysates of TGF-β-stimulated NIH3T3 cells infected with parental (control) or CCN3-expressing (CCN3) retroviral vector. Western blotting analysis was performed with 2 independent samples for each, and both results are shown. Left and right panels show the signals obtained by anti-CCN3 and anti-β-actin (loading control), respectively. Positions of molecular weight markers are depicted at the left in kilodaltons
Repression of TGF-β-induced fibrogenic phenotype in NIH3T3 by CCN3 overexpression
By utilizing murine embryonic skin fibroblast-derived NIH3T3 cells, we mimicked the biological situation occurring in fibrotic disorders by stimulating the cells with the profibrotic growth factor TGF-β. It is also widely known that NIH3T3 cells produce CCN2, the most important key molecule directing fibrosis, upon TGF-β stimulation (Grotendorst et al. 1996).
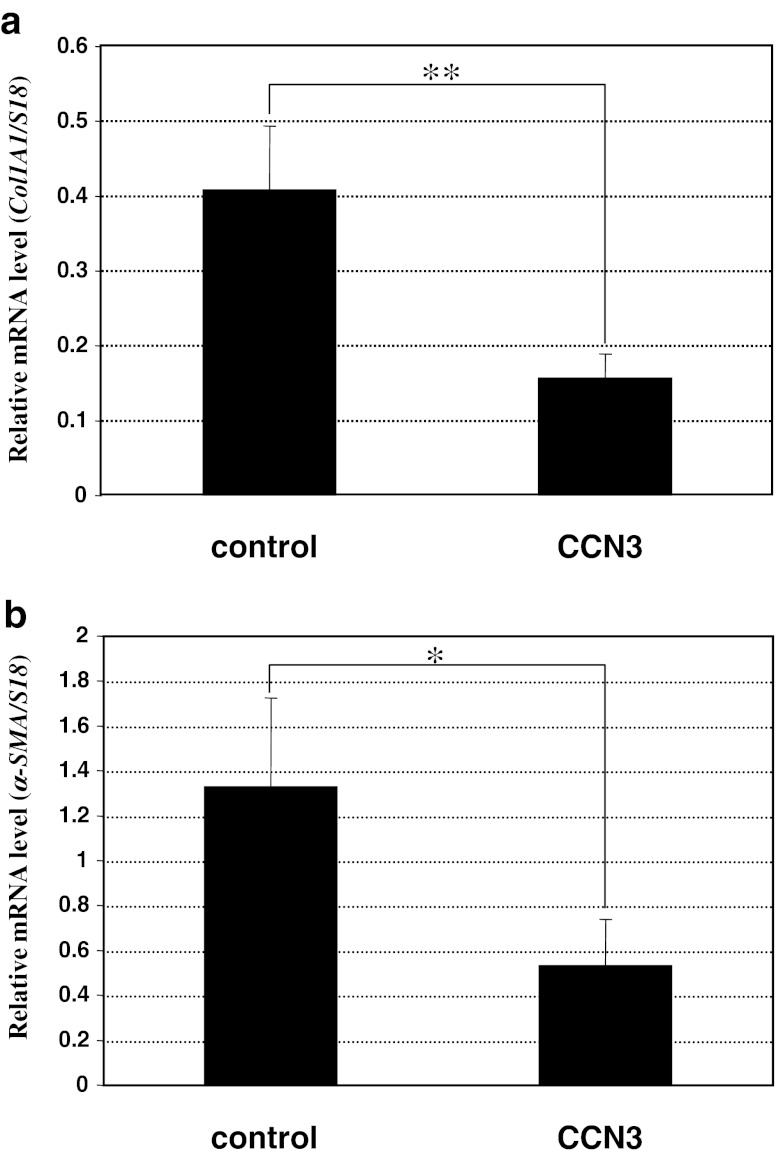
Using this in vitro model for fibrosis, we first evaluated the effect of CCN3 overexpression on the induced fibrotic phenotype. As expected, a significant level of type I collagen gene expression was observed after TGF-β treatment, which level was strikingly repressed by concomitant overexpression of CCN3 (Fig. 2a). Also, α-smooth muscle actin (α-SMA), which is expressed by the myofibroblasts involved in fibrotic tissue remodeling in a variety of tissues, was induced in the TGF-β-stimulated NIH3T3 cells. Albeit at a less remarkable level, statistically significant repression of α-SMA gene expression was observed by CCN3 overexpression (Fig. 2b). These results are consistent with previous findings obtained with renal and hepatic cells.
