Abstract
Background:
There are no validated markers that predict response in metastatic renal cell cancer (RCC) patients treated with sunitinib. We aim to study the impact of single-nucleotide polymorphisms (SNPs) that have recently been proposed as predictors of outcome to anti-VEGF-targeted therapy in metastatic RCC in an independent cohort of patients.
Methods:
We genotyped 16 key SNPs in 10 genes involved in sunitinib pharmacokinetics, pharmacodynamics and VEGF-independent angiogenesis in patients with metastatic clear-cell RCC treated with sunitinib as the first-line targeted therapy. Association between SNPs, progression-free survival (PFS) and overall survival (OS) were studied by multivariate Cox regression using relevant clinical factors associated with PFS and OS as covariates.
Results:
In a series of 88 patients, both PFS and OS were associated significantly with SNP rs1128503 in ABCB1 (P=0.027 and P=0.025), rs4073054 in NR1/3 (P=0.025 and P=0.035) and rs307821 in VEGFR3 (P=0.032 and P=0.011). Progression-free survival alone was associated with rs2981582 in FGFR2 (P=0.031) and rs2276707 in NR1/2 (P=0.047), whereas OS alone was associated with rs2307424 in NR1/3 (P=0.048) and rs307826 in VEGFR3 (P=0.013).
Conclusion:
Our results confirm former communications regarding the association between SNPs in ABCB1, NR1/2, NR1/3 and VEGFR3 and sunitinib outcome in clear-cell RCC. Prospective validation of these SNPs is now required.
Keywords: renal cell carcinoma, sunitinib, outcome, single-nucleotide polymorphisms
Inactivation of the von Hippel–Lindau (VHL) tumour-suppressor gene is the most frequent molecular alteration in clear-cell renal cell cancer (RCC). Inactivated VHL leads to elevated protein levels of hypoxia-induced factor-α that upregulates vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF) expression. Targeted therapies directed against some of these proteins have significantly improved the perspectives of patients with metastatic RCC. Sunitinib malate is an orally administered tyrosine kinase receptor inhibitor (TKI) that targets VEGF and PDGF receptors, KIT, FLT-3, colony stimulating factor-1 receptor and RET. In a randomised controlled trial, sunitinib significantly prolonged progression-free survival (PFS; 11 vs 5 months, P<0.001) as compared with interferon-α (Motzer et al, 2007, 2009). Median overall survival (OS) was 26.4 and 21.8 months, respectively (P=0.051). Sunitinib is a standard treatment option in clear-cell RCC, but other anti-VEGFR and anti-PDGFR-targeted TKIs such as sorafenib, pazopanib and axitinib are also used in different stages of the disease.
Although 50% of RCC patients receiving sunitinib experience an objective response and 43% achieve disease stabilisation, 7% will experience progressive disease (PD) at first evaluation, probably because of intrinsic resistance or other factors (Motzer et al, 2009). Moreover, even patients with an initial clinical benefit will finally progress because of acquired resistance or for other reasons. The identification of biomarkers able to predict intrinsic resistance could avoid unnecessary costs and side effects, guiding alternative treatment decisions. On the other hand, the identification of biomarkers for acquired resistance could provide novel directions to develop therapies that block these resistance pathways. Although different mechanisms of resistance have been proposed (Rini and Atkins, 2009), reliable biomarkers predictive of sunitinib sensitivity or primary/secondary resistance are still lacking.
Several clinical and biochemical markers for PFS and OS are available for sunitinib-treated patients (Heng et al, 2009; Patil et al, 2011). For PFS, these are baseline serum lactate dehydrogenase (LDH) level, the presence of two or more metastatic sites, no prior nephrectomy, Eastern Cooperative Oncology Group Performance Status (ECOG PS) and baseline platelet count. For OS, factors include presence of bone metastases, time between nephrectomy and start of systemic therapy, baseline serum LDH level, baseline haemoglobin, baseline calcium and baseline ECOG. The last five criteria are part of the Memorial Sloan Kettering Cancer Centre (MSKCC) score that categorises patients into a favourable-, intermediate- and poor-prognosis group (Motzer et al, 2004). These established clinical and biochemical markers are indicators of the general condition of the patient and the extension or stage of the disease. They do not take into account sunitinib pharmacokinetics (absorption, metabolisation) or pharmacodynamics (interaction of sunitinib with its molecular targets). Recently, a meta-analysis of pharmacokinetic data from 443 patients treated with sunitinib showed that higher plasma levels of sunitinib and its active metabolite SU12662 were associated with prolonged TTP and OS (Houk et al, 2010). Factors influencing the concentration of sunitinib in plasma are dose and schedule of the drug and patient compliance, but importantly, also the concentration of efflux pumps and metabolising enzymes. Moreover, sunitinib efficacy can be influenced by the expression level and variants of the molecular targets of the drug.
Recently, a number of studies have proposed that genetic variability in genes involved in sunitinib pharmacokinetics and pharmacodynamics alter the efficacy of sunitinib (van der Veldt et al, 2010; Garcia-Donas et al, 2011) or pazopanib (Xu et al, 2011a, 2011b) in metastatic RCC. As each of these studies investigated a different set of single-nucleotide polymorphisms (SNPs), these findings need to be validated independently. The aim of the present study is therefore to replicate association of these SNPs to sunitinib outcome by assessing an independent cohort of patients with metastatic clear-cell RCC treated with first-line sunitinib.
