Abstract
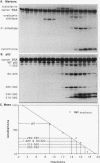
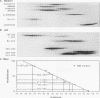
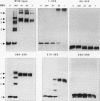
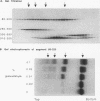
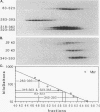
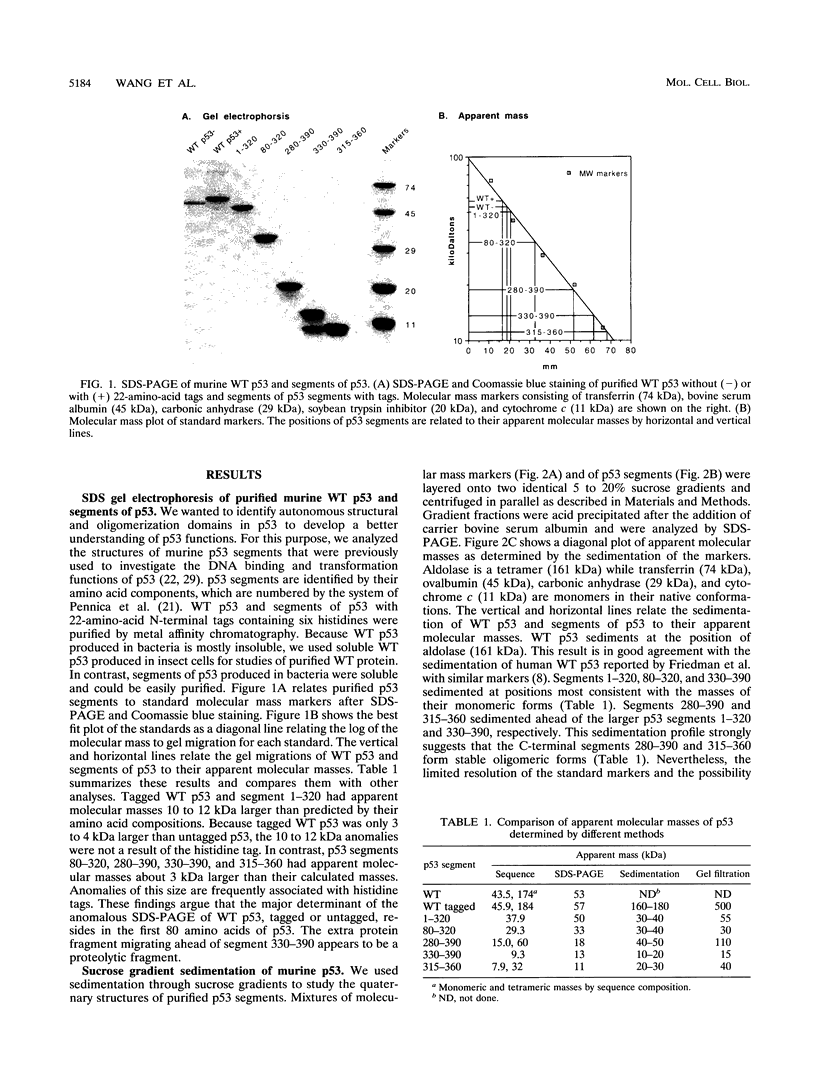
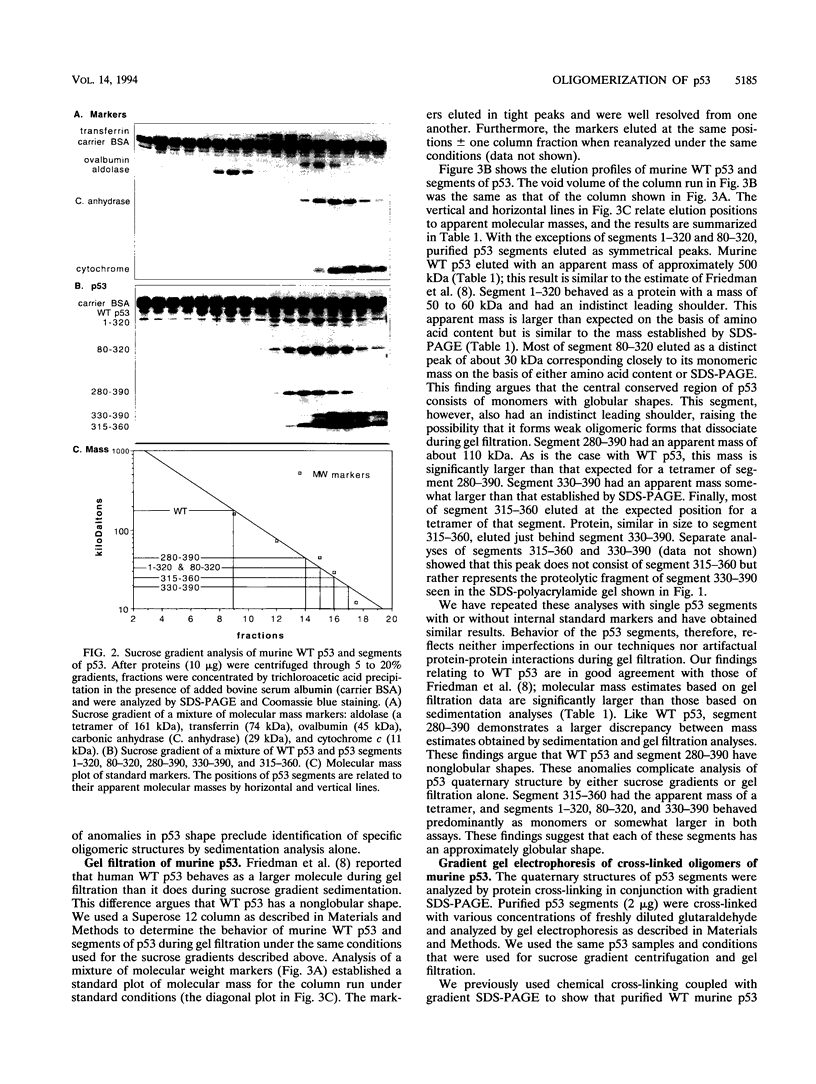
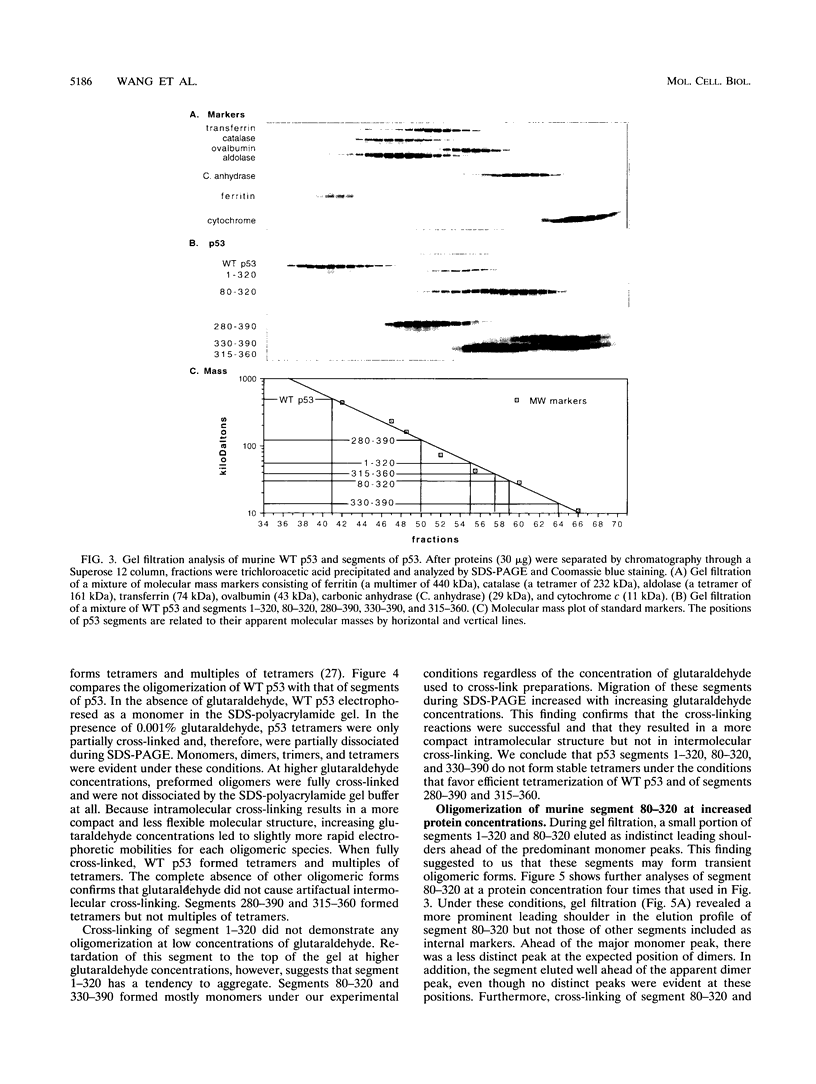
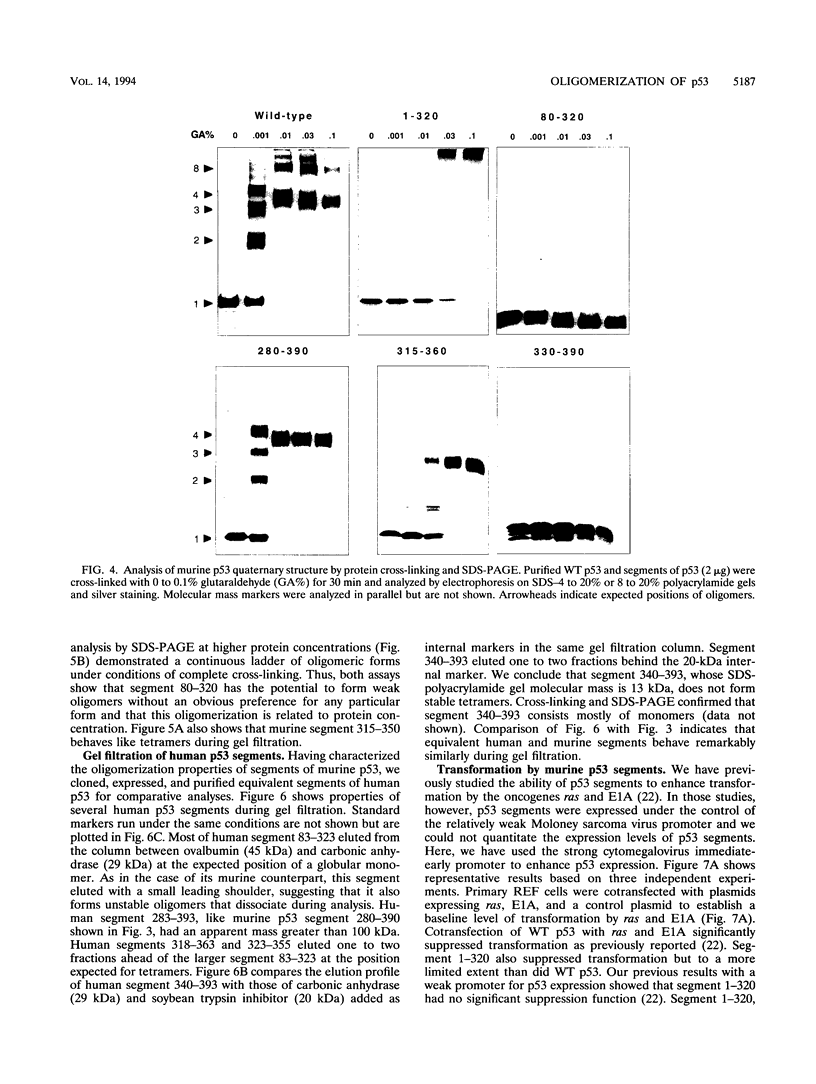
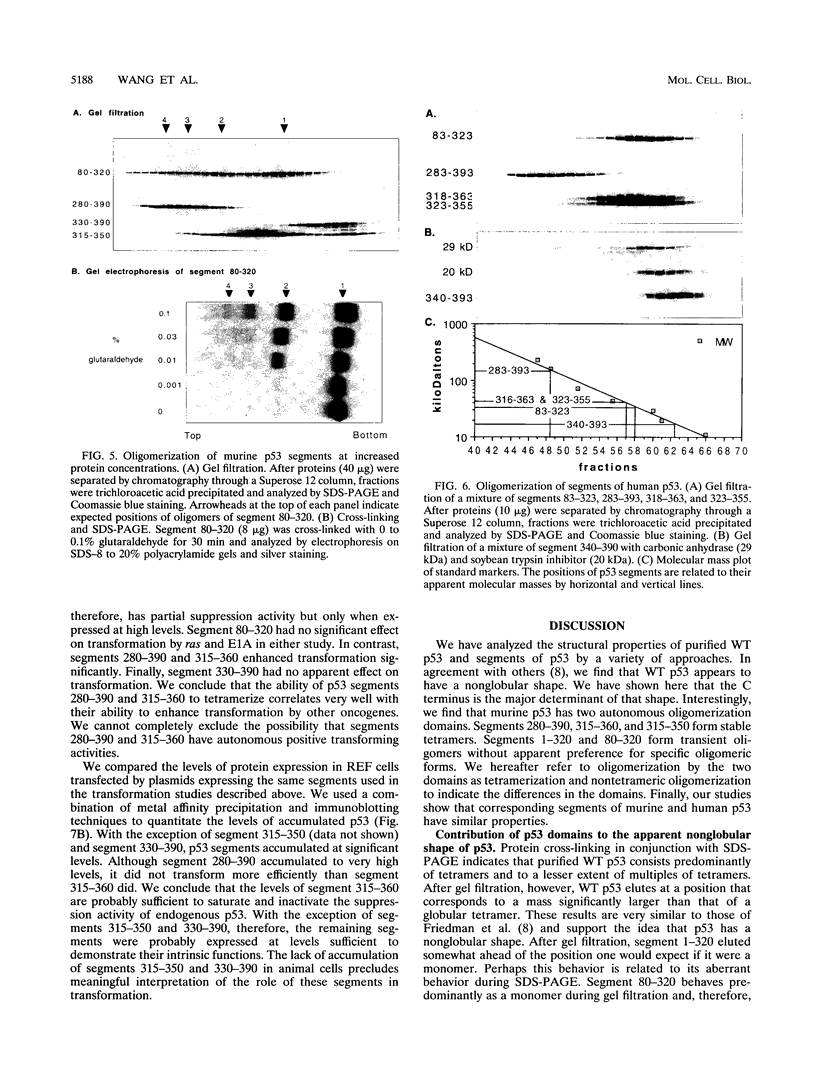
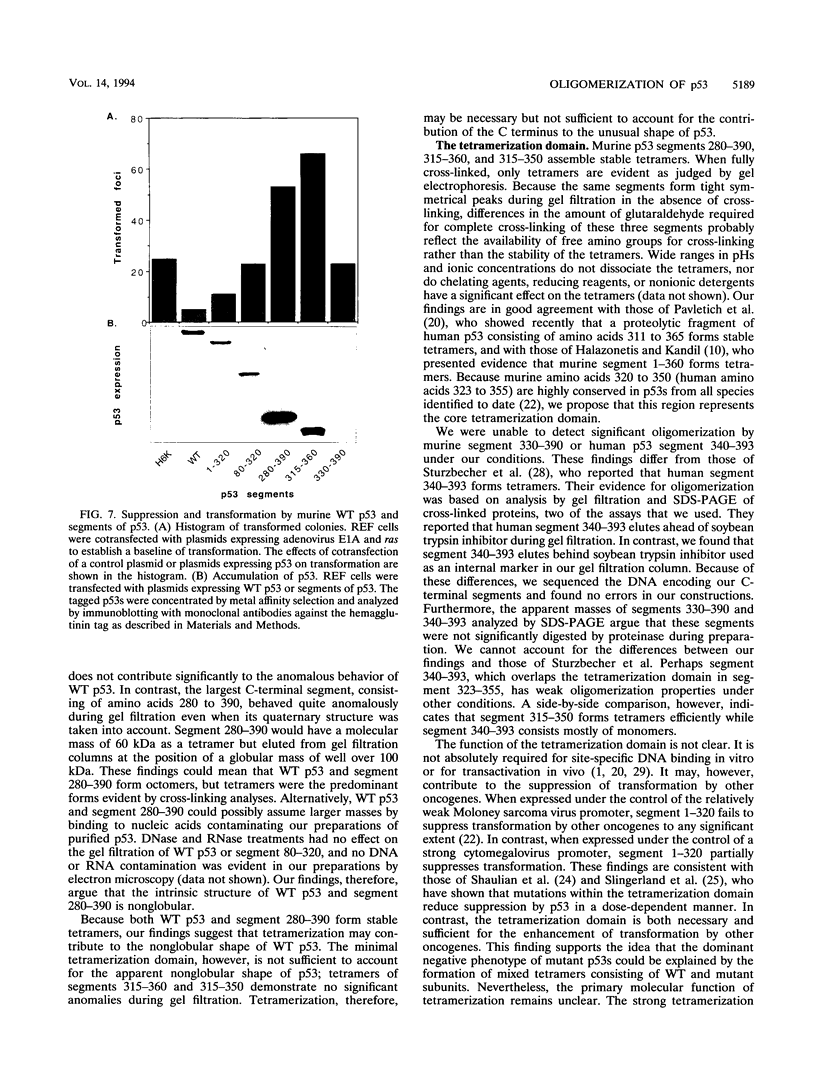
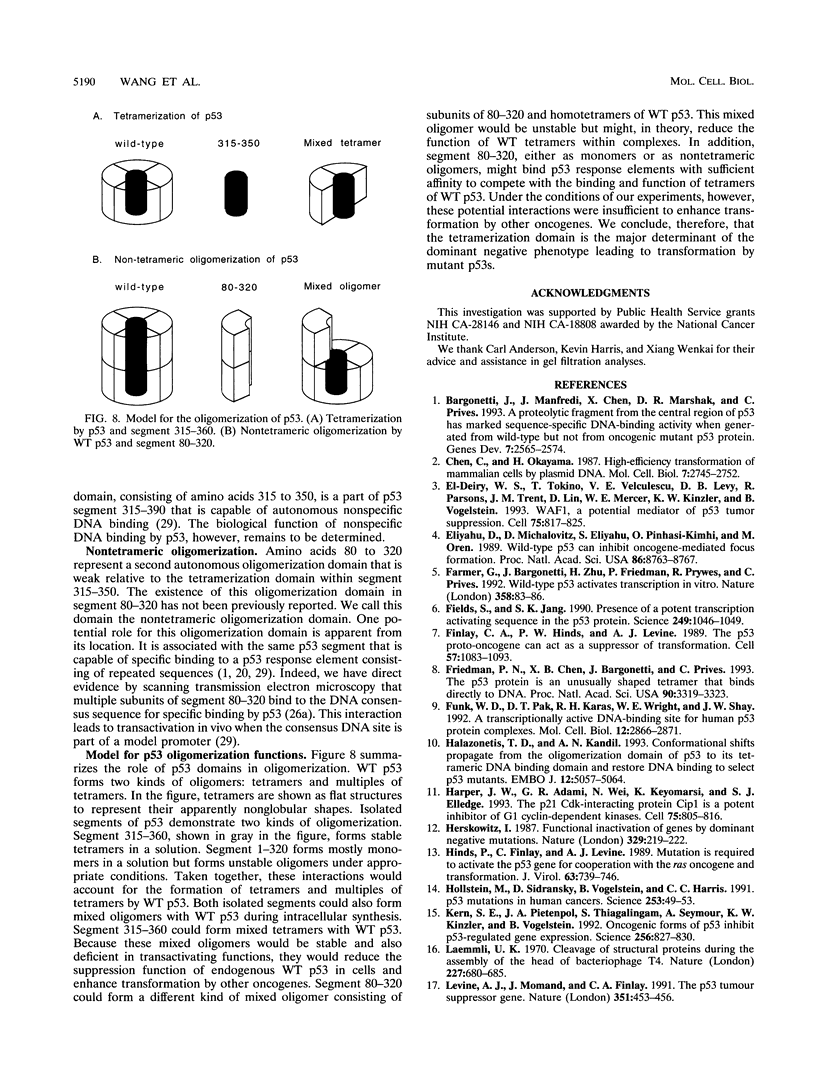
Wild-type p53 forms tetramers and multiples of tetramers. Friedman et al. (P. N. Friedman, X. B. Chen, J. Bargonetti, and C. Prives, Proc. Natl. Acad. Sci. USA 90:3319-3323, 1993) have reported that human p53 behaves as a larger molecule during gel filtration than it does during sucrose gradient sedimentation. These differences argue that wild-type p53 has a nonglobular shape. To identify structural and oligomerization domains in p53, we have investigated the physical properties of purified segments of p53. The central, specific DNA-binding domain within murine amino acids 80 to 320 and human amino acids 83 to 323 behaves predominantly as monomers during analysis by sedimentation, gel filtration, and gel electrophoresis. This consistent