Fig. 2.
Repression of TGF-β-induced fibrotic phenotype of NIH3T3 cells by CCN3. Significant repression of the gene expression of 2 fibrotic markers, type I collagen (a) and α-SMA (b), was observed following CCN3 overexpression. Relative mRNA levels were computed by standardizing the values against those of 18 s ribosomal RNA. Evaluation was performed 3 times, and the mean values are given with error bars representing standard deviations. Difference between the data indicated by brackets are statistically significant at p < 0.05 (*) and p = 0.01 (**)
Alteration of the expression profile of the CCN family genes by overexpressed CCN3
The distinct role of CCN2 in developing various fibrotic disorders is now widely recognized. Therefore, it would be suspected that the observed repressive effect of CCN3 on fibrosis was mediated by the negative action of CCN3 against CCN2. In this context, although the effect of CCN3 on the CCN2 gene expression has been evaluated in several types of cells to date, this issue still remains controversial.
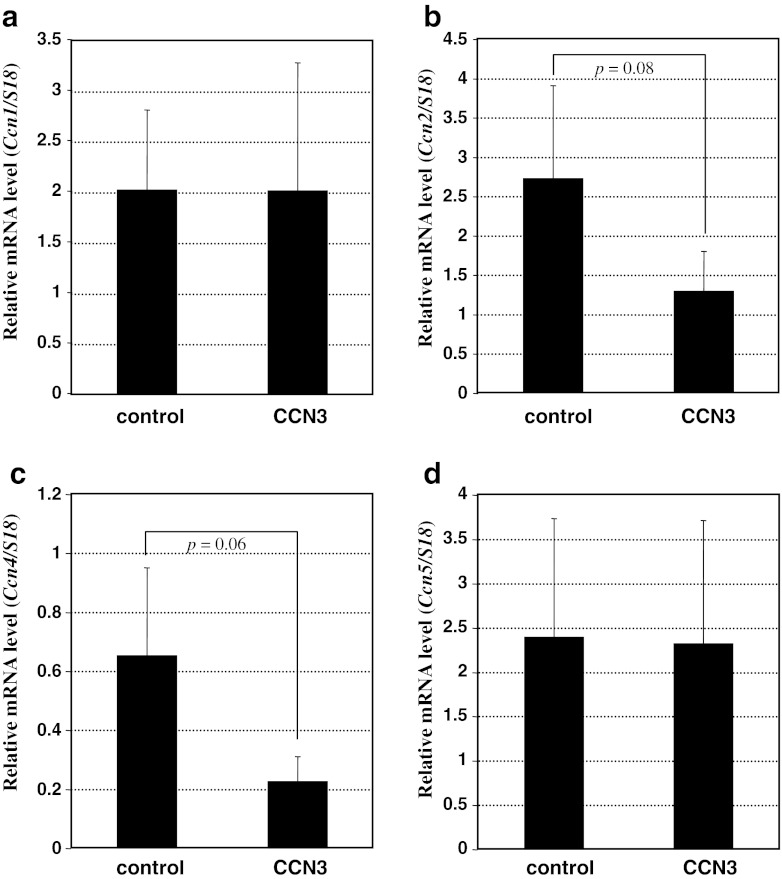
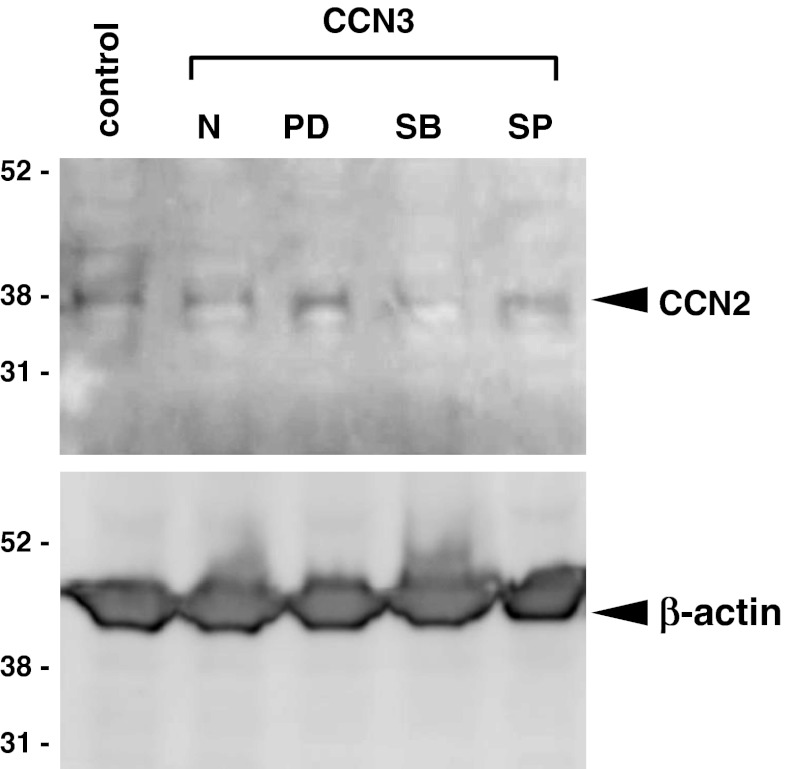
Nevertheless, the effect of CCN3 overproduction on the expression of the entire group of CCN family member genes upon fibrotic stimulation had not been evaluated. Therefore, next we analyzed the expression profile of all of the CCN family members by using the present in vitro model of fibrosis. As already shown in Fig. 1, the expression of endogenous Ccn3 was unaffected. Similarly, no appreciable effect on Ccn1 expression was detected upon CCN3 overexpression (Fig. 3a). Unexpectedly, overexpression of CCN3 caused only a modest decrease in the Ccn2 