Materials and methods
For this retrospective study, germline DNA samples were collected in the CIT-rein kidney tumour bank and in patients treated at the University Hospitals Leuven. The French-Belgian multicentric CIT-rein kidney tumour bank contains more than 250 frozen kidney tumour samples collected at 20 academic hospitals. We selected the samples of patients with pathologically confirmed clear-cell RCC treated in first line with sunitinib and for whom frozen normal kidney tissue was available. Eligible patients could have received cytokines as systemic treatment for kidney tumours before starting sunitinib as a monotherapy, but they could not have received any other TKI or mTOR (mammalian target of rapamycin) inhibitor before starting sunitinib. To make sure that the effect of sunitinib was accurately measured, patients had to take sunitinib during at least one complete cycle of 28 days and had to reach at least the first evaluation by CT scan. In the whole CIT-rein kidney tumour bank, 79 frozen normal kidney samples corresponded to these selection criteria. In order to extend the series, we added nine patients visiting the University Hospitals Leuven and complying to the same inclusion criteria. As no frozen normal kidney tissue was available for these patients, peripheral blood was sampled during out day clinic from July 2011 till December 2011.
The protocol was approved by the medical ethics review boards of all participating institutions, and signed consent was obtained from all patients. In some cases, we used frozen biologic material from patients who had already died and for whom a general positive advice for the utilisation of remaining tissue was foreseen by the institutional board.
All the patients were treated in routine clinical practice. Drug schedule, dose-reduction policy and timing of radiological assessments were left to the discretion of the attending doctors in accordance with current local practice guidelines. All the patients started their sunitinib therapy at the standard sunitinib dose of 50 mg day−1, 4 weeks on and 2 weeks off. The patient characteristics considered relevant for PFS and OS analysis were the five risk factors according to the MSKCC prognostic criteria and additional factors such as baseline neutrophil count, baseline platelet count, the presence or absence of liver metastases, the presence or absence of a component of sarcomatoid dedifferentiation and the presence or absence of bone metastases. The latter two parameters were associated to outcome on sunitinib in recent publications (Golshayan et al, 2009; Beuselinck et al, 2011; Patil et al, 2011).
The SNPs previously associated with TKI efficacy in RCC were selected from the literature (Table 1). These SNPs are located in genes affecting sunitinib pharmacokinetics (i.e., genes involved in sunitinib absorption, such as ABCB1, or metabolism, such as CYP3A5, NR1/2 and NR1/3), sunitinib pharmacodynamics (i.e., genes involved in PDGF- and VEGF-dependent angiogenesis such as HIF1A, PDGFRA, VEGFR2 and VEGFR3) or VEGF-independent alternative pro-angiogenic pathways (FGFR2, and IL8). DNA was isolated at INSERM U674 in Paris, France, from fresh frozen normal kidney tissue sampled in the nephrectomy specimen using the Qiaquick extraction kit (Qiagen, Valencia, CA, USA) and quantified by fluorometry (Fluoroskan Thermo Labsystems, Cergy-Pontoise, France). DNA was isolated from peripheral blood at the Vesalius Research Center in Leuven with the Qiagen DNA kit (Qiagen) and final DNA concentration quantified with Nanodrop (Nanodrop, Wilmington, DE, USA). High-throughput SNP genotyping was performed at the Vesalius Research Center in Leuven, Belgium, using the Sequenom MassArray platform (Sequenom, San Diego, CA, USA) (Reumers et al, 2011). Genotyping analysis was performed by investigators blinded for the clinical data. Overall, 16 SNPs were successfully genotyped, with success rates ⩾85% for each SNP and an overall average success rate of 96%. We failed to genotype SNP rs1126647 in IL-8 because of technical reasons. For most of the SNPs, genotypes were analysed in the same way as they were communicated in the original reports (i.e., according to dominant, recessive or co-dominant genetic models or in the context of a specific haplotype).
Table 1. SNPS linked to sunitinib outcome based on literature evidence.
| Gene | Polymorphism | SNP ID | Impact on outcome | Reference |
|---|---|---|---|---|
|
Genes involved in pharmacokinetics | ||||
| ABCB1 | 3435C>T | rs1045642 | PFS: 15.2 vs 8.4 months if a TCG copy was present in the ABCB1 haplotype composed of rs1045642, rs1128503 and rs2032582 (P=0.033) | (Van der Veldt et al, 2010) |
| 1236C>T | rs1128503 | |||
| 2677G>T or G>A | rs2032582 | |||
| CYP3A5 | 6986G>A | rs776746 | PFS: not reached for AA and AG genotypes vs 9.3 months for GG genotypes (P=0.032) | (Van der Veldt et al, 2010) |
| NR1/2 | 25385C>T | rs3814055 | PFS: 6.7 months for TT genotypes vs 10.8 months for CT and CC genotypes (P=0.025) OS: 10.2 months for TT genotypes vs 17.1 months for CT and CC genotypes (P=0.017) | (Van der Veldt et al, 2010) |
| OS: 29 vs 22 vs 23 months for the CC, CT and TT variants, respectively (P=0.03) | (Xu et al, 2011b) | |||
| 8055C>T | rs2276707 | PFS: 10.8 months for CC and CT genotypes vs 6.7 months for TT genotypes (P=0.025) | (Van der Veldt et al, 2010) | |
| NR1/3 | 5719C>T | rs2307424 | PFS: 13.3 vs 8.0 months if a CAT copy was absent in the NR1/3 haplotype composed of rs2307424, rs2307418 and r s4073054 (P=0.017) | (Van der Veldt et al, 2010) |
| 7738A>C | rs2307418 | |||
| |
7837T>G |
rs4073054 |
|
|
|
Genes involved in pharmacodynamics | ||||
| HIF1A | 1790G>A | rs11549467 | PFS: 44 months for GG genotypes vs 20 weeks for GA genotypes (P=0.03) | (Xu et al, 2011a) |
| PGDFRA | 1580T>C | rs35597368 | OS: 24.2 vs 14.8 months if a GCGT haplotype is present in both alleles of a PDGFRA haplotype composed of rs1800810, rs1800812, rs1800813 and rs35597368 vs patients with GCG–other or other–other haplotypes (P=0.002) | (Van der Veldt et al, 2010) |
| VEGFR2 | 1718T>A | rs1870377 | OS: 16.3 months for AA and AT genotypes vs 9.4 months for TT genotypes (P=0.016) | (Van der Veldt et al, 2010) |
| VEGFR3 | 3971G>T | rs307821 | PFS: 13.7 months for GG genotypes vs 6.7 months for GT genotypes (P=0.014) | (Garcia-Donas et al, 2011) |
| 1480A>G | rs307826 | PFS: 13.7 months for AA genotypes vs 3.6 months for AG genotypes (P=0.0079) | (Garcia-Donas et al, 2011) | |
| |
|
|
OS: 26, 23 and 3.2 months for the AA, AG and GG genotypes, respectively (P=0.04) |
(Xu et al, 2011b) |
|
Genes involved in alternative proangiogenic pathways | ||||
| FGFR2 | 906C>T | rs2981582 | OS: 28.0 months for CC genotypes vs 21.4 months for TT genotypes (P=0.009) | (Xu et al, 2011b) |
| IL8 | 251T>A | rs4073 | PFS: 49, 42 and 32 weeks for TT, AT and AA genotypes, respectively (P=0.01) | (Xu et al, 2011a) |
| 2767A>T | Rs1126647 | PFS: 48, 42 and 27 weeks for AA, AT and TT genotypes, respectively (P=0.009) | (Xu et al, 2011a) | |
Abbreviations: SNP=single-nucleotide polymorphism; PFS=progression-free survival; OS=overall survival.
