Abstract
Context:
Peroxisome proliferator-activated receptor-γ (PPARG) plays a pivotal role in adipogenesis and glucose homeostasis.
Objective:
We investigated whether PPARG gene variants were associated with type 2 diabetes (T2D) risk in the multiethnic Women's Health Initiative (WHI).
Research Design and Methods:
We assessed PPARG single-nucleotide polymorphisms (SNPs) in a case-control study nested in the prospective WHI observational study (WHI-OS) (1543 T2D cases and 2170 matched controls). After identifying 24 tagSNPs, we used multivariable logistic regression models and haplotype-based analyses to estimate these tagSNP-T2D associations. Single-SNP analyses were also conducted in another study of 5642 African American and Hispanic American women in the WHI SNP Health Association Resource (WHI-SHARe).
Results:
We found a borderline significant association between the Pro12Ala (rs1801282) variant and T2D risk in WHI-OS [odds ratio (OR) 0.51, 95% confidence interval (CI) 0.31–0.83, P = .01, combined group, additive model; P = .04, Hispanic American] and WHI-SHARe (OR 0.25, 95% CI 0.08–0.77, P = .02, Hispanic American) participants. In promoter region, rs6809631, rs9817428, rs10510411, rs12629293, and rs12636454 were also associated with T2D risk (range ORs 0.68–0.78, 95% CIs 0.52–0.91 to 0.60–1.00, P ≤ .05) in WHI-OS, in which rs9817428 was replicated in then WHI-SHARe Hispanic American group (P = .04).
Conclusions:
The association between PPARG Pro12Ala SNP and increased T2D susceptibility was confirmed, with Pro12 as risk allele. Additional significant loci included 5 PPARG promoter variants.
The peroxisome proliferator-activated receptor-γ (PPARG) is a ligand-activated transcription factor that plays an essential role in the regulation of lipid uptake, adipocyte differentiation, and energy balance (1). It also acts as an antiinflammatory molecule by hindering inflammatory reactions critical in the pathogenesis of type 2 diabetes (T2D) (2), which is also the target of the thiazolidinedione class of insulin-sensitizing drugs for glycemic control (3–5).
PPARG activates the expression of genes involved in glucose and lipid metabolism, which converts nutritional signals into metabolic consequences (5). PPARG is arguably the first gene identified for the complex late onset form of T2D via a candidate-gene approach. The PPARG Pro12Ala (rs1801282) has been the most vastly investigated single-nucleotide polymorphism (SNP), which was believed to alter transcriptional activity as a result of its location in the functional binding domain that has been associated with risk of T2D and its intermediate traits (6–13). Since its first report in 1997 assessing the effect of the Pro12Ala (rs1801282) variant on T2D risk, majority of the approximately 60 association studies have confirmed the relation of this particular variant to T2D risk, mainly in Caucasians. However, there appears to be some statistically significant heterogeneity across these studies [I2 = 37%, 95% confidence interval (CI) 9%–54%; P = .0028] (7). In particular, PPARG was not consistently confirmed to be significant in non-Caucasian populations, eg, a genome-wide association study (GWAS) conducted in African Americans (14), a multistage GWAS meta-analysis in East Asians (15), or a multistage GWAS meta-analysis in South Asians (16).
Therefore, we comprehensively assessed all common variants in the PPARG gene in relation to T2D risk in 2 independent studies in the Women's Health Initiative (WHI), a nested case-control study of postmenopausal women participating in the WHI Observation Study (WHI-OS) and the WHI SNP Health Association Resource (WHI-SHARe), both within and across ethnicity groups.
Materials and Methods
Study population
A detailed description of study participants of both WHI-OS and WHI-SHARe are given in Supplemental Materials and Methods (published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org). Briefly, the WHI-OS matched case-control participants included European Americans (n = 1922), African Americans (n = 1123), Hispanic Americans (n = 423), and Asian Americans (n = 245). A 1:1 matching ratio was used for European Americans and a 1:2 matching ratio was used for minorities to strengthen the power in these case groups with smaller sample sizes. For WHI-SHARe, after excluding those participants who also enrolled in WHI-OS, a total of 5642 participants were analyzed (3941 African Americans and 1701 Hispanic Americans).
