Abstract
The different steps of the human Top1 (topoisomerase I) catalytic cycle have been analysed in the presence of a pentacyclic-diquinoid synthetic compound. The experiments indicate that it efficiently inhibits the cleavage step of the enzyme reaction, fitting well into the catalytic site. Surprisingly the compound, when incubated with the binary topoisomerase–DNA cleaved complex, helps the enzyme to remove itself from the cleaved DNA and close the DNA gap, increasing the religation rate. The compound also induces the religation of the stalled enzyme–CPT (camptothecin)–DNA ternary complex. Analysis of the molecule docked over the binary complex, together with its chemical properties, suggests that the religation enhancement is due to the presence on the compound of two oxygen atoms that act as hydrogen acceptors. This property facilitates the deprotonation of the 5′ DNA end, suggesting that this is the limiting step in the topoisomerase religation mechanism.
Keywords: camptothecin, molecular docking, religation rate, topoisomerase I
Abbreviations: CPT, camptothecin; Et-KuQ, 1-ethylKuQuinone; TBS, Tris-buffered saline; Top1, topoisomerase I
INTRODUCTION
DNA topoisomerases are key enzymes that modulate the topological state of DNA through the breaking and rejoining of DNA strands. There are two classes of topoisomerases (types I and II), both characterized by a catalytic mechanism that involves a nucleophilic attack of a DNA phosphodiester bond by a tyrosyl residue from the enzyme, but type I cleaves a single DNA strand, whereas type II cleaves both strands. The human Top1 (topoisomerase I) enzyme is composed of 765 amino acids and has four distinct domains: the NH2-terminal domain (1–214), the core domain (215–635), the linker domain (636–712) and the COOH-terminal domain (713–765) [1,2]. The three-dimensional (3D) structure of reconstituted N-terminal truncated version of human Top1 in complex with a 22 bp DNA molecule shows the enzyme organized in multiple domains, which ‘clamp on’ the DNA molecule [3]. The active site tyrosine (Tyr723) starts the catalytic cycle of the enzyme through a nucleophilic attack on the DNA backbone, resulting in the breakage of one DNA strand with the enzyme covalently attached to the 3′-phosphate to form the cleavable complex. After changing the linking number a second nucleophilic attack, driven by the 5′-hydroxyl DNA end, restores an intact double-stranded DNA, and the enzyme is released. In both cases, to become a nucleophile the hydroxylic group belonging to the tyrosine or to the DNA end must donate a proton to a nearby group that has been proposed to be the lateral chain of Lys532[2,4].
DNA Top IB can be inhibited by several compounds that act through different mechanisms, such as prevention of DNA–topoisomerase binding, inhibition of DNA cleavage or stabilization of the cleavable complex. Inhibitors of Top1 are divided into two classes: poisons and catalytic inhibitors. Poisons include clinically used drugs, such as the derivatives of the natural compound CPT (camptothecin) that reversibly binds the covalent Top1–DNA complex slowing down the religation of the cleaved DNA strand, thus inducing cell death [5]. Two water-soluble CPT derivatives, topotecan and irinotecan have been approved by the FDA (Food and Drug Administration) for clinical use. The 3D structure of the topotecan–enzyme–DNA ternary complex has shown that topotecan mimics a DNA base-pair and binds at the site of DNA cleavage by intercalating between the upstream (−1) and downstream (+1) base-pairs [6] interacting also with the enzyme, acting as an interfacial uncompetitive inhibitor [5]. On the other hand, catalytic inhibitors act through inhibiting any other step of the Top1 enzymatic cycle. They include both natural and non-natural compounds [7] and they mainly act by inhibiting the cleavage [8–10] or the DNA binding [11,12], although some of them are able to inhibit both cleavage and religation [13,14]. The CPT derivatives are widely used in clinics; however, they have some intrinsic limits, the main one being linked to the equilibrium of the α-hydroxylactone E-ring of CPTs with the carboxylate form, which is inactive against Top1 and tightly binds to serum albumin. For this reason, other drugs are under development, the most promising one being indolocarbazoles and the indenoisoquinolines [5].
It has been shown that the ADP–ribose polymers, that is rapidly synthesized by PARP [poly(ADP–ribose) polymerase], activated by the occurrence of a DNA break, is also able to interact with human Topo I [15]. The interaction has been elegantly shown to produce two important effects, the first one being the cleavage inhibition, the second one consisting in the enhancement of the religation [16]. The second effect is quite astonishing and poly(ADP–ribose) has been up to now the only molecule able to induce this effect, although the molecular mechanism is still unclear.
In this work, in the search for a new topoisomerase poison, we have investigated the interaction of the enzyme with Et-KuQ (1-ethylKuQuinone) (Figure 1), a pentacyclic-diquinoid synthetic compound [17]. The results indicate that Et-KuQ inhibits cleavage and enhances religation in the presence of CPT. A mechanism for such a process is provided and it is proposed to also occur in the case of poly(ADP–ribose).
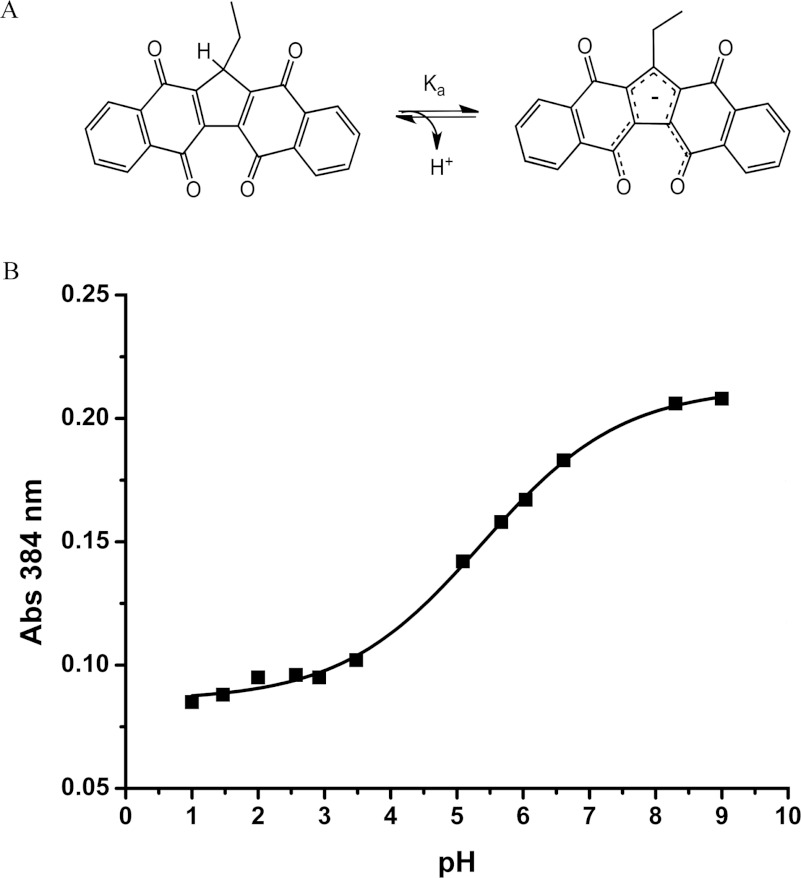
Figure 1. Structure of KuQ.