Clinical data were collected at 15 different sites in France and Belgium. The primary objective was PFS and OS, and the secondary objective was RR. We defined PFS as the time between the first day on sunitinib and the date of radiological PD or death. Patients who had not progressed at database closure were censored at last follow-up. Overall survival was defined as the time between the first day on sunitinib and the date of death or last date of follow-up. Objective response was assessed by the treating doctors and classified as complete response (CR), partial response (PR), stable disease (SD), or PD. Timing for assessments was dictated by individual institution policy.
All patient characteristics were tested in an univariate analysis for association with PFS and OS using Kaplan–Meier statistics and in a multivariate model using Cox proportional hazards. Fisher's exact tests and logistic regression were used to compare the incidence of poor-prognostic variants in patients with PD vs a group with SD, PR or CR as best response. The MSKCC score was used as a covariate in the multivariate analysis, as well as all other variables with a P⩽0.2 on univariate analysis that are not part of the MSKCC score. Results with a P-value of <0.05 were considered as significant in the multivariate analysis. Because this is a confirmatory rather than an exploratory study, SNPs were selected based on literature evidence and, hence, no correction for multiple testing was made.
Of the 11 clinical parameters assessed in the univariate analysis (for 88 patients), there were 13 missing values (1.3%). For the multivariate analysis, 82 patients with complete data could be included. Statistical analyses were conducted using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA) and XLSTAT software (Addinsoft, Paris, France).
Results
We enroled 88 patients who started sunitinib between November 2005 and July 2011 and closed the follow-up database in April 2012. Table 2 shows the clinical characteristics of enroled patients. Mean age at diagnosis was 59 years (range 38–84). The majority of patients (>94%) were of Caucasian origin. According to the MSKCC prognostic criteria, 15% of patients were categorised into the favourable risk group, and 56% had intermediate and 28% poor risk.
Table 2. Patient characteristics at diagnosis and at the start of sunitinib treatment and baseline clinical and biochemical parameters associated with PFS and OS.
| Total | Median PFS (months) | P-value, HR (95% CI) | 95% CI of median PFS | |
|---|---|---|---|---|
|
At initial diagnosis | ||||
| Male |
68% (60/88) |
— |
— |
— |
|
Ethnic origin | ||||
| Caucasian | 94% (83/88) | — | — | — |
| Unknown | 6% (5/88) | — | — | — |
| M1 (synchronous metastases) |
55% (46/84) |
— |
— |
— |
|
Fuhrman | ||||
| Grade 1–3 | 68% (58/85) | — | — | — |
| Grade 4 |
32% (27/85) |
— |
— |
— |
|
Sarcomatoid dedifferentiation | ||||
| Present | 9% (8/88) | 4 | 0.09 | 1–Not reached |
| Absent |
91% (80/88) |
18 |
0.37 (0.12–1.18) |
12–24 |
|
At the start of sunitinib | ||||
|
ECOG PS | ||||
| >0 | 49% (43/88) | 15 | 0.08 | 7–20 |
| 0 |
51% (45/88) |
21 |
0.63 (0.37–1.06) |
11–38 |
|
Neutrophils | ||||
| >4500 per mm3 | 40% (34/85) | 9.5 | 0.13 | 5–15 |
| <4500 per mm3 |
60% (51/85) |
19 |
0.65 (0.38–1.14) |
14–24 |
|
Platelets | ||||
| >400.000 per mm3 | 15% (13/88) | 11 | 0.25 | — |
| <400.000 per mm3 |
85% (75/88) |
18 |
— |
— |
|
Haemoglobin | ||||
| Low (<11.5 g dl−1 (women) or <13 g dl−1 (men)) | 42% (37/88) | 14 | 0.98 | — |
| Normal |
58% (51/88) |
18 |
— |
— |
|
LDH | ||||
| >1.5 ULN | 10% (8/84) | 10.5 | 0.09 | 4–19 |
| ⩽1.5 ULN |
90% (76/84) |
18.0 |
0.40 (0.14–1.16) |
12–25 |
|
Corrected calcium | ||||
| >10 mg dl−1 | 7% (6/84) | 22 | 0.9 | — |
| ⩽10 mg dl−1 |
93% (78/84) |
15 |
— |
— |
|
Time from nephrectomy to systemic treatment | ||||
| <12 months | 66% (58/88) | 18 | 0.30 | — |
| >12 months | 34% (30/88) | 15 | — | — |
