Abstract
Characterizing the spatial organization of the human mitochondrial proteome will enhance our understanding of mitochondrial functions at the molecular level and provide key insight into protein-disease associations. However, the sub-organellar location and possible association with mitochondrial diseases are not annotated for most mitochondrial proteins. Here, we characterized the functional and spatial organization of mitochondrial proteins by assessing their position in the Mitochondrial Protein Functional (MPF) network. Network position was assigned to the MPF network and facilitated the determination of sub-organellar location and functional organization of mitochondrial proteins. Moreover, network position successfully identified candidate disease genes of several mitochondrial disorders. Thus, our data support the use of network position as a novel method to explore the molecular function and pathogenesis of mitochondrial proteins.
The spatial information of proteins within the cell provides a foundation for elucidating protein-disease associations and their functional roles1. Proteins often form complexes and function in subcellular locations that offer the optimal chemical and physical environment2,3. We recently showed that integrating information about subcellular localization with the protein interaction network contributed greatly to advancing our understanding of the etiology and comorbidity of genetic diseases4.
Mitochondria are essential organelles that participate in important cellular processes, including ATP production, fatty acid metabolism, apoptosis, and aging. There is increasing evidence demonstrating that mitochondria are also involved in many human diseases, such as neurodegenerative diseases, cardiovascular disorders, cancers, and metabolic diseases5,6. Thus, establishing a comprehensive list of the mitochondrial proteome is essential for understanding the roles of this organelle in human disease. High-throughput approaches, such as mass spectrometry-based proteomics, epitope tagging combined with microscopy, and genome-wide predictions of protein subcellular localization, have been applied to investigate the mitochondrial protein inventory7,8,9.
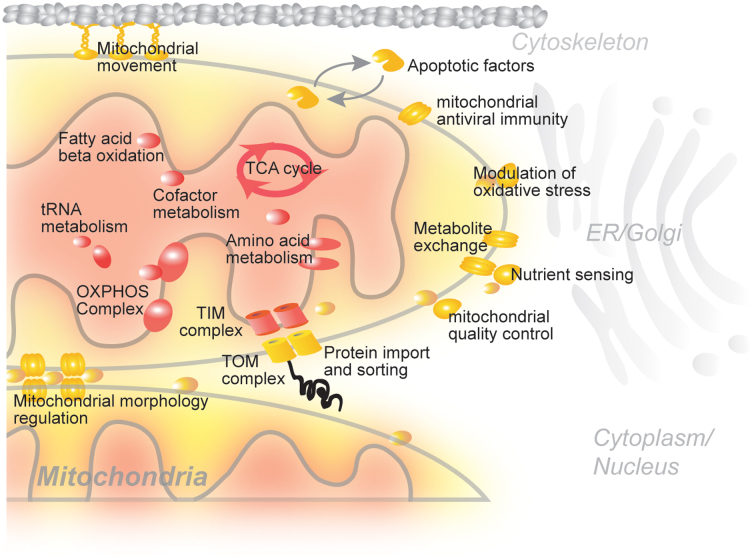
The spatial organization of mitochondrial proteins has been shown to correlate highly with protein function at the molecular level10,11,12. For example, enzymes involved in the TCA cycle and respiratory chain complexes are located within the mitochondrial matrix and inner membrane, whereas many regulatory proteins required for mitochondrial movement and apoptosis are found in the mitochondrial outer membrane (Figure 1). Despite our rapidly growing knowledge of the mitochondrial protein repertoire, our understanding of their spatial and functional organization within the organelle remains limited. In fact, the sub-compartmental localization of less than 30% of all mitochondrial proteins has been annotated.
Figure 1. Schematic representation of mitochondrial proteins and their sub-organellar locations.
Network position has also been applied to elucidate the architecture of social networks13. Network position also helps to identify the optimal location of servers to increase network traffic efficiency on the Internet14. In social network analyses, network position successfully classified nodes into the core and periphery categories according to their influence on communication traffic15,16.
Here, we developed a method to assign a network position to mitochondrial proteins based on information propagation in the Mitochondrial Protein Functional (MPF) network. We demonstrate that the network position of mitochondrial proteins corresponds to their spatial locations within the organelle. Furthermore, we employed our method to assign potential disease-associated proteins and confirmed that network position is an effective means for identifying candidate disease proteins.
Results
Assigning network positions of mitochondrial proteins
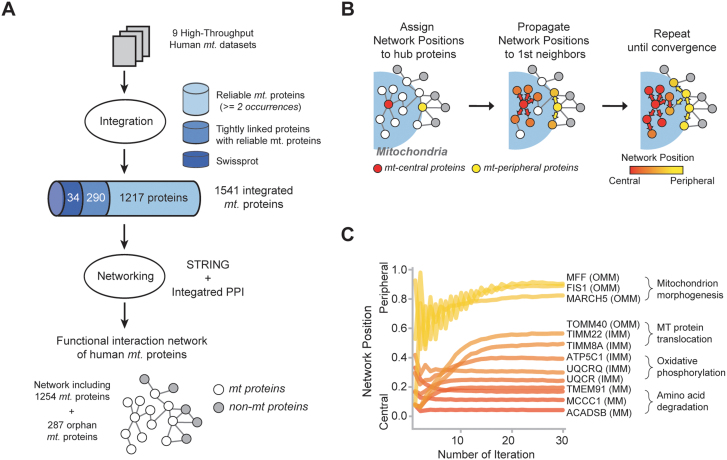
We constructed an MPF network and assigned a network position to every mitochondrial protein. This network was comprised of 1,254 mitochondrial proteins and 6,071 functional links. We integrated proteomic experiments, database annotations, and consensus localization information to compile the comprehensive mitochondrial proteome (Figure 2A). Nodes within the network were defined as proteins present more than twice in individual mitochondrial databases and that have functional links with each other. They cover more than 90% (435 among 469 proteins) of annotated mitochondrial proteins in the SwissProt database. Furthermore, we combined functional links that were supported by multiple lines of evidence, including protein-protein interactions, published literature, expression analysis, and genomic context (see Materials and Methods for more detail).