(A) Structure and dissociation equilibrium of Et-KuQ. (B) Spectrophotometric titration curve for the ionization of Et-KuQ in aqueous solution.
MATERIALS AND METHODS
Chemicals and pH titration
The Et-KuQ was synthesized as previously described [17]. CPT (Sigma) and Et-KuQ were dissolved in DMSO to a final concentration of 4 and 0.5 mg/ml, respectively, and stored at −20°C. Anti-FLAG M2 monoclonal affinity gel, FLAG peptide and anti-FLAG M2 monoclonal antibody were purchased from Sigma.
Et-KuQ acid-base titration
The spectrophothometric titration of a 3.5×10−5 mol/l aqueous solution of Et-KuQ was performed with a Shimadzu UV-2450 spectrophotometer. The various pH values were obtained by diluting HCl or NaOH mother solutions and measuring the desired pH value through a pH metre (Amel 334-B) equipped with a glass electrode CRISON5203. The electrode was checked with commercial standard buffer solutions at pH 4.01, 7.01 and 9.26. The titration curve for the acid ionization was obtained by recording the absorption value of Et-KuQ at 384 nm as a function of pH (Figure 1); a corresponding sigmoid fit of the data [18] provides the pKa value of 5.4.
Yeast strains and plasmids
The Saccharomyces cerevisiae strain EKY3 (ura3-52, his3Δ200, leu2Δ1, trp1Δ63, top1::TRP1, MATα) previously described [19], was used to express the human Top1 gene. Top1 is expressed in YCpGAL1-e-Top1 single copy plasmid [19,20]. The epitope-tagged construct, indicated as ‘e’ contains the N-terminal sequence DYKDDDY and is recognized by the M2 monoclonal antibody.
Top1 purification
Top1 was expressed under the galactose inducible promoter in a single copy plasmid, YCpGAL1-e-Top1, used for transformation of EKY3 cells, that were grown on SC-uracil plus 2% (w/v) dextrose and diluted 1:100 in SC-uracil plus 2% (w/v) raffinose. At a D595=1.0, the cells were induced with 2% (w/v) galactose for 6 h. Cells were then centrifuged, washed with ice-cold water and resuspended in 2 ml buffer/g of cells [50 mM Tris/HCl, pH 7.4, 1 mM EDTA, 1 mM EGTA, 10% (v/v) glycerol and protease inhibitors cocktail (Roche)], supplemented with 0.1 mg/ml sodium bisulfate, 0.8 mg/ml sodium fluoride, 1 mM PMSF and 1 mM DTT (dithiothreitol). After an addition of 0.5 vol of 425–600 mm diameter glass beads, the cells were disrupted by vortexing for 30 s alternating with 30 s on ice and then were centrifuged. The column, containing 2 ml of anti-FLAG M2 affinity gel, was washed with 20 column volumes of TBS (Tris-buffered saline; 50 mM Tris/HCl pH 7.4 and 150 mM KCl), prior to loading the lysate. Elution of FLAG-fusion-e-Top1, was performed by competition with five column volumes of a solution containing 100 mg/ml FLAG peptide in TBS. Fractions (500 μl) were collected and 40% (v/v) glycerol was added in all preparations, which were stored at −20°C [21]. Fractions were resolved by SDS/PAGE using the epitope-specific monoclonal antibody M2 protein. The protein concentration has been estimated by densitometry of the chosen fraction and compared with the purified Top I (purchased from Topogene). Our Top1 has a concentration of 10 ng/μl.
DNA relaxation assays
The activity of 1 μl of Top1 was assayed in 30 μl of reaction volume containing 0.5 μg of negatively supercoiled pBlue-Script KSII(+) DNA, that is present in both dimeric and monomeric forms and reaction buffer (20 mM Tris/HCl pH 7.5, 0.1 mM Na2EDTA, 10 mM MgCl2, 50 μg/ml acetylated BSA and 150 mM KCl). The effect of Et-KuQ on enzyme activity was measured by adding different concentrations of the compound; the dose-dependent analysis has been performed by simultaneously adding Top1 and the supercoiled plasmid in the presence of DMSO as control, or with different concentrations of Et-KuQ or pre-incubating the enzyme with various concentrations of Et-KuQ for 10 min at 37°C before adding the DNA; the reactions were stopped after 60 min. The relaxation activity of Top1, preincubated with 5 μM Et-KuQ, or with the same concentration of DMSO, before adding the supercoiled plasmid, was also analysed as a function of time at 37°C. Reactions were stopped with 0.5% SDS and the samples were resolved in a 1% (w/v) agarose gel in 48 mM Tris, 45.5 mM boric acid, 1 mM EDTA. The gel was stained with ethidium bromide (0.5 μg/ml), destained with water and photographed under UV illumination.
Cleavage kinetics
Oligonucleotide substrate CL14 (5′-GAAAAAAGACTTAG-3′) was radiolabelled with [γ-32P]ATP at the 5′ end. The CP25 complementary strand (5′-TAAAAATTTTTCTAAGTCTTTTTTC-3′) was 5′ phosphorylated with unlabelled ATP. The two strands were annealed at 2-fold molar excess of CP25 over CL14, creating the partial duplex suicide substrate. The cleavage reactions were carried out in 20 mM Tris/HCl pH 7.5, 0.1 mM Na2EDTA, 10 mM MgCl2, 50 μg/ml acetylated BSA and 150 mM KCl at 25°C, by incubating 20 nM CL14/CP25 and an excess of Top1 with DMSO, as no-drug control, or 50 μM Et-KuQ, or pre-incubating the enzyme and the compound for 10 min before adding the suicide substrate [22]. At various time points 5 μl aliquots were removed and the reaction stopped with 0.5% (w/v) SDS. After ethanol precipitation samples were resuspended in 5 μl of 1 mg/ml trypsin to degrade the protein and were analysed by denaturing 7 M urea/20% (w/v) PAGE in 48 mM Tris, 45.5 mM boric acid, 1 mM EDTA. However, a short trypsin-resistant peptide is always left, explaining why the Cl1 migrates slower than the Cl14 oligonucleotide [23]. The percentage of cleavage, determined by PhosphorImager and ImageQuant software, was calculated in each lane as the ratio between the amount of cleavage product and the total amount of radioactivity. The percentage of cleavage product, corresponding to the plateau observed for the experiment carried out with the substrate and the enzyme alone, was then considered as 100% of cleavage product and all the other measurements were normalized to this value.