| Immunotherapy before sunitinib |
28% (24/87) |
— |
— |
— |
|
Site of metastasis | ||||
| Lung | 84% (74/88) | — | — | — |
| Liver metastases | 18% (16/88) | 15 | 0.59 | — |
| No liver metastases | 82% (72/88) | 18 | — | — |
| Bone metastases | 35% (31/88) | 15 | 0.5 | — |
| No bone metastases | 65% (57/88) | 18 | — | — |
| Brain |
6% (5/88) |
— |
— |
— |
|
MSKCC prognosis | ||||
| Favourable | 15% (13/85) | Not reached | 0.21 | 8–Not reached |
| Intermediate | 56% (48/85) | 15 | 11–21 | |
| Poor | 28% (24/85) | 15 | 4–25 | |
| At initial diagnosis | Total | Median OS (months) | P-value, HR (95% CI) | 95% CI of median OS |
|---|---|---|---|---|
| Male |
68% (60/88) |
— |
— |
— |
|
Ethnic origin | ||||
| Caucasian | 94% (83/88) | — | — | — |
| Unknown | 6% (5/88) | — | — | — |
| M1 (synchronous metastases) |
55% (46/84) |
— |
— |
— |
|
Fuhrman | ||||
| Grade 1–3 | 68% (58/85) | — | — | — |
| Grade 4 |
32% (27/85) |
— |
— |
— |
|
Sarcomatoid dedifferentiation | ||||
| Present | 9% (8/88) | 16.5 | 0.19 | 5–Not reached |
| Absent |
91% (80/88) |
30 |
0.45 (0.13–1.50) |
23–42 |
|
At the start of sunitinib | ||||
|
ECOG PS | ||||
| >0 | 49% (43/88) | 23 | 0.08 | 17–34 |
| 0 |
51% (45/88) |
35 |
0.60 (0.34–1.06) |
23–Not reached |
|
Neutrophils | ||||
| >4.500 per mm3 | 40% (34/85) | 22 | 0.39 | — |
| <4.500 per mm3 |
60% (51/85) |
34 |
— |
— |
|
Platelets | ||||
| >400.000 per mm3 | 15% (13/88) | 27 | 0.45 | — |
| <400.000 per mm3 |
85% (75/88) |
29 |
— |
— |
|
Haemoglobin | ||||
| Low (<11.5 g dl−1 (women) or <13 g dl−1 (men)) | 42% (37/88) | 27 | 0.42 | — |
| Normal |
58% (51/88) |
34 |
— |
— |
|
LDH | ||||
| >1.5 ULN | 10% (8/84) | 24.5 | 0.19 | 19–34 |
| ⩽1.5 ULN |
90% (76/84) |
34 |
0.51 (0.19–1.38) |
23–45 |
|
Corrected calcium | ||||
| >10 mg dl−1 | 7% (6/84) | 42 | 0.98 | — |
| ⩽10 mg dl−1 |
93% (78/84) |
29 |
— |
— |
|
Time from nephrectomy to systemic treatment | ||||
| <12 months | 66% (58/88) | 27 | 0.13 | 22–35 |
| >12 months | 34% (30/88) | Not reached | 1.58 (0.88–2.85) | 19–Not reached |
| Immunotherapy before sunitinib |
28% (24/87) |
— |
— |
— |
|
Site of metastasis | ||||
| Lung | 84% (74/88) | — | — | — |
| Liver metastases | 18% (16/88) | 22 | 0.60 | — |
| No liver metastases | 82% (72/88) | 29 | — | — |
| Bone metastases | 35% (31/88) | 22 | 0.06 | 11–34 |
| No bone metastases | 65% (57/88) | 35 | 0.54 (0.29–1.03) | 24–Not reached |
| Brain |
6% (5/88) |
— |
— |
— |
|
MSKCC prognosis | ||||
| Favourable | 15% (13/85) | Not reached | 0.0097 | Not reached–not reached |
| Intermediate | 56% (48/85) | 24 | 20–41 | |
| Poor | 28% (24/85) | 27 | 19–42 | |
Abbreviations: PFS=progression-free survival; OS=overall survival; ECOG PS=Eastern Cooperative Oncology Group Performance Status; LDH=lactate dehydrogenase; ULN=upper limit of normal; MSKCC=Memorial Sloan Kettering Cancer Center; HR=hazard ratio; 95% CI=95% confidence interval.
In the univariate analysis, median PFS and median OS were estimated by Kaplan–Meier and P-values were derived from a log-rank test. The impact of the presence of lung metastases and previous immunotherapy was not assessed as these parameters have not been strongly linked to PFS or OS.
At the time of analysis, 57 (64.8%) patients had reached progression and 48 (54.5%) had died. The median follow-up was 46.0 months (range 1.0–73.0 months; 95% confidence interval (CI) 42.0–51.0 months) after the start of sunitinib. The median PFS was 15.0 months (95% CI 11.0–23.0 months) and the median OS was 29.0 months (95% CI 23.0–42.0 months). Best response assessment was available in 82 patients (in the 6 remaining patients, there was a clinical benefit, but response assessment was poorly defined in the medical records, and as a consequence, it was unclear whether the best response was either PR or SD in these 6 patients). In all, 6 out of 82 (7.3%) patients had a CR, 30 out of 82 (36.6%) patients a PR, 36 out of 82 (43.9%) SD and 10 out of 88 (11.4%) PD as best response.