mRNA level with marginal statistical significance (p = 0.08); whereas Ccn4 showed an even better negative response to CCN3 overexpression (p = 0.06; cf. Fig. 3b and c). Western blotting analysis of CCN2 in the cell lysates revealed the results consistent with these ones at RNA levels, showing a modest reduction in CCN2 production by CCN3-overexpressing cells. It should be noted that the decreased CCN2 production appeared to be retrieved, not by a p38 MAPK inhibitor, but by MEK1/2 and JNK inhibitors (Fig. 4), suggesting possible involvement of the MEK/ERK and JNK signaling pathways in this regulation. In addition, Ccn5 was expressed at significant levels in those cells, regardless of CCN3 overexpression (Fig. 3d); whereas no Ccn6 transcript was detected in either case (data not shown). These findings suggest a minor role of CCN3-induced down-regulation of the CCN2 gene expression in the anti-fibrotic function of CCN3 and also the possible contribution of CCN4 to the development of fibrosis.
Fig. 3.
Effect of CCN3 overexpression on the expression of the other CCN family members in TGF-β-stimulated NIH3T3 cells. The mRNA levels of Ccn1 (a), Ccn2 (b), Ccn4 (c), and Ccn5 (d), which were standardized against those of 18 s ribosomal RNA, in the cells infected with parental (control) or CCN3-expressing retroviral vector (CCN3) are comparatively shown. Ccn6 expression was not detectable in the cells. Data are represented as mean values computed from 3 independent experiments with error bars indicating standard deviations. Differences with marginal statistical significance between 2 data, as indicated by the bracket, are shown with the respective p values
Fig. 4.