Genetic data
We used a 2-stage approach. The first stage involved comprehensive SNP discovery to select a subset of tagSNPs representative of most common variants. The second stage involved genotyping 24 tagSNPs in the WHI-OS-matched case-control sample according to linkage disequilibrium patterns defined in first stage. Also, 8 of the 24 tagSNPs were included in the WHI-SHARe case-control sample. Details about tagSNP selection, genotyping methods, and quality control are given in the Supplemental Materials and Methods.
Statistical analysis
Hardy-Weinberg equilibrium was calculated using χ2 analyses. In WHI-OS-matched case-control sample, single-SNP and haplotype-based analyses of data were performed based on multivariate and conditional logistic regression models. In WHI-SHARe case-control sample, single-SNP analyses were performed based on the multivariate logistic regression models to independently validate results of WHI-OS, and pathway analysis was conducted using the Gene Set Enrichment Algorithm (GSEA). Details of statistical methods are given in the Supplemental Materials and Methods.
Results
Single-SNP analyses in WHI-OS
Except for rs10510411, rs12629293, and rs12636454, the genotype distributions of all other tagSNPs varied significantly across the 4 ethnic groups (P ≤ .0001) (Supplemental Table 1). Under additive model (Table 1), in conditional logistic regression with adjustment for risk factors for T2D, the Pro12Ala (rs1801282) was associated with T2D risk in Hispanic American women [odds ratio (OR) 0.27, 95% CI 0.08–0.93, P = .04)] and combined group (OR = 0.55, 95% CI 0.35–0.86, P = .01). In addition, rs6809631 (OR 0.76, 95% CI 0.58–0.99, P = .04), rs9817428 (OR 0.78, 95% CI 0.60–1.00, P = .05), rs10510411 (OR 0.68, 95% CI 0.52–0.91, P = .01), rs12629293 (OR 0.72, 95% CI 0.55–0.94, P = .02), and rs12636454 (OR 0.73, 95% CI 0.57–0.95, P = .02) also appeared to be associated with T2D risk in the combined group. To guard against type I error, we used the Benjamini-Hochberg procedure to obtain the false discovery rate (FDR) adjusted P values for these 6 SNPs. We further investigated the association of Pro12Ala (rs1801282) with T2D risk and found that among Hispanic Americans, CG genotype appeared to be at lower risk (OR 0.17, 95% CI 0.04–0.77, P = .02) compared to CC genotype (Supplemental Table 2). Results under dominant and recessive genetic models are shown in Supplemental Tables 3 and 4.
Table 1.
Single-SNP Association Studies of the 24 tagSNPs in the PPARG Gene With T2D Risk Under Additive Model in WHI-OS Nested Case-Control Sample (n = 3713)a
| dbSNP ID | European American (954/968)b | African American (369/754) | Hispanic American (141/282) | Asian American (79/166) | Combined (1543/2170) | P Value (Combined) |
|---|---|---|---|---|---|---|
| rs9878908 | 0.85 (0.55–1.32) | 0.75 (0.35–1.61) | 1.18 (0.45–3.06) | 1.00 (0.62–1.60) | 0.89 (0.65–1.21) | .46 |
| rs6798713 | 0.75 (0.47–1.17) | 0.98 (0.57–1.67) | 1.40 (0.52–3.74) | 1.06 (0.67–1.67) | 0.89 (0.66–1.20) | .44 |
| rs6809631 | 0.71 (0.46–1.10) | 0.78 (0.50–1.23) | 0.66 (0.32–1.38) | 0.80 (0.48–1.34) | 0.76 (0.58–0.99) | .04c |