For each of these 16 polymorphisms, the respective genotypes, allele frequencies and changes at the amino acid level are given in Table 3. The allele frequencies of the genotyped polymorphisms were similar as previously reported in the dbSNP database (dbSNP build 136) or 1000 Genomes Project, except for SNPs rs2276707 and rs307821. Their observed minor allele frequencies were slightly higher compared with their frequency reported in dbSNP. In the case of rs11549467, there was only one heterozygous patient. As a consequence, the impact of this SNP could not be analysed.
Table 3. Genotype and allele distribution of selected SNPs.
| Gene | RS ID | Polymorphism | Location or functional consequence | n | Wild-type/wild-type, n (%) | Wild-type/variant, n (%) | Variant/variant, n (%) | Observed minor allele frequency (%) | Minor allele frequency in dbSNP (%) |
|---|---|---|---|---|---|---|---|---|---|
|
Sunitinib pharmacokinetics | |||||||||
| ABCB1 | rs1045642 | 3435C>T | I1154I | 87 | 25 (29) | 43 (49) | 19 (22) | 46.6 | 53.4 |
| rs1128503 | 1236C>T | G412G | 88 | 38 (43) | 35 (40) | 15 (17) | 36.9 | 45.1 | |
| rs2032582 | 2677G>T or G>A | A893S | 80 | 32 (40) | 36 (45) | 12 (15) | 37.5 | 41.7 | |
| CYP3A5 | rs776746 | 6986G>A | Affecting splicing | 75 | 69 (92) | 6 (8) | 0 (0) | 4.0 | 3.6 |
| NR1/2 | rs3814055 | 25385C>T | UTR-5′ | 82 | 32 (39) | 35 (43) | 15 (18) | 39.6 | 33.6 |
| rs2276707 | 8055C>T | Intron | 83 | 57 (69) | 21 (25) | 5 (6) | 18.7 | 9.3 | |