Effect of CCN3 overexpression on the CCN2 protein production and its dependency on MAPK signaling. CCN2 protein in the TGF-β-stimulated NIH3T3 cells infected with parental (control) or CCN3-expressing retroviral vector (CCN3) in the absence (N) and presence of a MEK1/2 inhibitor (PD), p38 MAPK inhibitor (SB), or JNK inhibitor (SP) was detected by Western blotting. Signals representing β-actin are also displayed as loading controls. Experiments were repeated twice, yielding comparable results, and representative data are shown. Positions of molecular weight markers are depicted at the left in kilodaltons
Discussion
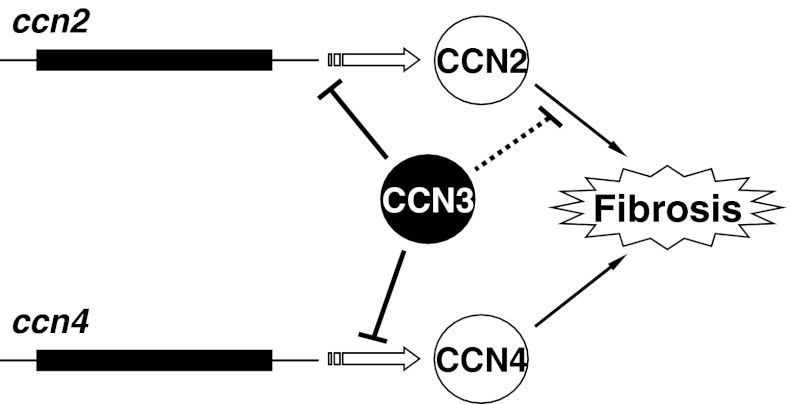
By utilizing a simple in vitro model, we found that overexpression of CCN3 distinctly acted against the development of fibrosis that was induced by TGF-β. Of note, expression of both CCN2 and CCN4 genes was modestly repressed during this process. Since accumulating evidence firmly indicates the critical role of CCN2 in a variety of fibrotic disorders, the repressive action of CCN3 on CCN2 gene expression itself was expected, as was clearly shown in a previous study with kidney cells (Riser et al. 2009). It is suspected that the marginal statistical significance observed here may well be due to type 2 error. Nevertheless, this effect on the CCN2 gene was not strong enough to ascribe the anti-fibrotic effect of CCN3 solely to the negative regulation of the CCN2 gene in our system. More interestingly, we found that the expression of another member of the CCN family was also affected by CCN3 expression. Although not so many reports are available as those for CCN2, the profibrotic property of CCN4 has also been described as well. Indeed, CCN4 is defined as a prohypertrophic and profibrotic growth factor for cardiac fibroblasts, based on a number of data obtained with mouse experimental cardiac hypertrophy model and primary cell culture systems (Colston et al. 2007). Elevated expression of the CCN4 gene is observed in the lungs of patients with idiopathic pulmonary fibrosis, and exogenous CCN4 promotes the production of ECM by lung fibroblasts (Königshoff et al. 2009). As it is also clear that CCN2 plays a significant role in the development of fibrosis both in heart and kidney, we may propose that CCN2 and CCN4 collaborate together during the development of cardiac and pulmonary fibrosis. CCN3 appears to act simultaneously against the gene expression of 2 key profibrotic factors, CCN2 and CCN4, thus which resulting in strong repression of the transition to the fibrotic phenotype (Fig. 5).
Fig. 5.
Possible mechanism of the anti-fibrotic action of CCN3 in fibroblasts. CCN3 represses the gene expression of CCN2 and CCN4, both of which are profibrotic CCN family members. Additionally, CCN3 was shown to interact directly with CCN2, which interaction may restrict the molecular action of CCN2 in the development of fibrosis
The mechanism as to how CCN3 repressed the gene expression of these 2 CCN family members is unclear. Although our Western blotting system for CCN4 was not sensitive enough to quantitatively evaluate the CCN4 protein in NIH3T3 cells (data not shown), our present data at least suggest that the MEK/ERK and JNK signaling pathways may play a role in the process of Ccn2 repression by CCN3. Additionally, we may speculate that CCN3 may be directly inhibiting the transcription of CCN2 and CCN4 genes. It is reported that CCN3 is present in the nucleus and behaves like a transcription factor therein (Planque et al. 2006). Of our interest, CCN5, another CCN family member, actually regulates gene expression in the nucleus (Sabbah et al. 2011). Investigation of the mechanism of this CCN3 action is of great scientific interest as well.
In addition to the effect of CCN3 at the level of gene expression, one may not overlook the direct and indirect molecular interaction of CCN3 with CCN2 and possibly, with CCN4. Such interactions may interfere with the molecular action of these profibrotic proteins, thus leading to attenuation of their fibrotic actions. Interestingly, a recent study revealed that CCN2 and CCN3 directly interact physically with each other, which interaction may have a significant impact on their functions (Hoshijima et al. 2012). If so, the anti-fibrotic action of CCN3 may be partly supported by the sequestration of CCN2 via direct binding to CCN3 (Fig. 5). Also, since all of the CCN family members are highly interactive, direct interaction of CCN3 with CCN4 is considerable and thus should be investigated as well. In addition, CCN2 and CCN3 are reported to differentially regulate chondrocytes and osteoblasts via direct interaction with a common binding counterpart, bone morphogenetic protein 2, which is a critical molecule in cartilage and bone development (Canalis 2007; Smerdel-Ramoya et al. 2008; Kawaki et al. 2008, 2011; Maeda et al. 2009). A similar molecular mechanism mediated by a common cofactor may thus be considered for understanding the regulation of fibrosis by CCN2 and CCN3.