Figure 2. Construction of human mitochondrial protein functional network and assignment of network positions.
(A) Building the human mitochondrial protein functional network. (B) Information propagation was used to assign network position to mitochondrial proteins. (C) Convergence of network positions after iterations of information propagation.
Our analysis is based on the logic that mitochondrial proteins localized in the center of the MPF network are connected mostly with other mitochondrial proteins, whereas proteins localized to the periphery of the network are predominantly linked to non-mitochondrial proteins. Mitochondria are not isolated organelles within the cell; instead they became incorporated components through evolution due to a symbiotic relationship with the host cell17,18. Therefore, information about connections to non-mitochondrial proteins was also used to assign the spatial information of mitochondrial proteins. The network position of each protein was assigned based on the fraction of mitochondrial protein partners among direct neighbors.
First, we assigned network positions to hub proteins since proteins with fewer partners could generate bias due to limited information from network connections. Then, we applied information propagation to assign the network positions of non-hub proteins (Figure 2B). The propagation steps delivered information about network position to distantly located nodes through network connections. In doing so, we could assign network positions to all 1,254 mitochondrial proteins (Figure S1). The network position of all the mitochondrial proteins can be accessed in Table S1.
Information propagation retrieved the spatial information of distantly located proteins in the network. We discovered that network position fluctuated at the initial stage but converged into a specific position with each iteration (Figure 2C and S1). The network positions of the 362 proteins with more than 30 linked partners were initially set based on direct neighboring proteins. However, the network positions of the 892 proteins with less than 30 neighbors eventually converged into their positions after 50 iterations. For instance, the network positions of methylcrotonoyl-CoA carboxylase 1 (MCCC1) and short/branched chain acyl-CoA dehydrogenase (ACADSB) converged to 0.115 and 0.062, respectively, and were defined as central proteins. Meanwhile, the network positions of mitochondrial fission factor and membrane-associated ring finger protein 5 (MARCH5) converged into 0.935 and 0.931, respectively, and were defined as peripheral proteins.
Network position and spatial organization of mitochondrial proteins
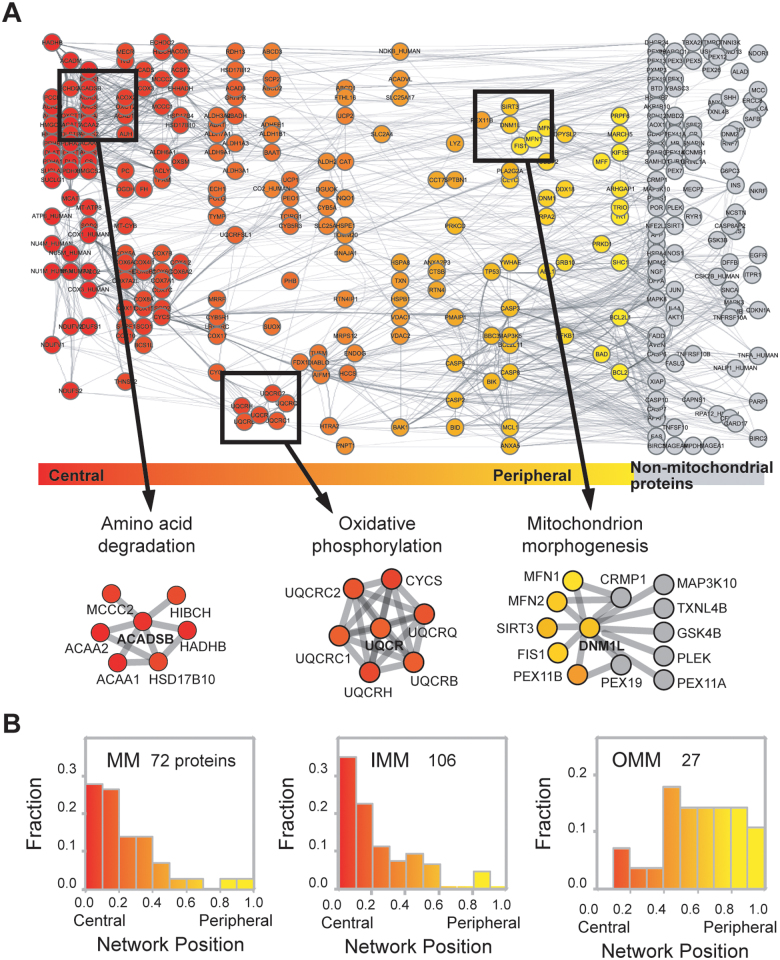
We sought to determine whether network position also represents the sub-compartmental organization of mitochondrial proteins. Our analysis revealed that proteins located at the network periphery tended to be outer mitochondrial membrane (OMM) proteins, whereas proteins located at the center of the network were inner mitochondrial membrane (IMM) or mitochondrial matrix (MM) proteins (Figure 3A). Examples of core mitochondrial functional modules found with a central network position are dehydrogenase complex and mitochondrial complex III, which exhibited average network positions of 0.080 and 0.170, respectively. Meanwhile, proteins that modulate mitochondrial morphogenesis were found at the network periphery with an average network position of 0.828.