| NR1/3 | rs2307424 | 5719C>T | P151P | 88 | 45 (51) | 32 (36) | 11 (12.5) | 30.7 | 33.6 |
| rs2307418 | 7738A>C | Intron | 86 | 61 (71) | 22 (26) | 3 (3) | 16.3 | 15.9 | |
| |
rs4073054 |
7837T>G |
Intron |
87 |
40 (46) |
35 (40) |
12 (14) |
33.9 |
40.7 |
|
Sunitinib pharmacodynamics | |||||||||
| HIF1A | rs11549467 | 1790G>A | A588T | 84 | 83 (99) | 1 (1) | 0 (0) | 0.6 | 1.7 |
| PDGFRA | rs35597368 | 1580T>C | S478P | 88 | 69 (78) | 18 (20) | 1 (1) | 11.3 | 13.3 |
| VEGFR2 | rs1870377 | 1718T>A | Q472H | 81 | 46 (57) | 28 (35) | 7 (9) | 25.9 | 27.5 |
| VEGFR3 | rs307821 | 3971G>T | R1324L | 88 | 64 (73) | 23 (26) | 1 (1) | 14.2 | 5.8 |
| |
rs307826 |
1480A>G |
T494A |
88 |
65 (74) |
22 (25) |
1 (1) |
13.6 |
10.2 |
|
Alternative VEGF-independent proangiogenic pathways | |||||||||
| FGFR2 | rs2981582 | 906C>T | Intron | 87 | 23 (26) | 52 (60) | 12 (14%) | 43.6 | 45.6 |
| IL8 | rs4073 | 251T>A | 5′ near gene | 79 | 25 (31) | 42 (53) | 12 (15%) | 41.8 | 42.5 |
Abbreviations: SNP=single-nucleotide polymorphism; VEGF=vascular endothelial growth factor; UTR=untranslated region; dbSNP=SNP database.
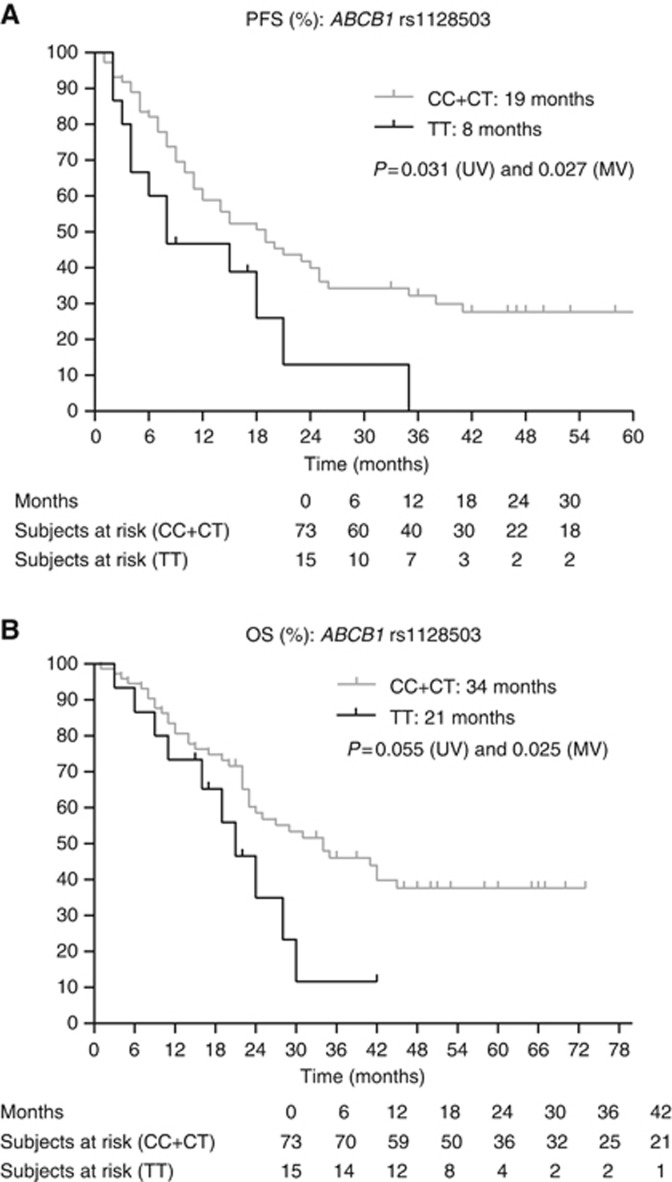
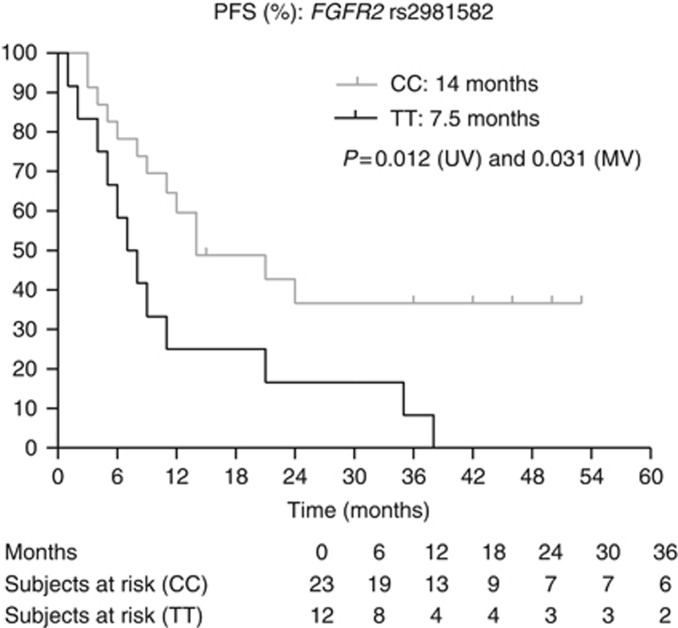
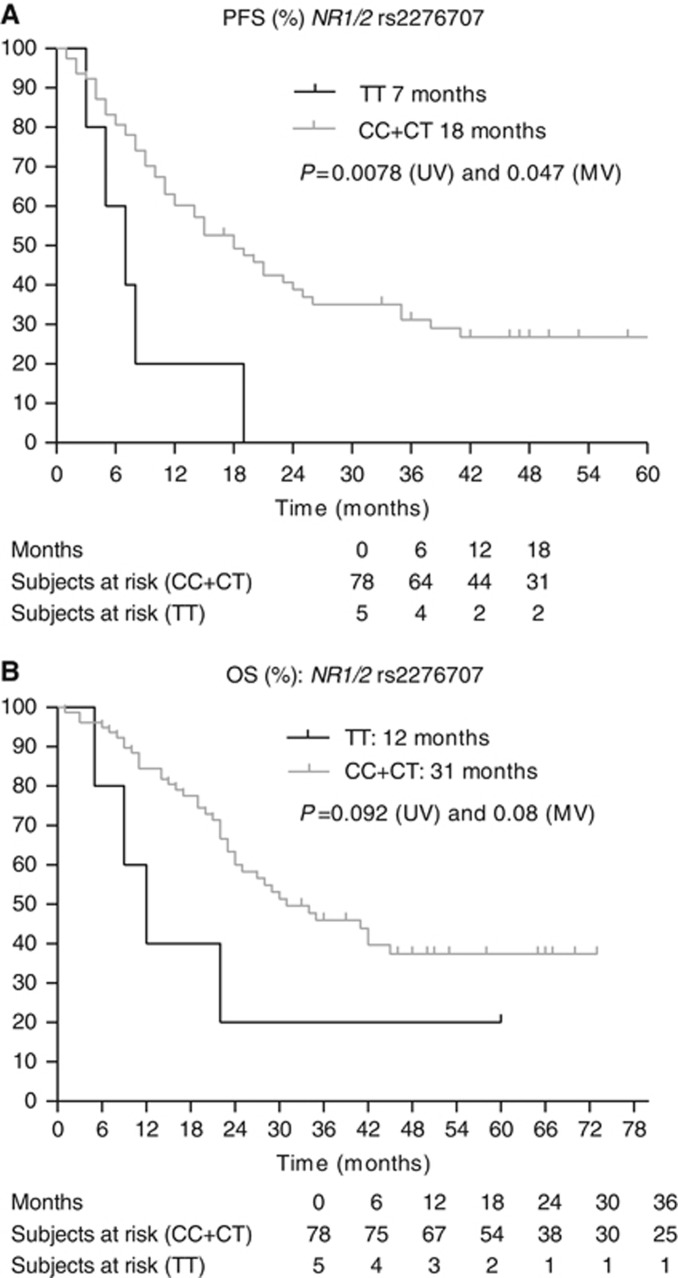
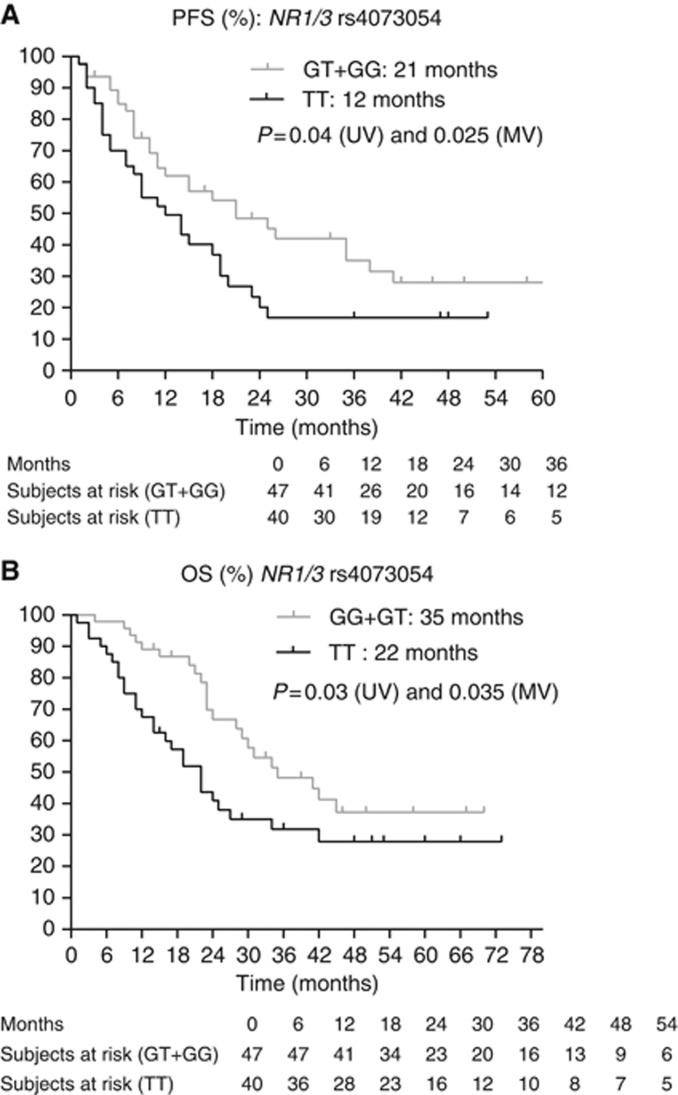
Next, we assessed the clinical and biochemical parameters associated with PFS and OS (Table 2). The MSKCC score, baseline neutrophil levels and the presence of a sarcomatoid component in the tumour were considered as covariates when assessing the effect