According to past studies, in which physiological effects of CCN family members were investigated, most CCN family proteins, except for CCN3 and CCN6, were found to promote certain aspects of cellular activities. In contrast, wild-type CCN3 has been mostly described as a novel molecule with repressive function except for its positive effect on articular cartilage development (Janune et al. 2011). Consistent with such findings, the present study suggests that CCN3 plays a distinct role as “yin”; whereas that of “yang” may be played by the collaboration of several other CCN family members. However, it should noted that a recent report indicated that CCN5, which is the only CCN family member lacking the CT module, functions to counteract CCN2 (Yoon et al. 2010). Thus, comprehensive analysis of all of the CCN members is of critical importance for better understanding of the functional properties of the CCN family members.
Acknowledgments
This study was supported by grants from the program Grants-in-aid for Scientific Research (S)(B) to M.T. and (C) to S.K. from the Japan Society for the Promotion of Science and by a research grant from Terumo Life Science Foundation to S.K. The authors would like to thank Matthew L. Springer at UCSF, who gave us the Ecotropic Phoenix cell line.
Contributor Information
Satoshi Kubota, Phone: +81-86-2356646, FAX: +81-86-2356649, Email: kubota1@md.okayama-u.ac.jp.
Masaharu Takigawa, Phone: +81-86-2356646, FAX: +81-86-2356649, Email: takigawa@md.okayama-u.ac.jp.
References
- Borkham-Kamphorst E, van Roeyen CR, Van de Leur E, Floege J, Weiskirchen R. CCN3/NOV small interfering RNA enhances fibrogenic gene expression in primary hepatic stellate cells and cirrhotic fat storing cell line CFSC. J Cell Commun Signal. 2012;6:11–25. doi: 10.1007/s12079-011-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigstock DR, Goldschmeding R, Katsube K, LamSCT LLF, Lyons K, Naus C, Perbal B, Riser B, Takigawa M, Yeger H. Proposal for a unified CCN nomenclature. Mol Pathol. 2003;56:127–128. doi: 10.1136/mp.56.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E. Nephroblastoma overexpressed (Nov) is a novel bone morphogenetic protein antagonist. Ann N Y Acad Sci. 2007;1116:50–58. doi: 10.1196/annals.1402.055. [DOI] [PubMed] [Google Scholar]
- Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41:771–783. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colston JT, de la Rosa SD, Koehler M, Gonzales K, Mestril R, Freeman GL, Bailey SR, Chandrasekar B. Wnt-induced secreted protein-1 is a prohypertrophic and profibrotic growth factor. Am J Physiol Heart Circ Physiol. 2007;293:H1839–H1846. doi: 10.1152/ajpheart.00428.2007. [DOI] [PubMed] [Google Scholar]
- Grotendorst GR, Okochi H, Hayashi N. A novel transforming growth factor beta response element controls the expression of the connective tissue growth factor gene. Cell Growth Differ. 1996;7:469–480. [PubMed] [Google Scholar]
- Hoshijima M, Hattori T, Aoyama E, Nishida T, Yamashiro T, Takigawa M. Role of heterotypic CCN2/CTGF-CCN3/NOV and homotypic CCN2-CCN2 interactions in expression of the differentiated phenotype of chondrocytes. FEBS J. 2012;279:3584–3597. doi: 10.1111/j.1742-4658.2012.08717.x. [DOI] [PubMed] [Google Scholar]
- Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janune D, Kubota S, Nishida T, Kawaki H, Perbal B, Iida S, Takigawa M. Novel effects of CCN3 that may direct the differentiation of chondrocytes. FEBS Lett. 2011;585:3033–3040. doi: 10.1016/j.febslet.2011.08.024. [DOI] [PubMed] [Google Scholar]
