Abstract
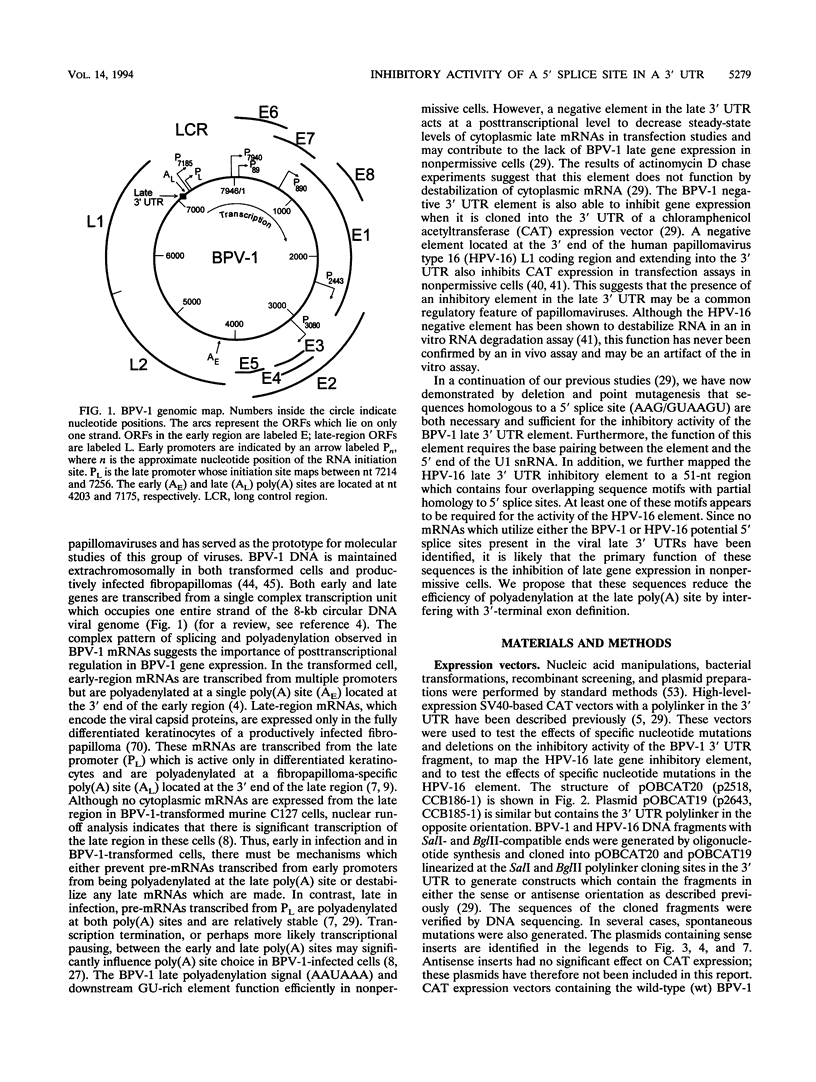
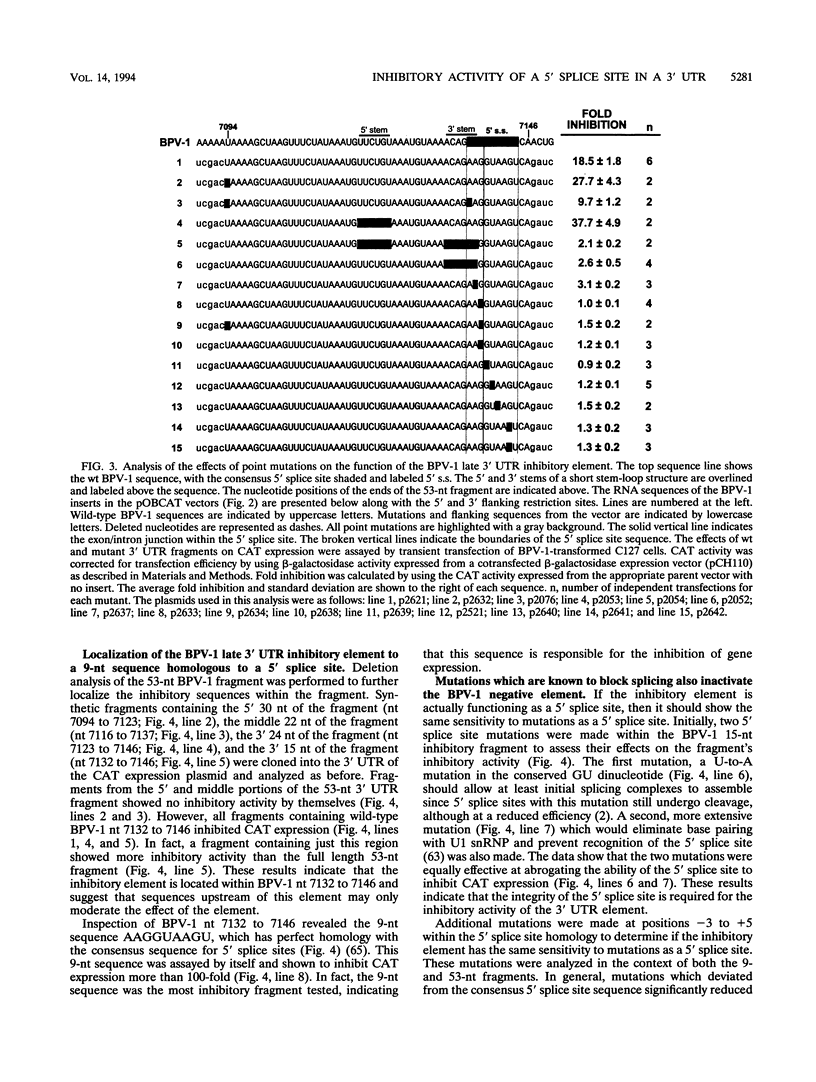
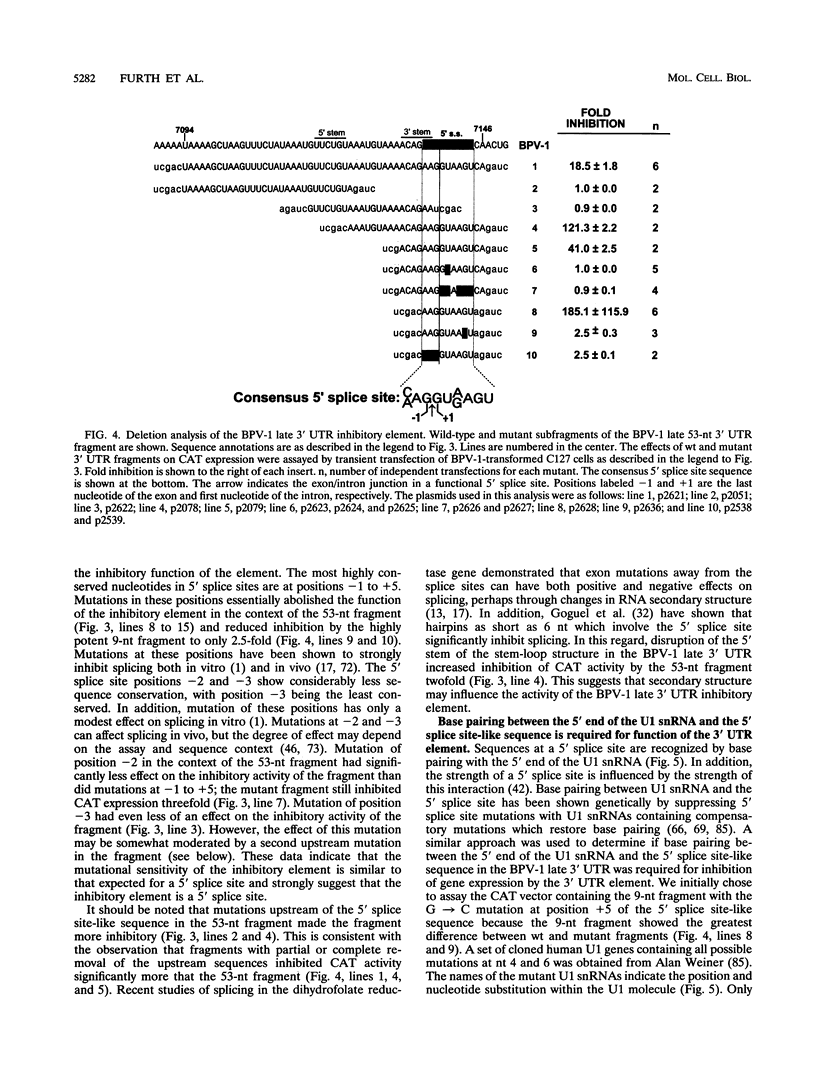
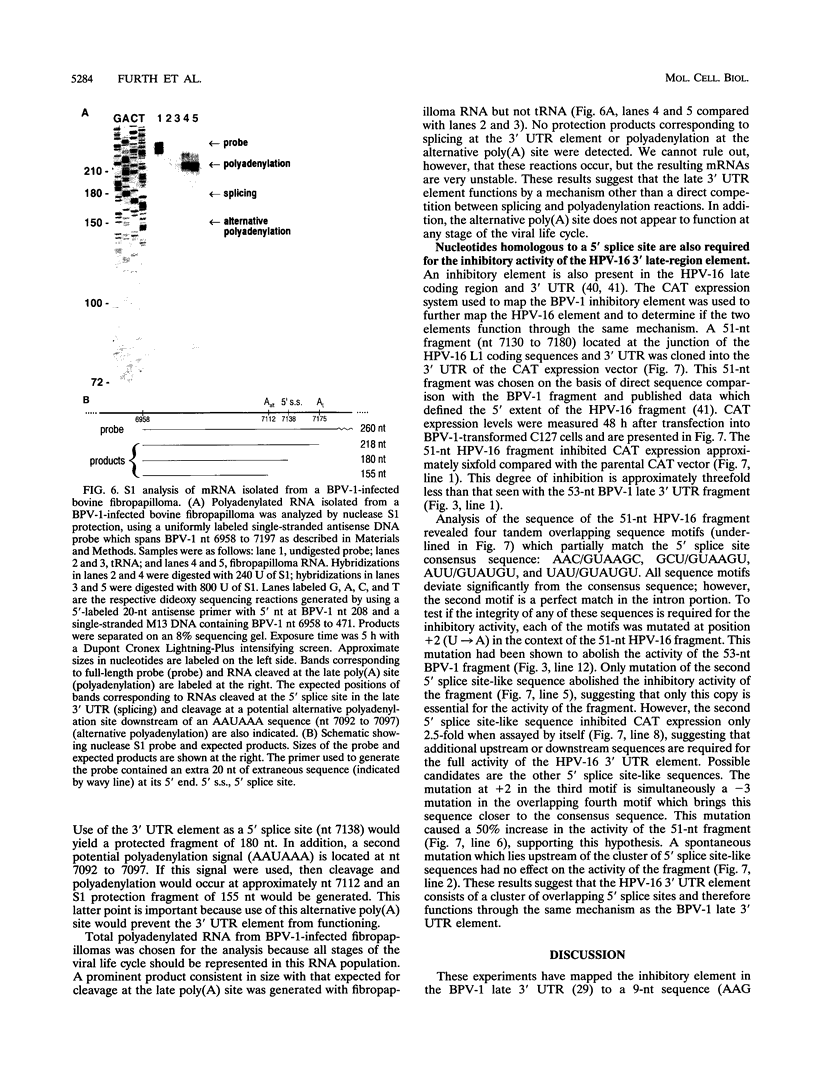
Expression of bovine papillomavirus type 1 (BPV-1) late genes is limited to terminally differentiated keratinocytes in an infected epithelium. We have previously shown that although the BPV-1 late polyadenylation site is functional in nonpermissive cells, a 53-nucleotide (nt) fragment of the late 3' untranslated region acts posttranscriptionally to reduce polyadenylated cytoplasmic RNA levels. This 53-nt fragment does not appear to function by destabilizing polyadenylated cytoplasmic RNA (P. A. Furth and C. C. Baker, J. Virol. 65:5806-5812, 1991). In this study, we used site-directed mutagenesis and deletion analysis to demonstrate that the sequence AAG/GUAAGU, which is identical to the consensus 5' splice site sequence, was both necessary and sufficient for the inhibitory activity of the 53-nt fragment. Furthermore, base pairing between the 5' end of the U1 small nuclear RNA and this 5' splice site-like sequence was shown to be required for the inhibitory activity in vivo. We have also further mapped the human papillomavirus type 16 late 3' inhibitory element (I. M. Kennedy, J. K. Haddow, and J. B. Clements, J. Virol. 65:2093-2097, 1991) to a 51-nt region containing four overlapping sequence motifs with partial homology to 5' splice sites. Mutation of each of these motifs demonstrated that only one of these motifs is required for the inhibitory activity. However, the presence of the other motifs may contribute to the full inhibitory activity of the element. No BPV-1 or human papillomavirus type 16 mRNAs which are spliced by using the potential 5' splice sites present in the viral late 3' untranslated regions have been identified. This suggests that the primary function of these 5' splice site-like sequences is the inhibition of late gene expression. The most likely mechanism of action of these elements is reduction of polyadenylation efficiency, perhaps through interference with 3'-terminal exon definition.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. C., Howley P. M. Differential promoter utilization by the bovine papillomavirus in transformed cells and productively infected wart tissues. EMBO J. 1987 Apr;6(4):1027–1035. doi: 10.1002/j.1460-2075.1987.tb04855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. C., Noe J. S. Transcriptional termination between bovine papillomavirus type 1 (BPV-1) early and late polyadenylation sites blocks late transcription in BPV-1-transformed cells. J Virol. 