| rs9817428 | 0.76 (0.49–1.17) | 0.80 (0.53–1.21) | 0.66 (0.32–1.37) | 0.78 (0.47–1.30) | 0.78 (0.60–1.00) | .05d |
| rs10510411 | 0.68 (0.44–1.06) | 0.64 (0.37–1.11) | 0.78 (0.38–1.60) | 1.06 (0.66–1.71) | 0.68 (0.52–0.91) | .01e |
| rs12629293 | 0.66 (0.43–1.04) | 0.78 (0.49–1.24) | 0.78 (0.38–1.58) | 1.05 (0.65–1.70) | 0.72 (0.55–0.94) | .02f |
| rs12636454 | 0.68 (0.44–1.06) | 0.79 (0.52–1.21) | 0.76 (0.37–1.54) | 1.10 (0.69–1.76) | 0.73 (0.57–0.95) | .02g |
| rs4518111 | 1.36 (0.91–2.03) | 1.06 (0.65–1.75) | 1.40 (0.70–2.82) | 1.18 (0.74–1.89) | 1.20 (0.94–1.54) | .15 |
| rs10510418 | 1.12 (0.74–1.69) | 0.75 (0.43–1.33) | 0.91 (0.40–2.06) | 0.69 (0.38–1.27) | 1.03 (0.79–1.36) | .81 |
| rs1801282 | 0.62 (0.35–1.12) | 0.59 (0.15–2.31) | 0.27 (0.08–0.93)h | 0.21 (0.04–1.04) | 0.55 (0.35–0.86) | .01i |
| rs1373640 | 1.17 (0.79–1.75) | 0.86 (0.49–1.49) | 1.14 (0.53–2.46) | 0.56 (0.27–1.12) | 1.11 (0.84–1.45) | .46 |
| rs2972162 | 1.23 (0.82–1.84) | 1.19 (0.79–1.80) | 1.02 (0.51–2.04) | 0.98 (0.58–1.67) | 1.17 (0.92–1.48) | .20 |
| rs10510419 | 1.45 (0.83–2.54) | 1.25 (0.69–2.26) | 0.98 (0.39–2.45) | 1.49 (0.19–11.51) | 1.22 (0.86–1.72) | .26 |
| rs2959272 | 1.22 (0.82–1.83) | 1.12 (0.75–1.68) | 0.99 (0.49–2.00) | 1.08 (0.64–1.80) | 1.15 (0.91–1.46) | .24 |
| rs709150 | 0.67 (0.44–1.03) | 0.80 (0.48–1.31) | 0.99 (0.50–1.96) | 0.94 (0.56–1.57) | 0.80 (0.62–1.03) | .08 |
| rs709157 | 1.32 (0.88–1.98) | 0.67 (0.34–1.31) | 0.96 (0.44–2.06) | –j | 1.08 (0.81–1.43) | .61 |
| rs1175540 | 1.12 (0.76–1.65) | 1.04 (0.68–1.60) | 0.97 (0.46–2.03) | 1.18 (0.69–2.02) | 1.08 (0.85–1.37) | .53 |
| rs1175544 | 1.21 (0.82–1.79) | 0.81 (0.45–1.47) | 0.99 (0.45–2.17) | 1.25 (0.74–2.10) | 1.11 (0.85–1.44) | .44 |
| rs1797912 | 1.18 (0.80–1.74) | 0.91 (0.56–1.49) | 1.09 (0.49–2.45) | 1.00 (0.60–1.65) | 1.12 (0.86–1.44) | .40 |
| rs1152002 | 0.93 (0.64–1.37) | 0.94 (0.62–1.43) | 2.01 (0.92–4.41) | 0.84 (0.51–1.37) | 1.06 (0.83–1.35) | .65 |
| rs3856806 | 0.93 (0.50–1.71) | 0.52 (0.22–1.22) | 0.31 (0.09–1.07) | 0.83 (0.47–1.48) | 0.71 (0.48–1.07) | .10 |
| rs1152003 | 1.09 (0.73–1.63) | 0.96 (0.65–1.41) | 1.72 (0.84–3.50) | 1.01 (0.62–1.64) | 1.07 (0.85–1.36) | .56 |
| rs1152007 | 1.15 (0.74–1.81) | 0.74 (0.45–1.24) | 1.10 (0.46–2.64) | 0.88 (0.56–1.37) | 0.92 (0.70–1.21) | .55 |
| rs709167 | 1.06 (0.73–1.55) | 1.09 (0.66–1.79) | 1.12 (0.49–2.54) | 0.86 (0.47–1.57) | 1.06 (0.82–1.36) | .68 |
Adjusted OR (95% CI) for each tagSNP was estimated under the additive genetic model, using conditional logistic regression models with adjustments for age, body mass index, ln(fasting insulin), ln(fasting glucose), cigarette smoking (never, past, and current), alcohol intake (never, past, and current), hormone replacement therapy usage (never, past, and current), T2D family history (presence/absence), and physical activity per week at baseline. Due to the small Asian population size, body mass index, ln(fasting insulin), and ln(fasting glucose) were excluded to cause the model to converge. Confirmatory T2D association results for rs9817428 and rs1801282 (both highlighted) under an additive model in WHI-SHARe case-control sample are shown in Supplemental Table 5. Associations with FDR-adjusted P ≤ .05 are shown in bold.