Religation kinetics
CL14/CP25 (20 nM) was incubated with an excess of Top1 for 60 min at 25°C followed by 30 min at 37°C in 20 mM Tris/HCl pH 7.5, 0.1 mM Na2EDTA, 10 mM MgCl2, 50 μg/ml acetylated BSA and 150 mM KCl. After the formation of the cleavable complex (Cl1) a 5-μl aliquot was removed and used as the time 0 point, then DMSO or 50 μM Et-KuQ were added and the religation reaction was started by adding a 200-fold molar excess of R11 oligonucleotide (5′-AGAAAAATTTT-3′) over the CL14/CP25 [21]. The KCl concentration was increased to 350 mM when R11 was added, to avoid the religated product being recleaved by the protein [24]. Where indicated, the compound was pre-incubated with the cleavable complex for 10 min at 37°C, before adding the R11 oligonucleotide. At 37°C 5 μl aliquots were removed at various time points, and the reaction stopped with 0.5% (w/v) SDS. After ethanol precipitation, samples were resuspended in 5 μl of 1 mg/ml trypsin and incubated at 37°C for 60 min. Samples were analysed by denaturing 7 M urea/20% (w/v) PAGE in 48 mM Tris, 45.5 mM boric acid, 1 mM EDTA. The religation percentage, determined by ImageQuant software, was plotted as a function of time and normalized to the plateau value.
Cleavage/religation equilibrium
Oligonucleotide CL25 (5′-GAAAAAAGACTTAGAAAAATTTTTA-3′) was 5′ radiolabelled with [γ-32P]ATP. The CP25 complementary strand (5′-TAAAAATTTTTCTAAGTCTTTTTTC-3′) was phosphorylated at its 5′ end with unlabelled ATP. The two strands were annealed at a 2-fold molar excess of CP25 over CL25. The assay was done by adding 20 nM duplex CL25/CP25 to Top1 alone (DMSO), in the presence of 50 μM CPT, in the presence of 50 μM Et-KuQ or after incubating the reaction mixture with 50 μM CPT for 1 min and then adding 50 μM Et-KuQ, or incubating with 50 μM Et-KuQ for 1 min and then adding 50 μM CPT. Reactions were performed at 25°C in 20 mM Tris/HCl pH 7.5, 0.1 mM Na2EDTA, 10 mM MgCl2, 50 mg/ml acetylated BSA and 150 mM KCl and were stopped at two different times: 1 and 2 min, by adding 0.5% (w/v) SDS, precipitating with ethanol and digesting with trypsin [25]. Products were resolved in 20% (w/v) PAGE/7 M urea gel and quantified using ImageQuant software. The values were calculated by the ratio between the cleavage amount and the total radioactivity of each lane, subtracting the control background.
Molecular docking
The deprotonated Et-KuQ molecular geometry in the gas phase was obtained at the B3LYP/6-31+G(d,p) level of theory using GAMESS-US-2011 ed. [26]. The molecular geometry of the Top1 non-covalent binary complex was obtained by modelling the 1A36 PDB entry; after removing the DNA, the same structure was used to obtain a model of the Top1 enzyme in the absence of DNA. The 1T8I PDB entry was used to obtain a model for the ternary complex. The MGLTools 1.5.6 rc1 was used to convert the PDB into PDBQT format, to calculate the Gasteiger-Marsilii charges and produce the input file. Grid was calculated using an in-house modified version of AutoGrid 4.2.2.1 using a 256×256×256 grid with step of 0.5 Å (1 Å = 0.1 nm) enclosing the whole Top1. 250 runs of docking were generated using the standard GA algorithm with AutoDock 4.2.2.1 [27] for Et-KuQ on each of the three systems. The complexes were clusterized on the basis of their RMSD (root-mean-square deviation), using a cut-off value of 2 Å. Figures 6–8 were generated with UCSF Chimera 1.9 (http://www.cgl.ucsf.edu/chimera).
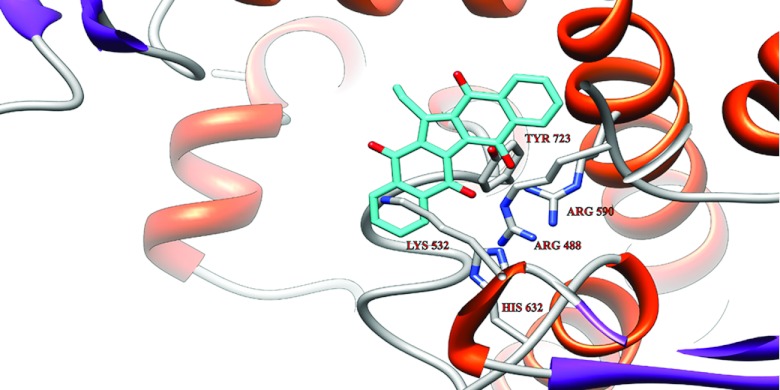
Figure 6. Representative structure of Et-KuQ docked on Top1 in the absence of DNA.
The Et-KuQ (cyan) and the residues of the catalytic pentad are represented as sticks, while the rest of the protein appears as a ribbon. The lateral chains of the residues of the catalytic pentad are explicitly represented and labelled.
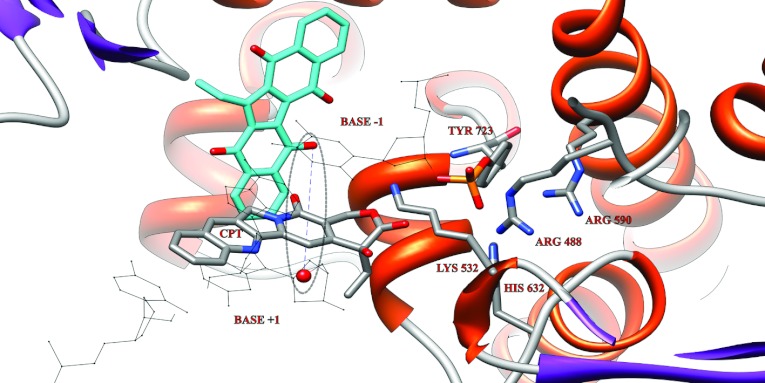
Figure 8. Representative structure of Et-KuQ docked on the CPT+Top1+DNA ternary complex.
The Et-KuQ is represented in cyan; the lateral chains of the residues of the catalytic pentad and the CPT are represented as sticks and labelled; the rest of the protein appears as a ribbon; the base-pairs in position +1 and −1 as black lines. A blue dashed line indicates the distance between the 5′ DNA end, represented as a red sphere, and the closest Et-KuQ oxygen acceptor.
RESULTS AND DISCUSSION
Et-KuQ physical-chemical properties
KuQuinones are a new class of pentacyclic di-quinoid compounds that have recently been discovered and synthesized with a facile one-pot reaction [17]. The polycyclic system can be considered a naphthoquinone derivative and its name is due to the structural similarities with vitamin K. In this work the Et-KuQ has been reacted with the human Top1 enzyme. The main feature of its structure is the presence of a pentacyclic skeleton (similarly to CPT) with a double-quinone functional group; the five rings consist of two naphthalene units fused through a 5-membered moiety that contains an alkyl chain (Figure 1A). Extended electronic conjugation is proved by the intense absorption spectrum with a broad band between 400 and 600 nm. These molecules were isolated as enol tautomers stabilized by the intramolecular hydrogen bond with the nearest carbonyl group, as shown in the crystal structure of the Et-KuQ derivative [17]. The strong hydrogen-bond interaction is evidenced by the very high value of the chemical shift (18 ppm) of the enolic hydrogen in the 1H NMR spectrum. On the other hand, the shift of the relative tautomeric equilibrium towards the keto form appears to be strongly sensitive to solvent polarity and hydrogen-bonding capacity. Therefore it can be reasonably assumed that the keto tautomer is favoured in aqueous media; a complete characterization of the equilibrium is under current investigation.