1989 Aug;63(8):3529–3534. doi: 10.1128/jvi.63.8.3529-3534.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barksdale S. K., Baker C. C. Differentiation-specific expression from the bovine papillomavirus type 1 P2443 and late promoters. J Virol. 1993 Sep;67(9):5605–5616. doi: 10.1128/jvi.67.9.5605-5616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M., Farrell P. J., Barrell B. G. Transcription and DNA sequence of the BamHI L fragment of B95-8 Epstein-Barr virus. EMBO J. 1984 May;3(5):1083–1090. doi: 10.1002/j.1460-2075.1984.tb01933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens W. C., Jansen E. J., van Venrooij W. J., Stripecke R., Mattaj I. W., Gunderson S. I. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993 Mar 26;72(6):881–892. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991 Jul 5;220(1):49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Grunberger D., Chasin L. A. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1993 Aug;13(8):5085–5098. doi: 10.1128/mcb.13.8.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell S., Alwine J. C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989 Oct;9(10):4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Messenger RNA transport and HIV rev regulation. Science. 1990 Aug 10;249(4969):614–615. doi: 10.1126/science.2143313. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989 Dec 1;59(5):789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Chen I. T., Chasin L. A. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol Cell Biol. 1993 Jan;13(1):289–300. doi: 10.1128/mcb.13.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. T., Chasin L. A. Large exon size does not limit splicing in vivo. Mol Cell Biol. 1994 Mar;14(3):2140–2146. doi: 10.1128/mcb.14.3.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington J., Ganem D. Regulation of polyadenylation in human immunodeficiency virus (HIV): contributions of promoter proximity and upstream sequences. EMBO J. 1992 Apr;11(4):1513–1524. doi: 10.1002/j.1460-2075.1992.tb05196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington J., Russnak R., Ganem D. Upstream sequences and cap proximity in the regulation of polyadenylation in ground squirrel hepatitis virus. J Virol. 1992 Dec;66(12):7589–7596. doi: 10.1128/jvi.66.12.7589-7596.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou H. C., Dabrowski C., Alwine J. C. Simian virus 40 late mRNA leader sequences involved in augmenting mRNA accumulation via multiple mechanisms, including increased polyadenylation efficiency. J Virol. 