Figure 3. Network position and spatial organization of mitochondrial proteins.
(A) Spatial organization of the MPF network according to network position. (B) Distribution of the network positions of MM, IMM, and OMM proteins.
Network position can also be used to separate OMM proteins from IMM and MM proteins. We investigated the correlation between network position and sub-organellar localization of mitochondrial proteins and discovered that most IMM and MM proteins had central network positions, whereas proteins associated with the OMM had peripheral network positions (Figure 3B). OMM and IMM proteins exhibited statistically significant (P = 3.7 × 10−10; Mann-Whitney U test) differences in network position, whereas IMM and MM proteins had similar network positions (P = 0.47; Mann-Whitney U test).
We compared the performance of our method with other centrality measures. We found that conventional centrality measures failed to properly distinguish “central” mitochondrial proteins from “peripheral” ones. For example, the peripheral proteins of mitochondrial protein network obtained by conventional centrality measures (betweenness centrality or closeness centrality) did not have more connections with non-mitochondrial proteins. However, our method separated central proteins from peripheral proteins with the better performance (P = 3.60 × 10−131; Mann-Whitney test, Figure S2) compared to betweenness centrality (P = 1.17 × 10−12; Mann-Whitney test) and closeness centrality measures (P = 1.68 × 10−19; Mann-Whitney test). Moreover, network position could separate central mitochondrial proteins (red-colored nodes) from non-mitochondrial proteins (black-colored nodes) better than other measures (Figure S3). This suggests that our approach integrated spatial information to the network position of mitochondrial proteins.
We also measured the potential bias of degree distribution to see if OMM proteins may have less numbers of links than IMM and MM proteins. We found that OMM proteins have comparable number of functional links and PPIs compared with IMM and MM proteins (Figure S4). Specifically, OMM proteins have more functional links (Average of 23.3 links) than IMM proteins (Average of 17.4 links, P = 0.026; Mann-Whitney test) but have less numbers of links compared to MM proteins (Average of 31.2 links, P = 4.78 × 10−5; Mann-Whitney test). Further, we compared the number of PPI partners and found that they have comparable number of PPI partners. OMM proteins have an average of 6.0 PPIs, IMM proteins have an average of 4.2 PPIs (P = 0.29; Mann-Whitney test), and MM proteins have an average of 5.3 PPIs (P = 0.41; Mann-Whitney test). These suggest that our results are not biased by degree distribution or a lack of association information of OMM proteins.
Robustness of the selection criterion for network position
To demonstrate the robustness of our criterion, we compared network positions calculated with varying criteria. For example, we applied the mitochondrial proteins from MitoCara to calculate network positions and found that the results were comparable with each other (r = 0.85, P = 1.03 × 10−155; Spearman r) (Figure S5). MitoCara is known to include a highly accurate mitochondrial proteome. Furthermore, we tested the network propagation with a varying degree cutoff and found that K value over 30 showed the better performance of distinguishing IMM and OMM proteins (P = 7.6 × 10−8; Mann-Whitney U test) than the procedure without propagation (P = 4.7 × 10−5; Mann-Whitney U test).
Next, we tested our method with other interaction dataset, HumanNet19, which is a probabilistic functional gene network of protein encoding genes of Homo sapiens. We obtained similar results (r = 0.57, P = 2.96 × 10−92; Spearman r) with the HumanNet dataset. The network position distinguished IMM proteins from OMM proteins with statistical significance (P = 1.15 × 10−11; Mann-Whitney U test). These suggest that our criterion could be generally applicable to slightly different network links supported by various interaction datasets.
We also checked the performance of network position with varying degree cutoff K. We found that, when the value of cutoff K increased, the distinguishing power of IMM and OMM proteins were increased, although there were fluctuations with different K values. For example, when K was 0, 10, or 100, P-value was 6.80 × 10−6, 5.64 × 10−8, or 1.08 × 10−10, respectively (Figure S6). Thus, we presumed that when K is infinity, all the nodes could find their network position and the network effects might be best reflected in the measure.