The pH dependence of the spectrum permits us to evaluate a pKa of 5–6 for the equilibrium between a protonated and a deprotonated form (Figure 1B). The unusual acidity of the compound compares well with that reported for the hipposudoric acid [28], a natural structurally related compound. In fact, the conjugate base of Et-KuQ, i.e. the enolate species, is particularly stable because of the stabilization of the negative charge within the molecule.
Et-KuQ inhibits the relaxation activity of Top1
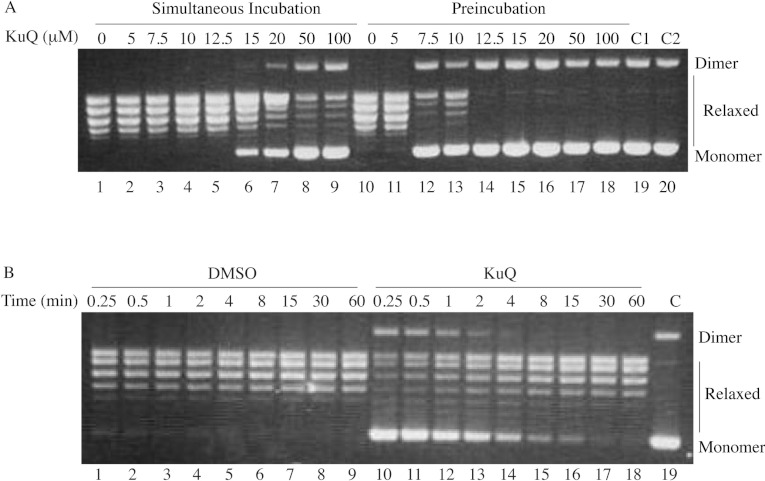
The relaxation activity of native Top1 has been assessed by incubating the enzyme in the presence of increasing concentration of Et-KuQ in a plasmid relaxation assay (Figure 2A). The simultaneous addition of enzyme, Et-KuQ and DNA never resulted in complete inhibition of the plasmid relaxation activity up to a concentration of 100 μM. The inhibitory effect is greater when the enzyme is pre-incubated with Et-KuQ before adding DNA and the compound fully inhibits the relaxation at a concentration of 12.5 μM (Figure 2A, lane 14). An additional control has been performed (C2 Figure 2A, lane 20) by adding the drug to the DNA plasmid, demonstrating that the Et-KuQ does not permit DNA migration and so does not bind to DNA. The relaxation assay carried out as function of time at an Et-KuQ concentration of 5 μM indicated that the inhibitory effect is abolished after 15 min (Figure 2B, lane 16), demonstrating that the binding of the compound is reversible. Since Et-KuQ is dissolved in DMSO, as a control, the reaction in presence of this solvent has been carried out and it shows that the enzyme, at the same DMSO concentration, maintains its activity over time (Figure 2B, lanes 1–9).
Figure 2. Relaxation of supercoiled DNA.
(A) Relaxation of negative supercoiled plasmid by Top1 incubated with increasing concentrations of Et-KuQ (lanes 2–9) or after pre-incubation for 10 min at 37°C with increasing concentrations of Et-KuQ (lanes 11–18). Lanes 1 and 10, no drug added; lane 19, no drug and protein added, lane 20, no protein added. (B) Relaxation of negative supercoiled plasmid in a time course experiment for DMSO (lanes 1–9), 5 μM Et-KuQ (lanes 10–18); lane 19, no protein added. The reaction products are resolved in an agarose gel and visualized with ethidium bromide. The two forms of the supercoiled plasmid DNA are indicated as ‘Dimer’ and ‘Monomer’.
Cleavage and religation analysis
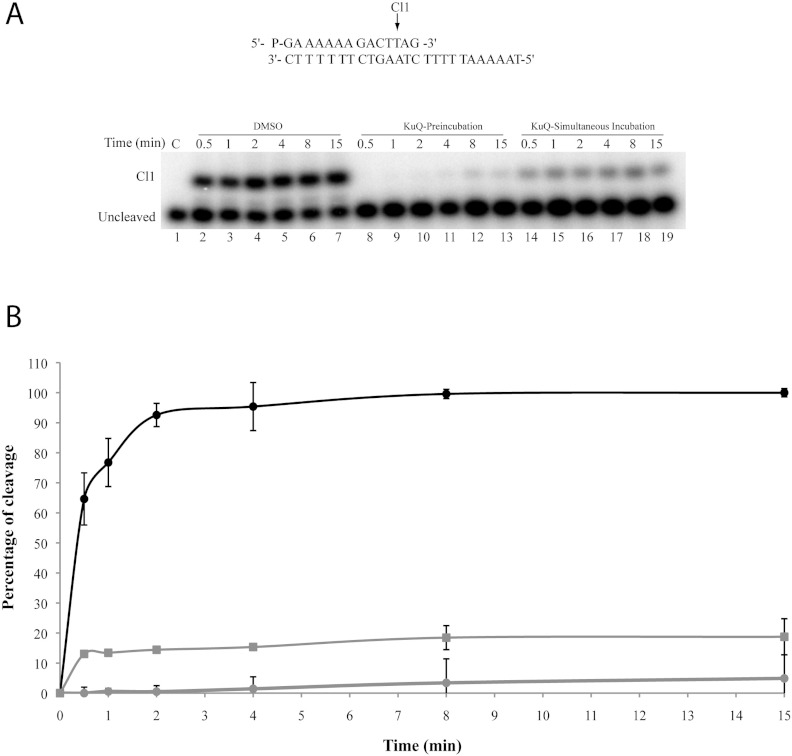
The time course of the cleavage of the wild-type enzyme in the presence and absence of Et-KuQ has been followed using a suicide cleavage substrate. In detail, a 5′ end radiolabelled oligonucleotide CL14 (5′-GAAAAAAGACTTAG-3′) was annealed to the CP25 (5′-TAAAAATTTTTCTAAGTCTTTT-TTC-3′) complementary strand, to produce a duplex with an 11-base 5′ single-strand extension that is preferentially cut by the enzyme at the site indicated by the arrow (Figure 3A, top). The religation step is precluded, because the dinucleotide, generated during cleavage, is too short to be religated, leaving the enzyme covalently attached to the 3′ end of the oligonucleotide. The data indicate that in the absence of the drug the wild-type protein shows a typical time-dependent cleavage of the fragment (Figure 3A, lanes 2–7), whereas pre-incubation of the enzyme with 50 μM Et-KuQ fully inhibits the cleavage reaction (lanes 8–13). The inhibitory effect of the compound, after a simultaneous incubation with both the enzyme and the suicide substrate, is also clear (lanes 14–19), although lower than that observed for the pre-incubation. These results can be well appreciated from the plot in Figure 3(B) where the percentage of cleavage is reported as a function of time. In detail in the absence of the drug (black line) the maximum of the cleaved product is observed after 4 min, while in the presence of the drug preincubated with enzyme (grey line with circles), no cleaved product is observed in the first 4 min and a small percentage is observed in the 8–15 min time window. This result unambiguously indicates that Et-KuQ is an inhibitor of cleavage and that its interaction with the enzyme is reversible due to the appearance of a small percentage of cleaved product at long times.