Sample size is presented as cases/controls.
P = .04, with FDR-adjusted P = .19 using the method of Benjamini and Hochberg.
P = .05, with FDR-adjusted P = .21 using the method of Benjamini and Hochberg.
P = .01, with FDR-adjusted P = .11 using the method of Benjamini and Hochberg.
P = .02, with FDR-adjusted P = .12 using the method of Benjamini and Hochberg.
P = .02, with FDR-adjusted P = .12 using the method of Benjamini and Hochberg.
P = .04, with FDR-adjusted P = .65 using the method of Benjamini and Hochberg.
P = .01, with FDR-adjusted P = .11 using the method of Benjamini and Hochberg.
Result is difficult to interpret because of small sample size within stratum.
Replicative analysis using WHI-SHARe and GSEA analysis
In the WHI-SHARe Hispanic American group, Pro12Ala (rs1801282) (OR 0.25, 95% CI 0.08–0.77, P = .02) and rs9817428 (OR 0.52, 95% CI 0.28–0.96, P = .04) were found associated with T2D risk (Supplemental Table 5), with respective FDR-adjusted P value of .12 and .15. Results under dominant and recessive genetic models are shown in Supplemental Tables 6 and 7, respectively.
In GSEA analysis, we identified 81 T2D-related biological pathways with at least a 10-gene size, ie, the peroxisome proliferator-activated receptor-α (PPARα)/retinoid X receptor-α activation pathway ranked first (nominal P = .0005, FDR adjusted P = .002) and the PPAR signaling pathway ranked fourth (nominal P = .23, FDR adjusted P = .18) among 7104 African American women in the WHI-SHARe cohort, whereas the PPAR signaling pathway ranked 12th (nominal P = .60, FDR adjusted P = .64) and the PPARα/ retinoid X receptor-α activation pathway ranked 69th (nominal P = .46, FDR adjusted P = 1.00) among 3253 Hispanic American women.
Haplotype analysis in WHI-OS
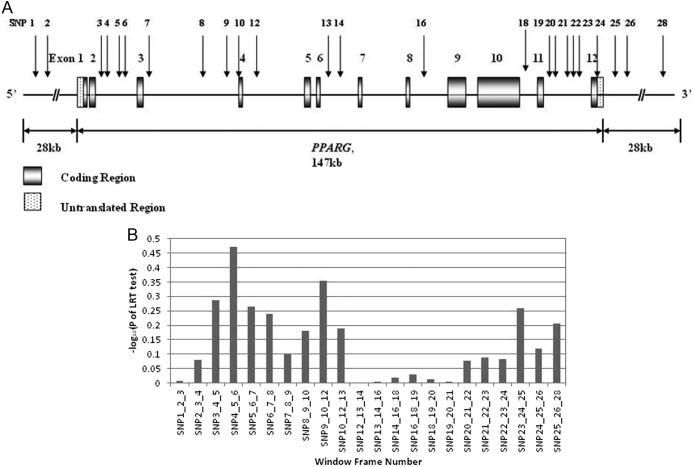
Figure 1A shows a schematic view of PPARG and selected tagSNPs. Two coding tagSNPs [rs1801282 (Pro12Ala) and rs3856806 (His477His)] are in exons 4 (formerly denoted exon B) and 12 (formerly denoted exon 6). Figure 1B depicts results of sliding-window (3 SNP) haplotype-based analyses in the combined sample. None reached a P < .0023 (Bonferroni corrected threshold).