Network position and function annotation of mitochondrial proteins
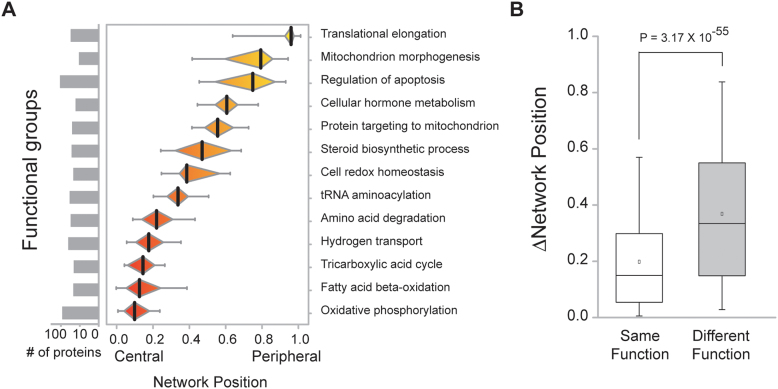
Mitochondrial proteins are engaged in diverse functions according to their sub-organellar compartments. We found that mitochondrial protein function can be inferred from network position (Figure 4A). Proteins associated with mitochondrial core functions, including oxidative phosphorylation, fatty acid beta-oxidation, TCA cycle, hydrogen transport, and amino acid degradation, were assigned central network positions. However, proteins related to mitochondrial regulatory functions, including translation elongation, mitochondrial morphogenesis, and regulation of apoptosis, were found in the periphery of the network (P = 4.4 × 10−48; Mann-Whitney U test). Additionally, we measured the network positions of proteins with roles in diverse functional groups and found that proteins in the same group tended to have more similar network positions compared to those with unrelated functions (P = 3.17 × 10−55; Mann-Whitney U test; Figure 4B). Nevertheless, determination of protein network position using a conventional distance measure alone could not distinguish central from peripheral functions (Figure S7). Thus, these results indicate that network position is also useful in annotating mitochondrial protein function.
Figure 4. Network position and function assignment of mitochondrial proteins.
(A) Distribution of the network positions of mitochondrial functional groups. (B) Network position differences were compared between protein pairs within the same functional group versus different functional groups.
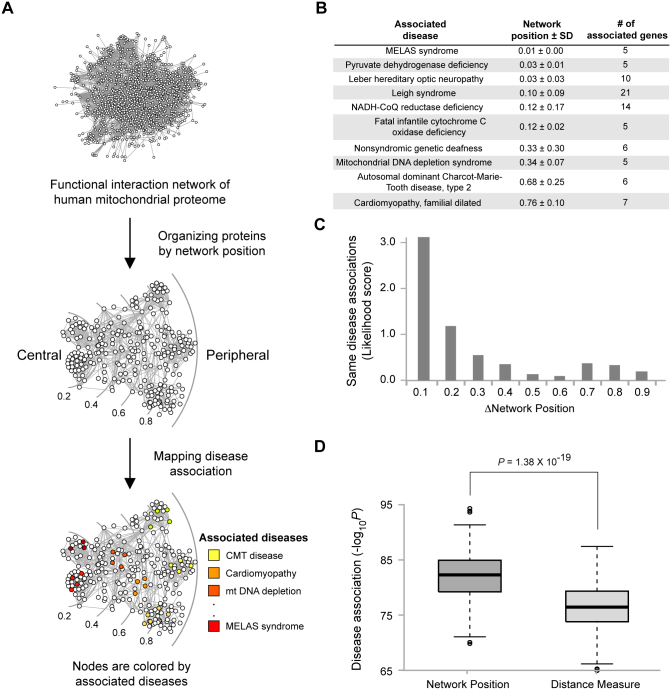
Network position helps identify proteins associated with disease
Information about a particular protein's localization has been used to uncover candidate disease proteins1,20. We investigated whether network position can be used to identify disease-associated proteins. We mapped disease-protein association onto the MPF network and analyzed the relationship of mitochondrial diseases and network positions. This analysis revealed that proteins associated with the same diseases tended to have similar network positions (Figure 5A and 5B). In contrast, proteins associated with different disease types possessed distant network positions. For example, proteins associated with Leigh syndrome had a central position (average network position = 0.10), while proteins associated with Charcot-Marie-Tooth disease type 2 were located at the network periphery (average network position = 0.68). These results are consistent with the fact that Leigh syndrome is caused by defective oxidative phosphorylation, which is a mitochondrial core function, whereas Charcot-Marie-Tooth disease type 2 is caused by a failure in axonal mitochondrial transport, which is a mitochondrial peripheral function.
Figure 5. Network position and disease association of mitochondrial proteins.
(A) Mapping mitochondrial disease-associated proteins into disease types. (B) Network positions of disease-associated proteins. (C) Differences between disease associations as measured by network positions. (D) Comparison of disease association prediction between network position and network distance as measured by the shortest path length.
We also found a strong correlation between the similarity of network positions and the likelihood that they are associated with the same disease. Likelihood was determined by measuring the probability of finding same disease-associated protein pairs compared to random chance. When the network positions of two proteins were similar (Δnetwork position < 0.2), the likelihood of their association with the same disease increased more than 2-fold (Figure 5C). As the similarity of network position decreased, the likelihood of association with the same disease also decreased.
Next, we tested whether network position performed better than conventional distance measurements in determining whether a protein is associated with a particular disease. Distance in biological networks has been used to identify disease-associated genes21,22. In this method, distance is defined by the shortest path length between two proteins. We compared the prediction performance of disease-protein associations by using network position and distance measure, confirmed that network position significantly improved the prediction of disease-protein association (Figure 5D). Our results demonstrate that proteins associated with the same diseases had closer network positions and a higher disease association than the distance measurement alone.