- Kawaki H, Kubota S, Suzuki A, Lazar N, Yamada T, Matsumura T, Ohgawara T, Maeda T, Perbal B, Lyons KM, Takigawa M. Cooperative regulation of chondrocyte differentiation by CCN2 and CCN3 shown by a comprehensive analysis of the CCN family proteins in cartilage. J Bone Miner Res. 2008;23:1751–1764. doi: 10.1359/jbmr.080615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaki H, Kubota S, Suzuki A, Suzuki M, Kohsaka K, Hoshi K, Fujii T, Lazar N, Ohgawara T, Maeda T, Perbal B, Takano-Yamamoto T, Takigawa M. Differential roles of CCN family proteins during osteoblast differentiation: Involvement of Smad and MAPK signaling pathways. Bone. 2011;49:975–989. doi: 10.1016/j.bone.2011.06.033. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Kubota S, Asaumi K, Kawaki H, Nishida T, Kawata K, Mitani S, Tabata Y, Ozaki T, Takigawa M. Promotion of bone regeneration by CCN2 incorporated into gelatin hydrogel. Tissue Eng Part A. 2008;14:1089–1098. doi: 10.1089/ten.tea.2007.0167. [DOI] [PubMed] [Google Scholar]
- Königshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, Günther A, Eickelberg O. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest. 2009;119:772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S (2012) CCN2 and orofacial tissue development and remodeling. Jpn Dent Sci Rev. in press, doi:10.1016/j.jdsr.2012.02.002
- Kubota S, Takigawa M. Role of CCN2/CTGF/Hcs24 in bone growth. Int Rev Cytol. 2007;257:1–41. doi: 10.1016/S0074-7696(07)57001-4. [DOI] [PubMed] [Google Scholar]
- Kubota S, Takigawa M. The role of CCN2 in cartilage and bone development. J Cell Commun Signal. 2011;5:209–217. doi: 10.1007/s12079-011-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A. Yin and Yang: CCN3 inhibits the pro-fibrotic effects of CCN2. J Cell Commun Signal. 2009;3:161–162. doi: 10.1007/s12079-009-0056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. 2006;119:4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- Leask A, Parapuram SK, Shi-Wen X, Abraham DJ. Connective tissue growth factor (CTGF, CCN2) gene regulation: a potent clinical bio-marker of fibroproliferative disease? J Cell Commun Signal. 2009;3:89–94. doi: 10.1007/s12079-009-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Nishida T, Aoyama E, Kubota S, Lyons KM, Kuboki T, Takigawa M. CCN family 2/connective tissue growth factor modulates BMP signalling as a signal conductor, which action regulates the proliferation and differentiation of chondrocytes. J Biochem. 2009;145:207–216. doi: 10.1093/jb/mvn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Kubota S, Fujisawa T, Sonoyama W, Kawaki H, Akiyama K, Shimono K, Oshima M, Nishida T, Yoshida Y, Suzuki K, Takigawa M, Kuboki T. Promotion of hydroxyapatite-associated, stem cell-based bone regeneration by CCN2. Cell Transplant. 2008;17:231–240. doi: 10.3727/000000008783907143. [DOI] [PubMed] [Google Scholar]
- Perbal B, Takigawa M. CCN Protein -A new family of cell growth and differentiation regulators- London: Imperial College Press; 2005. pp. 1–311. [Google Scholar]
- Planque N, Long Li C, Saule S, Bleau AM, Perbal B. Nuclear addressing provides a clue for the transforming activity of amino-truncated CCN3 proteins. J Cell Biochem. 2006;99:105–116. doi: 10.1002/jcb.20887. [DOI] [PubMed] [Google Scholar]
- Riser BL, Najmabadi F, Perbal B, Peterson DR, Rambow JA, Riser ML, Sukowski E, Yeger H, Riser SC. CCN3 (NOV) is a negative regulator of CCN2 (CTGF) and a novel endogenous inhibitor of the fibrotic pathway in an in vitro model of renal disease. Am J Pathol. 2009;174:1725–1734. doi: 10.2353/ajpath.2009.080241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah M, Prunier C, Ferrand N, Megalophonos V, Lambein K, De Wever O, Nazaret N, Lachuer J, Dumont S, Redeuilh G. CCN5, a novel transcriptional repressor of the transforming growth factor β signaling pathway. Mol Cell Biol. 2011;31:1459–1469. doi: 10.1128/MCB.01316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdel-Ramoya A, Zanotti S, Deregowski V, Canalis E. Connective tissue growth factor enhances osteoblastogenesis in vitro. J Biol Chem. 2008;283:22690–22699. doi: 10.1074/jbc.M710140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon PO, Lee MA, Cha H, Jeong MH, Kim J, Jang SP, Choi BY, Jeong D, Yang DK, Hajjar RJ, Park WJ. The opposing effects of CCN2 and CCN5 on the development of cardiac hypertrophy and fibrosis. J Mol Cell Cardiol. 2010;49:294–303. doi: 10.1016/j.yjmcc.2010.04.010. [DOI] [PubMed] [Google Scholar]