Figure 3. Cleavage kinetics.
(A) Time course (0.5–15 min) of the cleavage experiment between the CL14/CP25 suicide substrate and the purified Top1 alone (lanes 2–7), after 10 min pre-incubation with 50 μM Et-KuQ (lanes 8–13) or simultaneously incubated with 50 μM Et-KuQ (lanes 14–19). In lane 1 the protein has not been added. Cl1 represents the DNA substrate cleaved by Top1 at the preferred cleavage site, indicated by an arrow over the DNA sequence. (B) The percentage of cleavage, determined by PhosphorImager and ImageQuant software, was normalized to the total amount of radioactivity in each lane. The DMSO (black line), simultaneous incubation with Et-KuQ (grey line with squares) and the experiment performed pre-incubating the Top1 with Et-KuQ (grey line with circles) are shown.
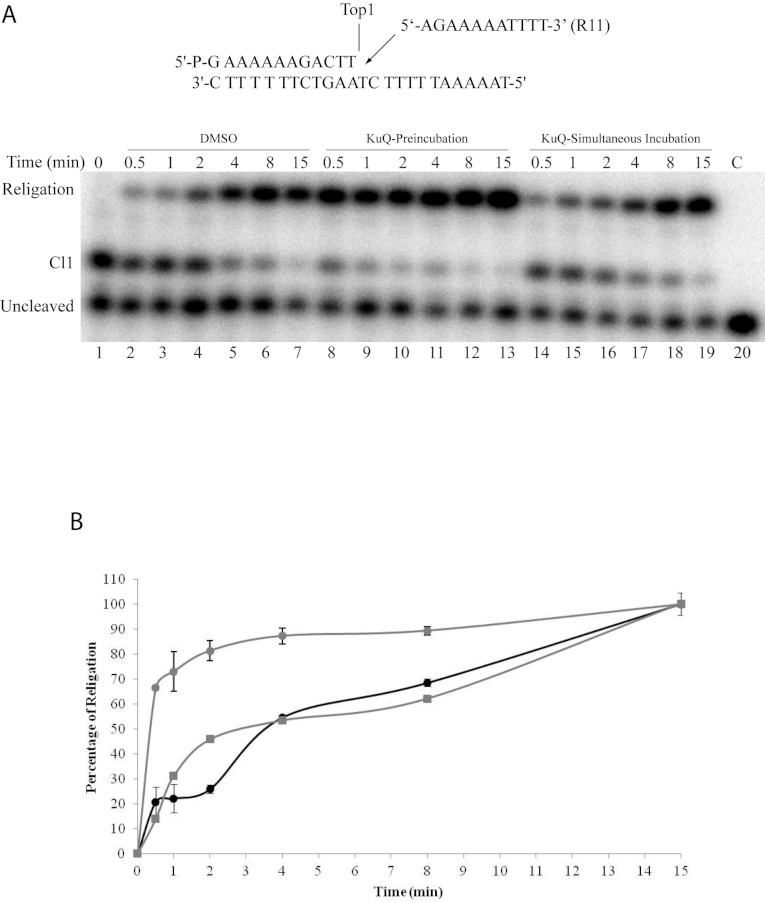
In order to analyse the effect of Et-KuQ on the religation reaction, Top1 was incubated with the suicide substrate, to allow the cleavage reaction (Figure 4A, lane 1). The religation is then followed in the absence of the compound (Figure 4A, lanes 2–7), or by pre-incubating the cleavable suicide complex with 50 μM Et-KuQ before adding 200-fold molar excess of the complementary R11 oligonucoleotide (Figure 4A, lanes 8–13) or adding Et-KuQ together with the religation substrate (Figure 4A, lanes 14–19). In all the experiments, the KCl concentration is maintained at 350 mM when R11 is added to the samples, in order to avoid the religated product being cleaved again by the protein, since cleavage reaction is inhibited at high salt concentration [24]. Aliquots have been removed at different times, the reaction stopped by addition of SDS and the products analysed by PAGE (Figure 4A). The Figure clearly shows that Et-KuQ, when preincubated for 10 min with the covalent complex, stimulates the rate of religation (lanes 8–13), the amount of product formed being much higher than in the reaction carried out in the absence of Et-KuQ, or without preincubating it with the covalent complex. A quantitative analysis of this result is shown in the graph in Figure 4(B). This is to our knowledge the first time that a small organic compound acts as a stimulator of the Top1 religation reaction. The same experiment, performed using an excess of Et-KuQ (200 μM), shows that, at this high Et-KuQ concentration, an increase of the rate of religation is observed also when the compound is added together with the R11 substrate (see Supplementary Figure S1 at http://www.bioscirep.org/bsr/033/bsr033e025add.htm).
Figure 4. Religation kinetics.
(A) Gel analysis of the religation kinetics (0.5–15 min) between the wild-type suicide covalent complex (Cl1) and the R11 complementary oligonucleotide (shown at the top of the Figure) in the absence of Et-KuQ (lanes 2–7), after pre-incubation with 50 μM Et-KuQ (lanes 8–13), or incubated together with 50 μM Et-KuQ (lanes 14–19). In lane 1 the cleaved complex, which represents time 0, has been loaded. In lane 20 no protein was added. ‘Cl1’ represents the DNA fragment cleaved at the preferred enzyme site; ‘Religation’ is the restored fully duplex oligonucleotide that is the final product of the reaction. (B) Percentage of religation, determined by ImageQuant software, was plotted as a function of time and normalized to the plateau value for the experiment with Top1 alone. Substrate and Top1 (black circles), substrate added to the enzyme pre-incubated with Et-KuQ (grey circles), simultaneous mixing of substrate, enzyme and drug (grey squares).