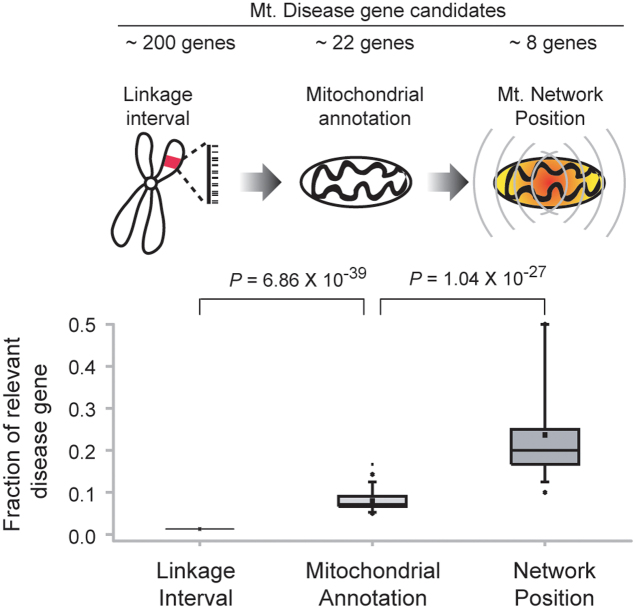
We also examined whether network position can reduce the number of candidate disease genes identified better than the linkage interval method, which was applied previously along with protein spatial information to discover disease-causing genes20. To address this, we compared the results from using network positional information and linkage interval in predicting disease involvement. We found a more than 10-fold increase in detection capability using network position compared to linkage interval (Figure 6). Network position effectively reduced the number of candidates without eliminating known disease-causing proteins (Table 1). For example, we successfully identified two genes known to associate with hepatic mtDNA depletion among 17 potential mitochondrial disease genes identified using linkage interval. One of them, MPV17, has been shown to be a cause of hepatic mtDNA depletion disease23. We also successfully reduced the candidate proteins associated with Charcot-Marie-Tooth disease type 2 including HSPB1 which has shown to be an important factor for the disease24.
Figure 6. Differences in disease association as measured by linkage information.
Fraction of relevant mitochondrial disease causing genes is increasing according to use of mitochondrial annotation and network position.
Table 1. Candidates of mitochondrial disease-causing genes.
| Disease (OMIM) | Linkage region | Gene loci | Mitochondrial candidates | Network position | Disease-causing gene candidates |
|---|---|---|---|---|---|
| Hepatic mtDNA depletion | D2S2373–D2S2259 | 186 | 17 | 2 | CAD, MPV17 |
| Charcot-Marie-Tooth disease type 2 (CMT2F) | 7q11–7q21 | 256 | 20 | 4 | CYP51A1, HSPB1, ELN, GTPBP10 |
| Charcot-Marie-Tooth disease type 2 (CMT2L) | D12S366–D12S1611 | 107 | 17 | 11 | DIABLO, TRIAP1, COQ5, MSI1, RPS2P5, RAB35, HSPB8, POP5, CLIP1, PEBP1, PLA2G1B |
| Cardiomyopathy, familial dilated. (FDC) | 9q13–9q22 | 214 | 22 | 14 | TDRD7, HSPBL2, PRKACG, RFK, VPS13A, ANXA1, C9orf89, SECISBP2, UBQLN1, NTRK2, NCBP1, NANS, HNRNPK, IARS |
| Cardiomyopathy, familial dilated. (FDC) | 10q21–10q23 | 313 | 43 | 18 | PSAP, RPS24, ACTA2, CYP26A1, MARCH5, PPA1, IDE, ANXA11, IFIT3, SLC25A16, SUPV3L1, PHYHIPL, NDST2, AIFM2, CYP2C19, MINPP1, ARID5B, LDB3 |
| mtDNA depletion syndrome (MDS) | 2p13 | 125 | 18 | 3 | HK2, SPR, DGUOK |
| mtDNA depletion syndrome (MDS) | 15q22–15q26 | 435 | 45 | 6 | GAPDHL6, CYP11A1, CLPX, ETFA, BCL2A1, RAB11A |
Bold texts represent the mitochondrial disease-causing genes with experimental evidences.