of SNPs on PFS. For OS, the MSKCC score, the presence of bone metastases and the presence of a sarcomatoid component in the tumour were considered as covariates. Table 4 and Figures 1, 2, 3, 4, 5, 6, 7 show the results of the univariate and multivariable analyses for each of the genotyped SNPs for both PFS and OS after correction for these covariates. In the multivariate analysis, PFS and OS were associated significantly with SNP rs1128503 in ABCB1 (P=0.027 and P=0.025), rs4073054 in NR1/3 (P=0.025 and P=0.035) and rs307821 in VEGFR3 (P=0.032 and P=0.011). Progression-free survival was associated with rs2981582 in FGFR2 (P=0.031) and rs2276707 in NR1/2 (P=0.047). Overall survival was associated with rs2307424 in NR1/3 (P=0.048) and rs307826 in VEGFR3 (P=0.013).
Table 4. Univariate and multivariate analyses: association between SNPs and outcome.
| Gene (a) SNP ID | Polymorphism | No. of pts | Median PFS (months) | P-value (UV) | P-value (MV) | HR | 95% CI of HR | 95% CI of median PFS (months) |
|---|---|---|---|---|---|---|---|---|
| ABCB1 rs1045642 3435C>T | CC | 25 | 14 | 0.67 | NA | NA | NA | NA |
| CT | 43 | 15 | NA | |||||
| |
TT |
19 |
18 |
|
|
|
|
NA |
| ABCB1 rs1128503 1236C>T | CT+CC | 73 | 19 | 0.031 | 0.027 | 0.464 | 0.234–0.918 | 11–25 |
| |
TT |
15 |
8 |
|
|
|
|
3–21 |
| ABCB1 rs2032582 2677G>T or G>A | GG | 32 | 14 | 0.45 | NA | NA | NA | NA |
| GT/GA | 36 | 19 | NA | |||||
| |
TT/TA |
12 |
15 |
|
|
|
|
NA |
| ABCB1 TCG copy | Present | 16 | 15 | 0.68 | NA | NA | NA | NA |