1991 Dec;65(12):6677–6685. doi: 10.1128/jvi.65.12.6677-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- DeZazzo J. D., Falck-Pedersen E., Imperiale M. J. Sequences regulating temporal poly(A) site switching in the adenovirus major late transcription unit. Mol Cell Biol. 1991 Dec;11(12):5977–5984. doi: 10.1128/mcb.11.12.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J., Parton A., Hartley K., Banks L., Crook T., Stanley M., Crawford L. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology. 1990 Sep;178(1):254–262. doi: 10.1016/0042-6822(90)90401-c. [DOI] [PubMed] [Google Scholar]
- Edery I., Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris P., Levitt N., Briggs D., Proudfoot N. J. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991 Jul;10(7):1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D., Mayeda A., Maniatis T., Krainer A. R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5' and 3' splice site selection. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth P. A., Baker C. C. An element in the bovine papillomavirus late 3' untranslated region reduces polyadenylated cytoplasmic RNA levels. J Virol. 1991 Nov;65(11):5806–5812. doi: 10.1128/jvi.65.11.5806-5812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin G. M., Fleming E. S., Oetjen J. Activation of HIV-1 pre-mRNA 3' processing in vitro requires both an upstream element and TAR. EMBO J. 1992 Dec;11(12):4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin G. M., Nevins J. R. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989 Dec;3(12B):2180–2190. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- Goguel V., Wang Y., Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol Cell Biol. 1993 Nov;13(11):6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson S. I., Beyer K., Martin G., Keller W., Boelens W. C., Mattaj L. W. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994 Feb 11;76(3):531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B. E., Grabowski P. J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3' splice site by a network of interactions spanning the exon. Genes Dev. 1992 Dec;6(12B):2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- Horabin J. I., Schedl P. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5' splice site. Mol Cell Biol. 1993 Dec;13(12):7734–7746. doi: 10.1128/mcb.