Discussion
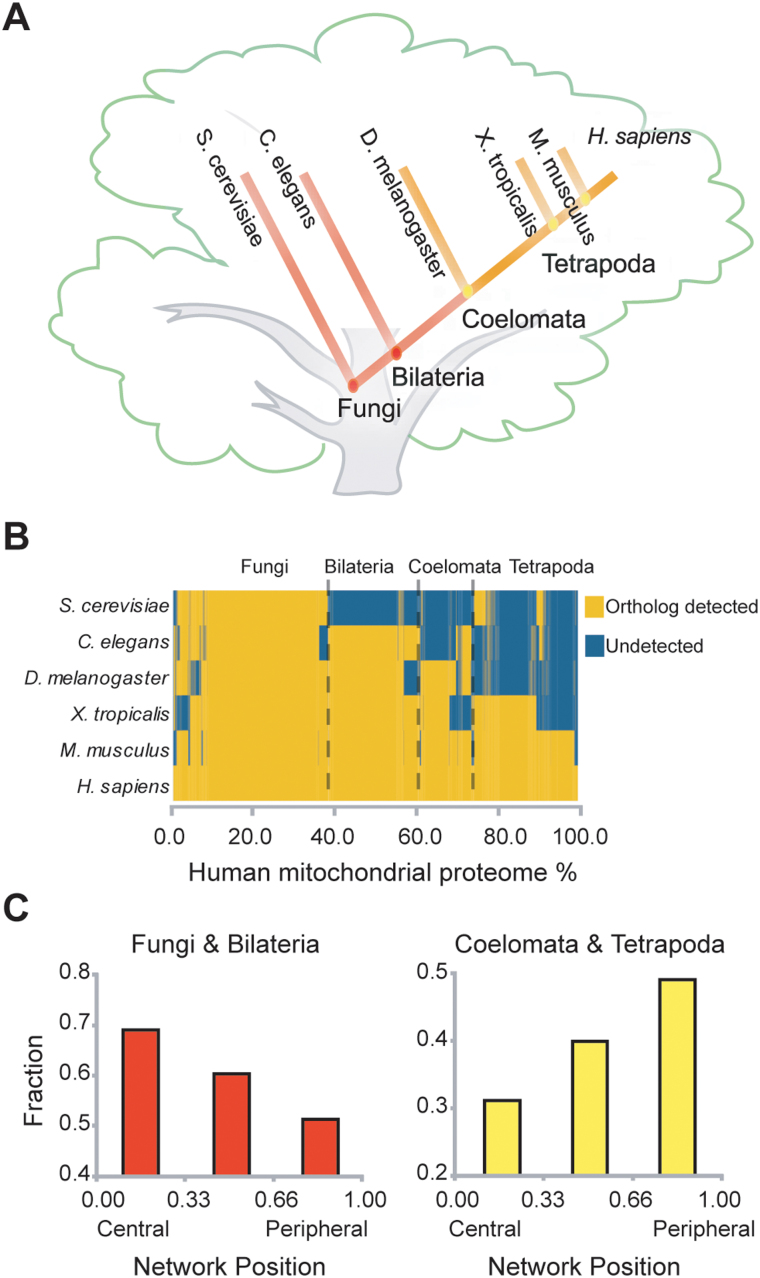
Mitochondria originated from distinct prokaryotic organisms and have coevolved symbiotically with the host cell for the last 1.5 billion years, all the while undergoing enormous changes in their proteome17,25. As a result, they play critical cellular roles aside from ATP production by interacting functionally with host proteins in higher eukaryotes26,27,28. We compared the network positions of mitochondrial proteins evolved from a prokaryotic ancestor with those from a eukaryotic host cell to understand how mitochondrial functions evolved. The evolutionary origin of mitochondrial proteins was analyzed by phylogenetic profiling29 and they were classified according to their network position. We found that recently evolved mitochondrial proteins tended to be located in the periphery of the MPF network (Figure 7). Moreover, the majority of peripheral proteins do not possess prokaryotic homologues30. Typical functions of the peripheral proteins we identified were apoptosis and hormone metabolism, which are usually found in higher eukaryotes17. These data suggest that recently evolved peripheral proteins may be involved in more communicative functions with mitochondria, maintaining the organelle's intimate relationship with the host cell.
Figure 7. Analysis of mitochondrial protein evolution based on network position.
(A) Phylogenetic tree of model species. (B) Phylogenetic profile of mitochondrial proteins. (C) Evolution of mitochondrial proteins according to their network positions.
Peripheral proteins of mitochondria participate in inter-compartmental communication in eukaryotic cells. During eukaryotic evolution, compartmentalization played a major role in increasing cellular complexity; thus, the interconnection between different subcellular organelles was important31,32. In particular, the relationship between mitochondria and other cellular compartments has become a principal feature in controlling cellular physiology33,34. For example, interaction between the endoplasmic reticulum and mitochondria is crucial for the synthesis and intracellular transport of phospholipids, as well as for intracellular Ca2+ homeostasis35. We found that network position is an effective measure for distinguishing OMM proteins from IMM and MM proteins. This is particularly important because contemporary methods for determining sub-mitochondrial localization are ineffective in identifying OMM proteins11,36. Moreover, large-scale proteomics studies are deficient in identifying OMM proteins due to difficulties in separating many pre-translocating matrix and IMM proteins from OMM proteins10.
In this study, we applied network position to investigate the spatial and functional organization of mitochondrial proteins. With the advancement of proteomics technology, large quantities of mitochondrial proteins were identified. However, generating experimental evidences for the sub-mitochondria compartmental location of proteins are laborious and expensive. Thus, our network approach would provide a method to assign the sub-mitochondrial location of proteins and maximize the utility of the other proteomics approaches. Furthermore, we proved that network position can effectively reduce the number of candidate disease genes identified into a manageable number. Hence, network position will be a useful tool for facilitating our understanding of the function of mitochondrial proteins and discovering novel disease-related genes relevant to mitochondrial pathogenesis.
Methods
Collection of mitochondrial proteome data
We collected 2,374 mitochondrial proteins from nine reference data sets: MitoRes37, Locate38, MitoProteome39, MitoP240, MitoCarta41, Maestro20, ConLoc1, HMPDb (http://bioinfo.nist.gov/hmpd), and human orthologs of yeast mitochondrial proteins. We downloaded the protein sequences of yeast from NCBI. We mapped the human orthologs of yeast mitochondrial proteins using Inparanoid8,42 and identified 1,217 primary mitochondrial proteins that were detected more than twice from the individual database (Figure 2).