Cleavage/religation equilibrium assay
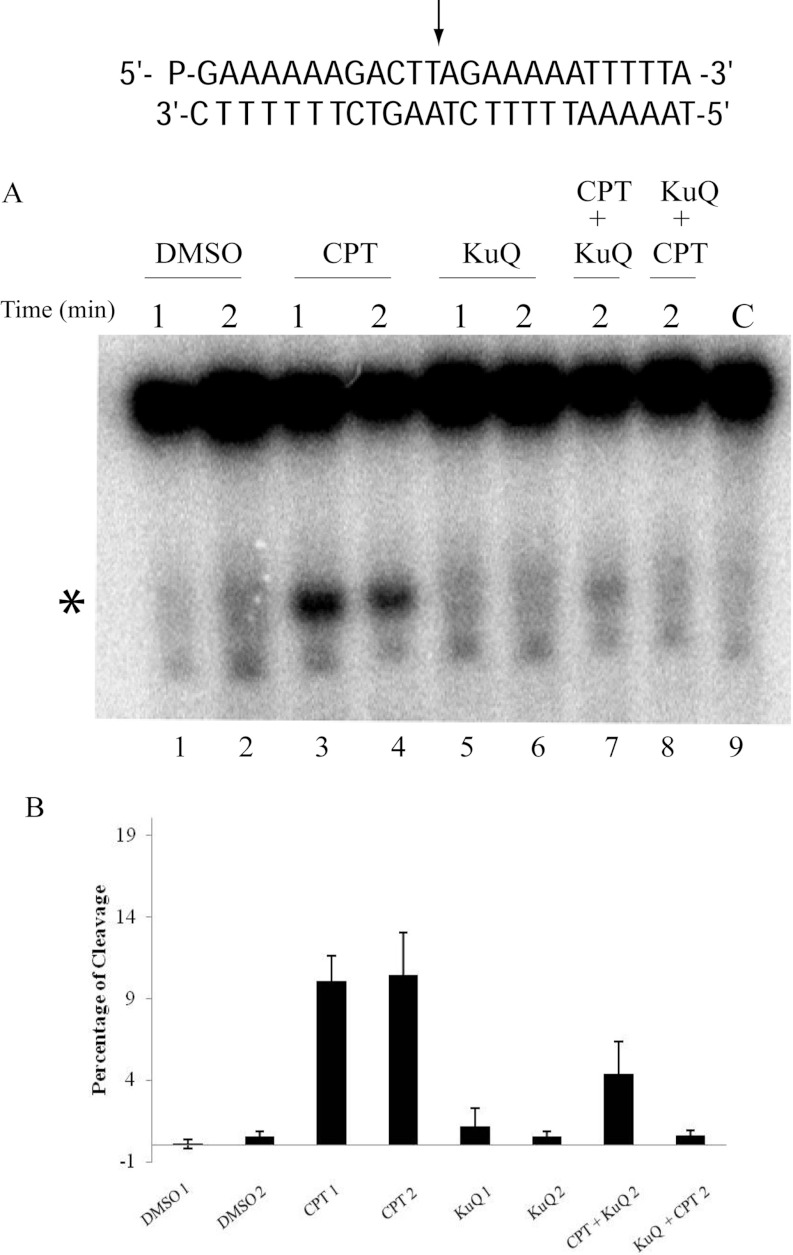
To understand the effect of Et-KuQ on the stability of the covalent DNA–Top1 complex, a cleavage/religation equilibrium experiment on the 25-mer full duplex oligonucleotide has also been performed (Figure 5A). The assay was done at two different times: 1 and 2 min, respectively, for the substrate and the enzyme alone (lanes 1 and 2), in the presence of 50 μM CPT (lanes 3 and 4), or with 50 μM Et-KuQ (lanes 5 and 6), or after incubating the reaction mixture with 50 μM CPT and then adding 50 μM Et-KuQ and stopping the reaction after another minute (lane 7), or incubating with 50 μM Et-KuQ for 1 min and then adding 50 μM CPT and stopping the reaction after another minute (lane 8). The reactions were stopped with SDS, the samples digested with trypsin and the products analysed by PAGE (Figure 5A). In the presence of DMSO, the cleavage/religation equilibrium is shifted towards religation and a small level of cleavage of the labelled DNA strand is detected (Figure 5A, lanes 1 and 2). When the enzyme is in the presence of 50 μM CPT the cleavage/religation equilibrium is shifted towards cleavage, since CPT reversibly binds to the covalent DNA–enzyme intermediate, stabilizing it and slowing down the religation rate, as indicated by the band indicated by the asterisk corresponding to the substrate cleaved at the preferred site (Figure 5A, lanes 3 and 4) [5,29,30]. The band of the cleavage complex is not observed when Et-KuQ is added to the enzyme together with the substrate (Figure 5A, lanes 5 and 6), confirming that the compound inhibits the cleavage step. In line with this, incubation for 1 min of the enzyme, the substrate and the Et-KuQ, followed by addition and incubation of CPT for another minute, shows a band (Figure 5A, lane 8) similar to that observed in the presence of Et-KuQ alone, confirming that Et-KuQ is an inhibitor of cleavage. The most striking result is obtained upon addition of Et-KuQ to the enzyme previously reacted with the substrate in the presence of CPT. The Et-KuQ addition brings to 50% destabilization of the cleaved complex stabilized by CPT (Figure 5A, lane 7 compared with lane 4). This result indicates that Et-KuQ is able to stimulate the religation even in the presence of CPT, overcoming the poisoning effect of the drug on inhibiting the religation. A quantification of the cleaved complex observed in the different assays is shown in the histogram of Figure 5(B), that indicates the ability of Et-KuQ to reduce the CPT stabilization of the cleaved complex (compare lane 4 with lanes 7 and 8). These data provide evidence that Et-KuQ has a dual effect, inhibiting the cleavage and surprisingly promoting the religation, even in the presence of CPT.
Figure 5. Cleavage/religation equilibrium.
(A) Gel electrophoresis of the products from the incubation of Top1 with the [γ -32P] end-labelled duplex DNA (CL25/CP25), shown at the top of the Figure. The arrow indicates the preferred cleavage site. The duplex was incubated for 1 or 2 min with the enzyme (lanes 1 and 2), in presence of CPT (lanes 3 and 4), Et-KuQ (lanes 5 and 6), after a pre-incubation with CPT and then addition of Et-KuQ (lane 7) and after pre-incubation with Et-KuQ and then addition of CPT (lane 8). Lane 9, no enzyme added. The band corresponding to the enzyme-substrate cleaved complex is indicated by an asterisk. (B) Percentage of the cleavable complex normalized to the total radioactivity of each lane.
Molecular docking
Three molecular docking experiments were performed on the Et-KuQ deprotonated form (Figure 1A), in order to study the molecular mechanism underlying the observed inhibition of cleavage and the enhancement of religation. The first-docking experiment was performed using as a receptor the crystal structure of the Top1–DNA non-covalent complex [3] after DNA removal. The complexes obtained have been subjected to clusterization and the largest cluster shows that Et-KuQ interacts with the Top1 catalytic pentad with an energy of −9.6 kcal/mol (1 kcal = 4.184 kJ) (Figure 6). In this structure the Et-KuQ deprotonated oxygen molecules are at hydrogen bonding distance with the side-chains of three residues of the catalytic pentad: Arg488, Arg590 and Tyr723. This conformation is consistent with the observed inhibition of the DNA cleavage, and with the increase of the inhibition in the case of pre-incubation (Figure 3).