behavior argues that the central region of p53 is globular in shape. Under appropriate conditions, however, this segment can form transient oligomers without apparent preference for a single oligomeric structure. This region does not enhance transformation by other oncogenes. The biological implications of transient oligomerization by this central segment, therefore, remain to be demonstrated. Like wild-type p53, the C terminus, consisting of murine amino acids 280 to 390 and human amino acids 283 to 393, behaves anomalously during gel filtration and apparently has a nonglobular shape. Within this region, murine amino acids 315 to 350 and human amino acids 323 to 355 are sufficient for assembly of stable tetramers. The finding that murine amino acids 315 to 360 enhance transformation by other oncogenes strongly supports the role of p53 tetramerization in oncogenesis. Amino acids 330 to 390 of murine p53 and amino acids 340 to 393 of human p53, which have been implicated by Sturzbecher et al. in tetramerization (H.-W. Sturzbecher, R. Brain, C. Addison, K. Rudge, M. Remm, M. Grimaldi, E. Keenan, and J. R. Jenkins, Oncogene 7:1513-1523, 1992), do not form stable tetramers under our conditions. Our findings indicate that p53 has at least two autonomous oligomerization domains: a strong tetramerization domain in its C-terminal region and a weaker oligomerization domain in the central DNA binding region of p53. Together, these domains account for the formation of tetramers and multiples of tetramers by wild-type p53. The tetramerization domain is the major determinant of the dominant negative phenotype leading to transformation by mutant p53s.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bargonetti J., Manfredi J. J., Chen X., Marshak D. R., Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993 Dec;7(12B):2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliyahu D., Michalovitz D., Eliyahu S., Pinhasi-Kimhi O., Oren M. Wild-type p53 can inhibit oncogene-mediated focus formation. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8763–8767. doi: 10.1073/pnas.86.22.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer G., Bargonetti J., Zhu H., Friedman P., Prywes R., Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992 Jul 2;358(6381):83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- Fields S., Jang S. K. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990 Aug 31;249(4972):1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- Finlay C. A., Hinds P. W., Levine A. J. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989 Jun 30;57(7):1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- Friedman P. N., Chen X., Bargonetti J., Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk W. D., Pak D. T., Karas R. H., Wright W. E., Shay J. W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992 Jun;12(6):2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993 Dec 15;12(13):5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993 Nov 19;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987 Sep 17;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Hinds P., Finlay C., Levine A. J. Mutation is required to activate the p53 gene for cooperation with the ras oncogene and transformation. J Virol. 1989 Feb;63(2):739–746. doi: 10.1128/jvi.63.2.739-746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Kern S. E., Pietenpol J. A., Thiagalingam S., Seymour A., Kinzler K. W., Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992 May 8;256(5058):827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine A. J., Momand J., Finlay C. A. The p53 tumour suppressor gene. Nature. 1991 Jun 6;351(6326):453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Milner J., Medcalf E. A., Cook A. C. Tumor suppressor p53: analysis of wild-type and mutant p53 complexes. Mol Cell Biol. 1991 Jan;11(1):12–19. doi: 10.1128/mcb.11.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Chambers K. A., Pabo C. O. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993 Dec;7(12B):2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- Pennica D., Goeddel D. V., Hayflick J. S., Reich N. C., Anderson C. W., Levine A. J. The amino acid sequence of murine p53 determined from a c-DNA clone. Virology. 1984 Apr 30;134(2):477–482. doi: 10.1016/0042-6822(84)90316-7. [DOI] [PubMed] [Google Scholar]
- Reed M., Wang Y., Mayr G., Anderson M. E., Schwedes J. F., Tegtmeyer P. p53 domains: suppression, transformation, and transactivation. Gene Expr. 1993;3(1):95–107. [PMC free article] [PubMed] [Google Scholar]
- Shaulian E., Zauberman A., Ginsberg D., Oren M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992 Dec;12(12):5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E., Zauberman A., Milner J., Davies E. A., Oren M. Tight DNA binding and oligomerization are dispensable for the ability of p53 to transactivate target genes and suppress transformation. EMBO J. 1993 Jul;12(7):2789–2797. doi: 10.1002/j.1460-2075.1993.tb05940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingerland J. M., Jenkins J. R., Benchimol S. The transforming and suppressor functions of p53 alleles: effects of mutations that disrupt phosphorylation, oligomerization and nuclear translocation. EMBO J. 1993 Mar;12(3):1029–1037. doi: 10.1002/j.1460-2075.1993.tb05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Roth J. A., Maxwell S. A. Sequence-specific interaction of a conformational domain of p53 with DNA. Cancer Res. 1993 Nov 15;53(22):5361–5364. [PubMed] [Google Scholar]
- Stenger J. E., Mayr G. A., Mann K., Tegtmeyer P. Formation of stable p53 homotetramers and multiples of tetramers. Mol Carcinog. 1992;5(2):102–106. doi: 10.1002/mc.2940050204. [DOI] [PubMed] [Google Scholar]
- Stürzbecher H. W., Brain R., Addison C., Rudge K., Remm M., Grimaldi M., Keenan E., Jenkins J. R. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992 Aug;7(8):1513–1523. [PubMed] [Google Scholar]
- Wang Y., Reed M., Wang P., Stenger J. E., Mayr G., Anderson M. E., Schwedes J. F., Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993 Dec;7(12B):2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993 Nov 19;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]