With the “more than twice” criterion, we could collect primary mitochondrial protein set with high specificity (0.97) and high sensitivity (0.93) (Table S2), which is comparable with the MitoCarta specificity of 0.98, but contains more mitochondrial proteins than MitoCarta. This implies that the applied criterion efficiently reduced the false positives and false negatives. We checked how many yeast orthologs of mitochondrial proteins were included in our dataset. Among 807 human orthologs of yeast mitochondrial proteins, 429 proteins (53%) were included in the primary mitochondria set by the “more than twice” criterion and the rest of the orthologs (47%) were not included. We listed the overlap ratio between human and yeast orthologs of mitochondrial proteins in the supplementary data (Table S3).
We then added 290 mitochondrial proteins that have more than two interaction partners with the primary nodes to consider transient interaction partners. It has been successfully applied to include network nodes that are rarely detected but interact transiently with primary nodes in the protein interactome studies43. The resulting mitochondrial proteome showed high specificity (0.95) and sensitivity (0.93). We provide the list of proteins in the MPF network with the sources of mitochondrial databases and localization annotations (Table S4).
Construction of the MPF network
We mapped 1,541 reliable mitochondrial proteins into a functional network that connects proteins from diverse data sources via their physical and functional interactions, including functional associations from STRING44 and integrated protein-protein interactions. We combined functional associations and protein-protein interactions to build MPF network. Functional associations include direct physical binding and indirect associations, such as two proteins sharing a substrate in metabolic pathway, co-regulating by same transcription factors, or participating in the same protein complexes. STRING database have collected this information through compiling experimental repositories, curated pathway database, literature-mining resources, and computational predictions. For physical interactions, we used CRG-integrated human protein interactions45 that integrated a total of 21 existing protein interaction databases (Table S5). We removed low-confidence interactions that were not found in multiple databases. Ultimately, the MPF network was composed of 6,071 links between 1,254 mitochondrial proteins.
Assigning network positions of mitochondrial proteins
To assign the network position of mitochondrial proteins, we calculated the ratio of mitochondrial versus non-mitochondrial partners for each protein. Central mitochondrial proteins connected predominantly with mitochondrial proteins. However, peripheral mitochondrial proteins had more links with non-mitochondrial proteins than mitochondrial ones. If a protein had less than 30 links, we applied the information propagation algorithm. We propagated the degree values through network connections by assigning degree values to neighboring proteins based on following equation:
where NP(x) represented the network position of mitochondrial protein x and M(x) represented the set of neighboring proteins of x. |A| is the number of proteins in any set A. We converted the NP(x) values to percentile scores after each iteration. The percentile score ranged from 0 to 1, where 1 indicated functional interaction with non-mitochondrial proteins. Through information propagation, we obtained the converged NP(x) values of all mitochondrial proteins.
Collection of sub-mitochondrial localization information
We collected information regarding the sub-mitochondrial localization of each protein from Swiss-Prot database 54.8 (http://www.ebi.ac.uk/swissprot). We only used ‘Homo sapiens' annotation and eliminated terms such as ‘possible', ‘potential', ‘probable', ‘putative', or ‘by similarity' to gather a high-quality localization data set. To validate the performance of network position prediction, we collected an independent data set with sub-mitochondrial annotations from the updated Swiss-Prot 57.2 database (Table S6). Among the updated 24 mitochondrial proteins, there were eight each of outer mitochondrial membrane (OMM), inner mitochondrial membrane (IMM) and mitochondrial matrix (MM) proteins.
Phylogenetic profile of mitochondrial proteins
We used the human orthologs of five model organisms to investigate the evolutionary relationship of mitochondrial proteins. Human orthologs of S. cerevisiae, C. elegans, D. melanogaster, X. tropical, and M. musculus proteins were gathered using Inparanoid42. Each model organism represented the respective clade to which it belonged. We clarified the origin of genes by assessing the appearance of genes among model organisms. We divided the model organisms into two groups according to the period between C. elegans and D. melanogaster to distinguish newly evolved proteins after Coelomata in higher eukaryotes.
Disease-protein association data
We mapped disease associations to each protein using the OMIM database, which lists gene-disease associations for 2,929 disease types defined by the Morbid Map and 1,777 genes associated with particular disease types. Disease types were further categorized into 1,340 distinct diseases by consolidating subtypes into a single disease if similar names were used.
Author Contributions
J.S.Y. designed the whole work, produced all the data, and wrote the paper. J.K., S.P., J.J. and Y.E.S. helped to analyze the data. S.K. supervised the whole work and contributed to the manuscript preparation. All authors read and corrected the manuscript before the submission.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to the members of our laboratories. This work was supported in part by Korean National Research Foundation grants (2012002568, 20110027840, 2012054744, and R312012000101 of the World Class University program).