Figure 1.
A, The schematic presentation showed the positions of 24 tagSNPs (with SNP identifications) that spanned the PPARG genomic region. B, Sliding-window (3 SNP) haplotype-based analysis of 24 PPARG tagSNPs using additive genetic model in combined multiethnic WHI-OS case-control sample. The haplotype effects were estimated using an omnibus likelihood ratio test. The x-axis denoted the sliding window frames, and the y-axis denoted the −log10(P value). A −log10(P value) greater than 2.64 was used as the global significance threshold using Bonferroni correction for the 22 window frames, and none met the criterion.
Discussion
We confirmed a modest but significant association between well-studied Pro12Ala SNP and T2D risk in WHI-OS and WHI-SHARe participants. We also found that 5 promoter SNPs (rs6809631, rs9817428, rs10510411, rs12629293, and rs12636454) to be significantly associated with T2D risk. In WHI-SHARe, PPAR signaling pathway was ranked among the top 5 in African Americans and top 15 pathways in Hispanic Americans.
Interpreting the results with an appropriate balance between type I and type II statistical errors has always been a challenging task. According to Rothman (17), a way to minimize both type I and type II errors is to disregard significance testing and focus on confidence intervals instead. In our analysis, we adjusted for multiple comparisons using FDR and Bonferroni correction. We recognized that the large number of statistical tests has inflated the type I error, whereas the conventional 0.05 level for Bonferroni or FDR adjusted P values is too conservative. We therefore took a balanced approach when interpreting effect estimates and their relevant confidence intervals emanating from our analysis.
Because haplotype-based analysis is arguably more powerful than single-marker analysis (18), we performed sliding-window (3 SNP), haplotype-based analyses. The 3-SNP windows with the highest and the second highest −log10(P value) values (SNP4_5_6 and SNP9_10_12) encompass rs9817428 and rs1801282, respectively, which correspond to the 2 significant T2D risk variants revealed in single-SNP analysis, indicating consistency.
A comprehensive review and meta-analysis showed a moderate level of heterogeneity attributable to genuine variation in gene effect size for the PPARG Pro12Ala genetic variant, which reflected variation observed between ethnic populations as well as differences in body mass index (7). This may be explained by the role played by ethnicity and differences in dietary and habitual factors (19). Because both under- and overnutrition during pregnancy has been associated with the later development of obesity and diabetes, epigenetic modifications that affect glucose regulation and insulin secretion may also play a significant role in the pathophysiology of T2D. Our multiethnic WHI-OS participants, which consisted of European, African, Hispanic, and Asian Americans with comprehensive assessment of demographic and lifestyle variables, may further validate the earlier published body of evidence in the literature regarding the association between common variants of the PPARG gene and T2D risk.
Among 5 promoter SNPs detected by additive model rs6809631 (P = .04), rs9817428 (P = .05), rs10510411 (P = .01), and rs12629293 (P = .02) are located in the PPARγ3 promoter, and rs12636454 (P = .02) is located in the in PPARγ2 promoter. However, only rs9817428 was included in the 8 tagSNPs genotyped in WHI-SHARe (Supplemental Table 5), and this promoter SNP was associated with T2D risk in WHI-SHARe Hispanic American group (P = .04, Hispanic American). Therefore, our results did provide novel insights beyond the widely acclaimed PPARγ2 rs1801282 association with T2D.
In summary, the established association of PPARG Pro12Ala SNP with T2D susceptibility was confirmed in both WHI-OS (combined group and Hispanic American group) and WHI-SHARe Hispanic American group. Of the 5 additional PPARG promoter variants revealed in the WHI-OS combined group, rs9817428 was detected in the WHI-SHARe Hispanic American group. These SNPs deserve further confirmation in diverse ethnic groups (particularly Hispanic American).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant RO1-DK062290 and RO1-DK066401 (to S.L.). K.H.K.C. and S.L. are supported by the Burroughs Wellcome Fund Inter-School Training Program in Metabolic Diseases. T.N. is supported by a start-up fund of Tulane University. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through Contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. A list of WHI investigators is available in the Supplemental Data Information.