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. T., Gorman C. M. Intervening sequences increase efficiency of RNA 3' processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990 Feb 25;18(4):937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale M. J., DeZazzo J. D. Poly(A) site choice in retroelements: deja vu all over again? New Biol. 1991 Jun;3(6):531–537. [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy I. M., Haddow J. K., Clements J. B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J Virol. 1991 Apr;65(4):2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy I. M., Haddow J. K., Clements J. B. Analysis of human papillomavirus type 16 late mRNA 3' processing signals in vitro and in vivo. J Virol. 1990 Apr;64(4):1825–1829. doi: 10.1128/jvi.64.4.1825-1829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Lamond A. I. ASF/SF2: a splice site selector. Trends Biochem Sci. 1991 Dec;16(12):452–453. doi: 10.1016/0968-0004(91)90176-v. [DOI] [PubMed] [Google Scholar]
- Lancaster W. D. Apparent lack of integration of bovine papillomavirus DNA in virus-induced equine and bovine tumor cells and virus-transformed mouse cells. Virology. 1981 Jan 30;108(2):251–255. doi: 10.1016/0042-6822(81)90433-5. [DOI] [PubMed] [Google Scholar]
- Law M. F., Lowy D. R., Dvoretzky I., Howley P. M. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc Natl Acad Sci U S A. 1981 May;78(5):2727–2731. doi: 10.1073/pnas.78.5.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear A. L., Eperon L. P., Wheatley I. M., Eperon I. C. Hierarchy for 5' splice site preference determined in vivo. J Mol Biol. 1990 Jan 5;211(1):103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Linz U., Baker C. C. Promoters of bovine papillomavirus type 1: in vitro activity and utilization. J Virol. 1988 Aug;62(8):2537–2543. doi: 10.1128/jvi.62.8.2537-2543.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Carmichael G. G. Splice site choice in a complex transcription unit containing multiple inefficient polyadenylation signals. Mol Cell Biol. 1991 Oct;11(10):5291–5300. doi: 10.1128/mcb.11.10.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C. S., Alwine J. C. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994 Mar 1;8(5):576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- Mangin M., Ares M., Jr, Weiner A. M. U1 small nuclear RNA genes are subject to dosage compensation in mouse cells. Science. 1985 Jul 19;229(4710):272–275. doi: 10.1126/science.2409601. [DOI] [PubMed] [Google Scholar]
- Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991 Jan 4;251(4989):33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Helfman D. M., Krainer A. R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993 May;13(5):2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. T., Stoltzfus C. M. Two distant upstream regions containing cis-acting signals regulating splicing facilitate 3'-end processing of avian sarcoma virus RNA. J Virol. 1992 Jul;66(7):4242–4251. doi: 10.1128/jvi.66.7.4242-4251.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985 Jul;41(3):845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- Niwa M., MacDonald C. C., Berget S. M. Are vertebrate exons scanned during splice-site selection? Nature. 1992 Nov 19;360(6401):277–280. doi: 10.1038/360277a0. [DOI] [PubMed] [Google Scholar]
- Niwa M., Rose S. D., Berget S. M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990 Sep;4(9):1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- Pandey N. B., Chodchoy N., Liu T. J., Marzluff W. F. Introns in histone genes alter the distribution of 3' ends. Nucleic Acids Res. 1990 Jun 11;18(11):3161–3170. doi: 10.1093/nar/18.11.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Bryman M. B., Peiter M., Cowan C. Exon size affects competition between splicing and cleavage-polyadenylation in the immunoglobulin mu gene. Mol Cell Biol. 1994 Jan;14(1):77–86. doi: 10.1128/mcb.14.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Séraphin B. Who's on first? The U1 snRNP-5' splice site interaction and splicing. Trends Biochem Sci. 1991 May;16(5):187–190. doi: 10.1016/0968-0004(91)90073-5. [DOI] [PubMed] [Google Scholar]
- Schek N., Cooke C., Alwine J. C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol. 1992 Dec;12(12):5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Sherman L., Alloul N., Golan I., Durst M., Baram A. Expression and splicing patterns of human papillomavirus type-16 mRNAs in pre-cancerous lesions and carcinomas of the cervix, in human keratinocytes immortalized by HPV 16, and in cell lines established from cervical cancers. Int J Cancer. 1992 Feb 1;50(3):356–364. doi: 10.1002/ijc.2910500305. [DOI] [PubMed] [Google Scholar]
- Siebel C. W., Fresco L. D., Rio D. C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5' splice site control U1 snRNP binding. Genes Dev. 1992 Aug;6(8):1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Stenlund A., Zabielski J., Ahola H., Moreno-Lopez J., Pettersson U. Messenger RNAs from the transforming region of bovine papilloma virus type I. J Mol Biol. 1985 Apr 20;182(4):541–554. doi: 10.1016/0022-2836(85)90240-2. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantravahi J., Alvira M., Falck-Pedersen E. Characterization of the mouse beta maj globin transcription termination region: a spacing sequence is required between the poly(A) signal sequence and multiple downstream termination elements. Mol Cell Biol. 1993 Jan;13(1):578–587. doi: 10.1128/mcb.13.1.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A., Schek N., Alwine J. C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992 Sep;12(9):3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A., Zeichner S., Carswell S., Alwine J. C. The human immunodeficiency virus type 1 polyadenylylation signal: a 3' long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman K. M., Steitz J. A. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 1993 Apr;7(4):647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- Wawra E., Pöckl E., Müllner E., Wintersberger E. Effect of sodium butyrate on induction of cellular and viral DNA syntheses in polyoma virus-infected mouse kidney cells. J Virol. 1981 Jun;38(3):973–981. doi: 10.1128/jvi.38.3.973-981.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E. A., Gilmartin G. M., Nevins J. R. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991 Jan;10(1):215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990 Jul;15(7):277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- Wintersberger E., Mudrak I., Wintersberger U. Butyrate inhibits mouse fibroblasts at a control point in the G1 phase. J Cell Biochem. 1983;21(3):239–247. doi: 10.1002/jcb.240210306. [DOI] [PubMed] [Google Scholar]
- Yang Y. C., Okayama H., Howley P. M. Bovine papillomavirus contains multiple transforming genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1030–1034. doi: 10.1073/pnas.82.4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo C. Y., Weiner A. M. A U1 small nuclear ribonucleoprotein particle with altered specificity induces alternative splicing of an adenovirus E1A mRNA precursor. Mol Cell Biol. 1989 Aug;9(8):3429–3437. doi: 10.1128/mcb.9.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A. M., Neugebauer K. M., Lane W. S., Roth M. B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993 Apr 9;260(5105):219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelder C. W., Gunderson S. I., Jansen E. J., Boelens W. C., Polycarpou-Schwarz M., Mattaj I. W., van Venrooij W. J. A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J. 1993 Dec 15;12(13):5191–5200. doi: 10.1002/j.1460-2075.1993.tb06214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]