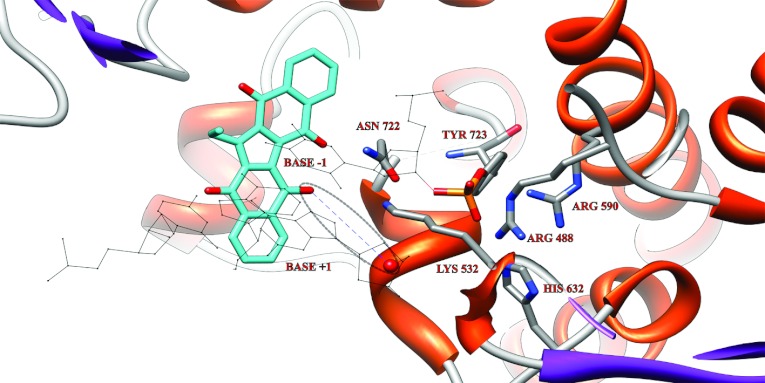
The second docking experiment was performed on the Top1–DNA covalent complex [3]. Owing to the presence of DNA covalently bound to the catalytic Tyr723, Et-KuQ cannot bind in the position previously observed in Figure 6 for the docking with the protein alone. When the docking is made with the Top1–DNA covalent complex, Et-KuQ has a binding site in proximity of the catalytic pentad with energy of −9.8 kcal/mole (Figure 7). In this complex the ethyl group of the drug and its aromatic scaffold show van der Waals interactions with the DNA major grove at the level of the bases in positions −1 and +1, while the deprotonated oxygen molecules of Et-KuQ form an hydrogen bond with the lateral chain of Asn722 (Figure 7). This structure permits us to provide an interpretation of the effect of the compound during the religation process. In fact, the deprotonated oxygen molecules of Et-KuQ that represent a strong hydrogen acceptor site [17] and that can then help the deprotonation process that the hydroxyl group of the 5′ DNA terminal must undergo to reseal the cleaved DNA strand, are at an average distance from the DNA O5′ of 6.7 Å (Figure 7). In the crystal structure of the covalent enzyme–DNA binary complex the distance between the DNA O5′ and the Nζ Lys532, that is considered the proton acceptor site, is 7.2 Å [4,6]. The relatively short distance between the DNA O5′ and a proton acceptor site provides an explanation for the increase of the religation rate observed in the presence of Et-KuQ, shown in Figure 4.
Figure 7. Representative structure of Et-KuQ docked on the covalent Top1+DNA binary complex.
The Et-KuQ is represented in cyan; the lateral chains of the residues of the catalytic pentad and of the Asn722 are represented as sticks and labelled, the rest of the protein is represented as ribbon. The base-pairs in position +1 and −1 are represented in black. A blue dashed line indicates the distance between the 5′ DNA end, represented as a red sphere and the closest Et-KuQ oxygen acceptor.
In order to confirm that this mechanism can be involved in the displacement of CPT by Et-KuQ (Figure 5), a third docking experiment was performed on the CPT+TopoI+DNA ternary complex. In this complex the CPT interfacial inhibitor intercalates between the cleaved DNA bases, making the distance between the O5′ DNA end and the Lys532 Nζ proton acceptor as long as 11.9 Å [6]. Docking experiments indicate that Et-KuQ can bind in proximity of the Top1 catalytic pentad with an energy of −10.7 kcal/mol (Figure 8), forming a complex where the Et-KuQ deprotonated oxygen molecules are at distance of 7.4 Å from the hydroxyl of the 5′ DNA end, i.e. the same distance observed between the 5′ end and the Lys532 in the X-ray covalent binary complex [3]. In the docked complex, Et-KuQ makes contacts with the DNA bases in positions +1 and −1 through its carbon scaffold and forms a hydrogen bond with the Lys755 of the CTD (C-terminal domain) of Top1 through the deprotonated oxygen molecules. The relatively short distance between the Et-KuQ deprotonated oxygen molecules and the cleaved DNA 5′-terminal (about 7 Å) makes possible the 5′ deprotonation process, permitting the release of the cleaved complex even in the presence of CPT as experimentally observed (Figure 5).
The chemical properties of Et-KuQ, characterized by the presence of two deprotonated oxygen molecules that in solution display a pKa=5.5 (Figure 1B), whose value can be modulated by the protein environment, together with the docking experiment, provide a molecular explanation for the facilitated religation in the presence of the compound. The Et-KuQ oxygen molecules may, in fact, act as hydrogen acceptors facilitating the religation process.
Conclusion
The planar pentacyclic structure of Et-KuQ permits a favourable interaction of the compound inside the human Top1 catalytic site as shown by the docking experiments with the enzyme alone (Figure 6), explaining the inhibition of the relaxation and of the cleavage displayed by the compound (Figures 2 and 3).
On the other hand, the proximity between the oxydryl of the DNA 5′end and the two close oxygen atoms (Figure 7), acting as efficient hydrogen acceptors, explains the ability of the compound to act as an enhancer of the enzyme religation rate (Figure 4). The Et-KuQ pKa defining the equilibrium between the protonated and deprotonated form is 5.6 in solution (Figure 1) and this value can be slightly modulated by the protein environment, facilitating the acceptance of an hydrogen by the two oxygen molecules that are at an average distance of 6.7 Å from the oxydryl group of the DNA 5′end in the docked complex with the covalent enzyme–DNA structure (Figure 7). This distance is shorter than the one observed in the X-ray diffraction structure between the O5′ and the lateral chain of Lys532 (3), that is supposed to be the proton acceptor in the reaction in the absence of any external compound [4]. This short distance, coupled with the hydrogen acceptor property of the two oxygen molecules, explains the increased religation rate promoted by Et-KuQ.
The same characteristics can be proposed to explain the Et-KuQ ability in displacing the stalled CPT–DNA–enzyme ternary complex and promoting the religation (Figure 5). In this case the Et-KuQ oxygen molecules-O5′ DNA distance, observed in the docked structure, is about 7 Å (Figure 8), permitting the deprotonation of the DNA oxydryl group. In this latter case two possible mechanisms can be envisaged. (1) The DNA oxydryl group releases its proton to the Et-KuQ oxygen molecules and the deprotonated 5′ end forces the CPT displacement promoting the religation reaction. (2) The DNA hydroxyl group releases its proton to the Et-KuQ oxygen molecules and the reversible binding of CPT finds the deprotonated 5′ ready to religate during the CPT off process. In both cases, the final result is that the religation occurs and CPT is displaced.
Recently it has been proposed that poly(ADP–ribose) displays a similar effect: it inhibits the cleavage, but it also reverses the preformed covalent Top–DNA complex, stimulating the religation activity of the enzyme [15,16]. It is probable that the same mechanism proposed for Et-KuQ can also work for the poly(ADP–ribose) polymer. The complex structure of the poly(ADP–ribose), made by many phosphate groups, may facilitate the deprotonation of the 5′ DNA end, again facilitating the religation. It can then be concluded that facilitation of the deprotonation of the 5′ DNA end is the crucial process to promote religation.
AUTHOR CONTRIBUTION
Barbara Arnò carried out the majority of the experimental part of the study, helped by Cinzia Tesauro and Laura Zuccaro. Sara Lentini, Pierluca Galloni, Valeria Conte and Barbara Floris performed the Et- KuQ synthesis and the Et-KuQ acid–base titration. Andrea Coletta carried out all of the docking analysis. Paola Fiorani and Alessandro Desideri designed, co-ordinated the studies and co-wrote the paper. All authors read and approved the paper.
FUNDING
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) [project 10121 (to A.D.)].