Disclosure Summary: The authors have nothing to declare.
Footnotes
- CI
- confidence interval
- FDR
- false discovery rate
- GSEA
- Gene Set Enrichment Algorithm
- GWAS
- genome-wide association study
- OR
- odds ratio
- PPARα
- peroxisome proliferator-activated receptor-α
- PPARG
- peroxisome proliferator-activated receptor-γ
- SNP
- single-nucleotide polymorphism
- T2D
- type 2 diabetes
- WHI
- Women's Health Initiative
- WHI-OS
- WHI observational study
- WHI-SHARe
- WHI SNP Health Association Resource.
References
- 1. Szanto A, Nagy L. The many faces of PPARγ: anti-inflammatory by any means? Immunobiology. 2008;213:789–803 [DOI] [PubMed] [Google Scholar]
- 2. Ahmed W, Ziouzenkova O, Brown J, et al. PPARs and their metabolic modulation: new mechanisms for transcriptional regulation? J Intern Med. 2007;262:184–198 [DOI] [PubMed] [Google Scholar]
- 3. Blackburn GL. From bench to bedside: novel mechanisms and therapeutic advances through the development of selective peroxisome proliferator-activated receptor γ modulators. Am J Clin Nutr. 91:251S–253S [DOI] [PubMed] [Google Scholar]
- 4. Cheatham WW. Peroxisome proliferator-activated receptor translational research and clinical experience. Am J Clin Nutr. 91:262S–266S [DOI] [PubMed] [Google Scholar]
- 5. Ryan KK, Li B, Grayson BE, Matter EK, Woods SC, Seeley RJ. A role for central nervous system PPAR-γ in the regulation of energy balance. Nat Med. 17:623–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gaulton KJ, Willer CJ, Li Y, et al. Comprehensive association study of type 2 diabetes and related quantitative traits with 222 candidate genes. Diabetes. 2008;57:3136–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gouda HN, Sagoo GS, Harding AH, Yates J, Sandhu MS, Higgins JP. The association between the peroxisome proliferator-activated receptor-γ2 (PPARG2) Pro12Ala gene variant and type 2 diabetes mellitus: a HuGE review and meta-analysis. Am J Epidemiol. 171:645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herder C, Rathmann W, Strassburger K, et al. Variants of the PPARG, IGF2BP2, CDKAL1, HHEX, and TCF7L2 genes confer risk of type 2 diabetes independently of BMI in the German KORA studies. Horm Metab Res. 2008;40:722–726 [DOI] [PubMed] [Google Scholar]
- 9. Lin Y, Li P, Cai L, et al. Association study of genetic variants in eight genes/loci with type 2 diabetes in a Han Chinese population. BMC Med Genet. 11:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232 [DOI] [PubMed] [Google Scholar]
- 11. Meigs JB, Manning AK, Fox CS, et al. Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC Med Genet. 2007;8(suppl 1):S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanghera DK, Ortega L, Han S, et al. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wen J, Ronn T, Olsson A, et al. Investigation of type 2 diabetes risk alleles support CDKN2A/B, CDKAL1, and TCF7L2 as susceptibility genes in a Han Chinese cohort. PLoS One. 5:e9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palmer ND, McDonough CW, Hicks PJ, et al. A genome-wide association search for type 2 diabetes genes in African Americans. PLoS One. 2012;7:e29202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kooner JS, Saleheen D, Sim X, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rothman KJ. Curbing type I and type II errors. Eur J Epidemiol. 2010;25:223–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He Y, Li C, Amos CI, Xiong M, Ling H, Jin L. Accelerating haplotype-based genome-wide association study using perfect phylogeny and phase-known reference data. PLoS One. 6:e22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luan J, Browne PO, Harding AH, et al. Evidence for gene-nutrient interaction at the PPARγ locus. Diabetes. 2001;50:686–689 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.