References
- Park S., Yang J. S., Jang S. K. & Kim S. Construction of functional interaction networks through consensus localization predictions of the human proteome. J Proteome Res 8, 3367–76 (2009). [DOI] [PubMed] [Google Scholar]
- Au C. E. et al. Organellar proteomics to create the cell map. Curr Opin Cell Biol 19, 376–85 (2007). [DOI] [PubMed] [Google Scholar]
- Yook S. H., Oltvai Z. N. & Barabasi A. L. Functional and topological characterization of protein interaction networks. Proteomics 4, 928–42 (2004). [DOI] [PubMed] [Google Scholar]
- Park S. et al. Protein localization as a principal feature of the etiology and comorbidity of genetic diseases. Mol Syst Biol 7, 494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfe C. et al. Mapping gene associations in human mitochondria using clinical disease phenotypes. PLoS Comput Biol 5, e1000374 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienaar I. S., Dexter D. T. & Burkhard P. R. Mitochondrial proteomics as a selective tool for unraveling Parkinson's disease pathogenesis. Expert Rev Proteomics 7, 205–26. [DOI] [PubMed] [Google Scholar]
- Huh W. K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–91 (2003). [DOI] [PubMed] [Google Scholar]
- Perocchi F. et al. Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet 2, e170 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs M. A. et al. Directing experimental biology: a case study in mitochondrial biogenesis. PLoS Comput Biol 5, e1000322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi R. P. et al. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell 17, 1436–50 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P. & Li Y. Prediction of protein submitochondria locations by hybridizing pseudo-amino acid composition with various physicochemical features of segmented sequence. BMC Bioinformatics 7, 518 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler A. M., Kerner J. & Hoppel C. L. Proteomics of mitochondrial inner and outer membranes. Proteomics 8, 4066–82 (2008). [DOI] [PubMed] [Google Scholar]
- Borgatti S. P., Mehra A., Brass D. J. & Labianca G. Network analysis in the social sciences. Science 323, 892–5 (2009). [DOI] [PubMed] [Google Scholar]
- TSE N. & H Z. A network positioning system for the Internet. In Proc USENIX Conference (2004). [Google Scholar]
- Mullen B., Johnson C. & Salas E. Effects of communication network structure: Components of positional centrality. Social Networks 13, (1991). [Google Scholar]
- Clifton A., Turkheimer E. & Oltmanns T. F. Personality Disorder in Social Networks: Network Position as a Marker of Interpersonal Dysfunction. Soc Networks 31, 26–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T. & Huynen M. A. From endosymbiont to host-controlled organelle: the hijacking of mitochondrial protein synthesis and metabolism. PLoS Comput Biol 3, e219 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M. A., de Hollander M. & Szklarczyk R. Mitochondrial proteome evolution and genetic disease. Biochim Biophys Acta 1792, 1122–9 (2009). [DOI] [PubMed] [Google Scholar]
- Lee I., Blom U. M., Wang P. I., Shim J. E. & Marcotte E. M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res 21, 1109–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S. et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet 38, 576–82 (2006). [DOI] [PubMed] [Google Scholar]
- George R. A. et al. Analysis of protein sequence and interaction data for candidate disease gene prediction. Nucleic Acids Res 34, e130 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S., Bauer S., Horn D. & Robinson P. N. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet 82, 949–58 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. J. et al. Mutations in the MPV17 gene are responsible for rapidly progressive liver failure in infancy. Hepatology 46, 1218–27 (2007). [DOI] [PubMed] [Google Scholar]
- Evgrafov O. V. et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet 36, 602–6 (2004). [DOI] [PubMed] [Google Scholar]
- Marcotte E. M., Xenarios I., van Der Bliek A. M. & Eisenberg D. Localizing proteins in the cell from their phylogenetic profiles. Proc Natl Acad Sci U S A 97, 12115–20 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F. et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A. & Schreiber S. L. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci U S A 106, 22229–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B. et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulais J. et al. Molecular characterization of the evolution of phagosomes. Mol Syst Biol 6, 423 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N., Wiedemann N., Meisinger C. & Lithgow T. Assembling the mitochondrial outer membrane. Nat Struct Mol Biol 11, 1044–8 (2004). [DOI] [PubMed] [Google Scholar]
- Ginger M. L., McFadden G. I. & Michels P. A. Rewiring and regulation of cross-compartmentalized metabolism in protists. Philos Trans R Soc Lond B Biol Sci 365, 831–45 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodbeen R. & van Hest J. C. Synthetic cells and organelles: compartmentalization strategies. Bioessays 31, 1299–308 (2009). [DOI] [PubMed] [Google Scholar]
- Ferri K. F. & Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol 3, E255–63 (2001). [DOI] [PubMed] [Google Scholar]
- Rizzuto R. & Pozzan T. Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol Rev 86, 369–408 (2006). [DOI] [PubMed] [Google Scholar]
- Lebiedzinska M., Szabadkai G., Jones A. W., Duszynski J. & Wieckowski M. R. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int J Biochem Cell Biol 41, 1805–16 (2009). [DOI] [PubMed] [Google Scholar]
- Zeng Y. H. et al. Using the augmented Chou's pseudo amino acid composition for predicting protein submitochondria locations based on auto covariance approach. J Theor Biol 259, 366–72 (2009). [DOI] [PubMed] [Google Scholar]
- Catalano D. et al. MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa. BMC Bioinformatics 7, 36 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger J. et al. LOCATE: a mammalian protein subcellular localization database. Nucleic Acids Res 36, D230–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter D., Guda P., Fahy E. & Subramaniam S. MitoProteome: mitochondrial protein sequence database and annotation system. Nucleic Acids Res 32, D463–7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokisch H. et al. MitoP2: the mitochondrial proteome database--now including mouse data. Nucleic Acids Res 34, D705–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini D. J. et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–23 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund A. C., Sjolund E., Ostlund G. & Sonnhammer E. L. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res 36, D263–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart L. M. et al. A systems biology analysis of the Drosophila phagosome. Nature 445, 95–101 (2007). [DOI] [PubMed] [Google Scholar]
- von Mering C. et al. STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 35, D358–62 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi A. & Lehner B. Tissue specificity and the human protein interaction network. Mol Syst Biol 5, 260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information