References
- 1.Wang J. C. DNA topoisomerases. Ann. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Champoux J. J. DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Redinbo M. R., Stewart L., Kuhn P., Champoux J. J., Hol W. G. J. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 4.Interthal H., Quigley P. M., Hol W. G. M., Champoux J. J. The role of lysine 532 in the catalytic mechanism of human topoisomerase I. J. Biol. Chem. 2004;279:2984–2992. doi: 10.1074/jbc.M309959200. [DOI] [PubMed] [Google Scholar]
- 5.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staker B. L., Hjerrild K., Feese M. D., Behnke C. A., Burgin A. B., Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl Acad. Sci. U.S.A. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castelli S., Coletta A., D’Annessa I., Fiorani P., Tesauro C., Desideri A. Interaction between natural compounds and human topoisomerase I. Biol. Chem. 2012;393:1327–1340. doi: 10.1515/hsz-2012-0240. [DOI] [PubMed] [Google Scholar]
- 8.Montaudon D., Palle K., Rivory L. P., Robert J., Douat-Casassus C., Quideau S., Bjornsti M. A., Pourquier P. Inhibition of topoisomerase I cleavage activity by thiol-reactive compounds. Importance of vicinal cysteine 504 and 505. J. Biol. Chem. 2007;282:14403–14412. doi: 10.1074/jbc.M611673200. [DOI] [PubMed] [Google Scholar]
- 9.Castelli S., Campagna A., Vassallo O., Tesauro C., Fiorani P., Tagliatesta P., Oteri F., Falconi M., Majumder H. K., Desideri A. Conjugated eicosapentaenoic acid inhibits human topoisomerase IB with a mechanism different from camptothecin. Arch. Biochem. Biophys. 2009;486:103–110. doi: 10.1016/j.abb.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Castelli S., Vassallo O., Katkar P., Che C. M., Wai-Yin Sun R., Desideri A. Inhibition of human DNA topoisomerase IB by a cyclometalated gold III compound: analysis on the different steps of the enzyme catalytic cycle. Arch. Biochem. Biophys. 2011;516:108–112. doi: 10.1016/j.abb.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Boege F., Straub T., Kehr A., Boesenberg C., Christiansen K., Andersen A., Jakob F., Kohrle J. Selected novel flavones inhibit the DNA binding or the DNA religation step of eukaryotic topoisomerase I. J. Biol. Chem. 1996;271:2262–2270. doi: 10.1074/jbc.271.4.2262. [DOI] [PubMed] [Google Scholar]
- 12.Minderman H., Wrzosek C., Cao S., Utsugi T., Kobunai T., Yamada Y., Rustum Y. M. Mechanism of action of the dual topoisomerase-I and -II inhibitor TAS-103 and activity against (multi)drug resistant cells. Cancer Chemother. Pharmacol. 2000;45:78–84. doi: 10.1007/PL00006747. [DOI] [PubMed] [Google Scholar]
- 13.Tesauro C., Fiorani P., D’Annessa I., Chillemi G., Turchi G., Desideri A. Erybraedin C, a natural compound from the plant Bituminaria bituminosa, inhibits both the cleavage and religation activities of human topoisomerase I. Biochem. J. 2010;425:531–539. doi: 10.1042/BJ20091127. [DOI] [PubMed] [Google Scholar]
- 14.Castelli S., Katkar P., Vassallo O., Falconi M., Linder S., Desideri A. A natural anticancer agent thaspine targets human topoisomerase IB. Anticancer Agents Med. Chem. 2012;13:356–363. doi: 10.2174/1871520611313020021. [DOI] [PubMed] [Google Scholar]
- 15.Malanga M., Althaus F. R. Poly(ADP-ribose) reactivates stalled DNA topoisomerase I and induces DNA strand break resealing. J. Biol. Chem. 2004;279:5244–5248. doi: 10.1074/jbc.C300437200. [DOI] [PubMed] [Google Scholar]
- 16.Malanga M., Althaus F. R. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 17.Coletti A., Lentini S., Conte V., Floris B., Bortolini O., Sforza F., Grepioni F., Galloni P. Unexpected one-pot synthesis of highly conjugated pentacyclic diquinoid compounds. J. Org. Chem. 2012;77:6873–6879. doi: 10.1021/jo300985x. [DOI] [PubMed] [Google Scholar]
- 18.Iglesias E. Enolization of Benzoylacetone in aqueous surfactant Solutions: a novel method for determining enolization constants. J. Phys. Chem. 1996;100:12592–12599. [Google Scholar]
- 19.Bjornsti M. A., Benedetti P., Viglianti G. A., Wang J. C. Expression of human DNA topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989;49:6318–6323. [PubMed] [Google Scholar]
- 20.Kauh E. A., Bjornsti M. A. SCT1 mutants suppress the camptothecin sensitivity of yeast cells expressing wild-type DNA topoisomerase I. Proc. Natl Acad. Sci. U.S.A. 1995;92:6299–6303. doi: 10.1073/pnas.92.14.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiorani P., Tesauro C., Mancini G., Chillemi G., D’Annessa I., Graziani G., Tentori L., Muzi A., Desideri Alessandro. Evidence of the crucial role of the linker domain on the catalytic activity of human topoisomerase I by experimental and simulative characterization of the Lys681Ala mutant. Nuleic Acid Res. 2009;37:6849–6858. doi: 10.1093/nar/gkp669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z., Champoux J. J. Reconstitution of enzymatic activity by the association of the cap and catalytic domains of human topoisomerase I. J. Biol. Chem. 2002;277:30815–30823. doi: 10.1074/jbc.M205302200. [DOI] [PubMed] [Google Scholar]
- 23.Stewart L., Ireton G. C., Champoux J. J. A functional linker in human topoisomerase I is required for maximum sensitivity to campthotecin in a DNA relaxation assay. J. Biol. Chem. 1999;274:32950–32960. doi: 10.1074/jbc.274.46.32950. [DOI] [PubMed] [Google Scholar]
- 24.Elban M. A., Sun W., Eisenhauer B. M., Gao R., Hecht S. M. Synthesis and biological evaluation of 10,11methylenedioxy-14-azacamptothecin. Org. Lett. 2006;16:3513–3516. doi: 10.1021/ol0611604. [DOI] [PubMed] [Google Scholar]
- 25.Fiorani P., Bruselles A., Falconi M., Chillemi G., Desideri A., Benedetti P. Single mutation in the linker domain confers protein flexibility and camptothecin resistance to human topoisomerase I. J. Biol. Chem. 2003;278:43268–43275. doi: 10.1074/jbc.M303899200. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt M. W., Baldridge K. K., Boatz J. A., Elbert S. T., Gordon M. S., Jensen J. H., Koseki S., Matsunaga N., Nguyen K. A., Su S., et al. General atomic and molecular electronic structure system. J. Comp. Chem. 1993;14:1347–1363. [Google Scholar]
- 27.Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsel D. S., Olson A. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comp. Chem. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto K., Saikawa Y., Nakata M. Studies on the red sweat of the Hippopotamus amphibious. Pure Appl. Chem. 2007;79:507–517. [Google Scholar]
- 29.Pommier Y., Pourquier P., Fan Y., Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:83–105. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 30.Liu L. F., Desai S. D., Li T. K., Mao Y., Sun M., Sim S. P. Mechanism of action of camptothecin. Ann. NY Acad. Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]