| |
Absent |
64 |
19 |
|
|
|
|
NA |
| CYP3A5 rs776746 6986G>A | GG | 69 | 18 | 0.36 | NA | NA | NA | NA |
| |
AG |
6 |
21 |
|
|
|
|
NA |
| NR1/2 s3814055 25385C>T | CC+CT | 67 | 18 | 0.26 | NA | NA | NA | NA |
| |
TT |
15 |
19 |
|
|
|
|
NA |
| NR1/2 rs2276707 8055C>T | CC+CT | 78 | 18 | 0.0078 | 0.047 | 2.978 | 1.012–8.761 | 12–25 |
| |
TT |
5 |
7 |
|
|
|
|
3–19 |
| NR1/3 rs2307424 5719C>T | CC | 45 | 20 | 0.18 | 0.155 | 1.513 | 0.856–2.675 | 11–38 |
| |
CT+TT |
43 |
15 |
|
|
|
|
9–21 |
| NR1/3 rs2307418 7738A>C | AA | 61 | 14 | 0.45 | NA | NA | NA | NA |
| |
AC+CC |
27 |
28 |
|
|
|
|
NA |
| NR1/3 rs4073054 7837T>G | TT | 40 | 12 | 0.04 | 0.025 | 1.864 | 1.082–3.210 | 8–19 |
| |
TG+GG |
47 |
21 |
|
|
|
|
12–38 |
| NR1/3 CAT copy | Present | 51 | 15 | 0.67 | NA | NA | NA | NA |
| |
Absent |
36 |
15 |
|
|
|
|
NA |
| FGFR2 rs2981582 906C>T | TT | 12 | 7.5 | 0.012 | 0.031 | 2.669 | 1.094–6.511 | 5–11 |
| |
CC |
23 |
14 |
|
|
|
|
11–Not reached |
| IL8 rs4073 251T>A | TT | 25 | 8 | 0.22 | NA | NA | NA | NA |
| |
AA |
12 |
21 |
|
|
|
|
NA |
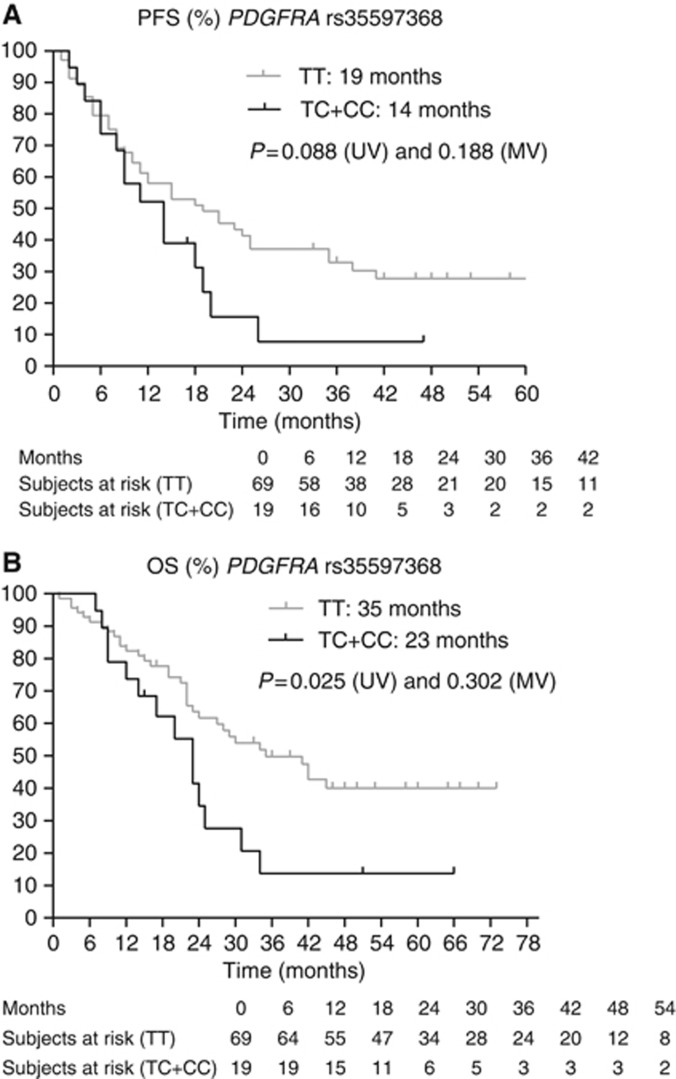
| PDGFRA rs35597368 1580T>C | TT | 69 | 19 | 0.088 | 0.188 | 1.528 | 0.813–2.870 | 11–25 |
| |
TC+CC |
19 |
14 |
|
|
|
|
8–19 |
| VEGFR2 rs1870377 1718T>A | TT | 48 | 15 | 0.76 | NA | NA | NA | NA |
| |
TA+AA |
40 |
19 |
|
|
|
|
NA |
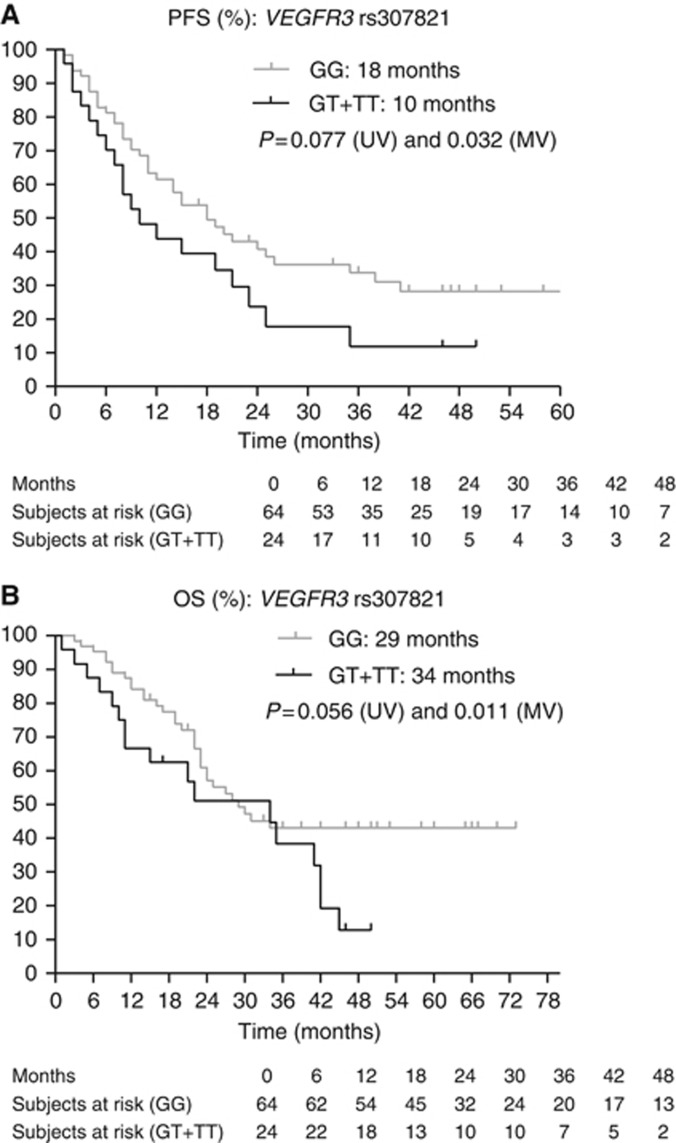
| VEGFR3 (b) rs3078213971G>T | GT+TT | 24 | 10 | 0.077 | 0.032 | 1.981 | 1.060–3.702 | 7–21 |
| |
GG |
64 |
18 |
|
|
|
|
12–26 |
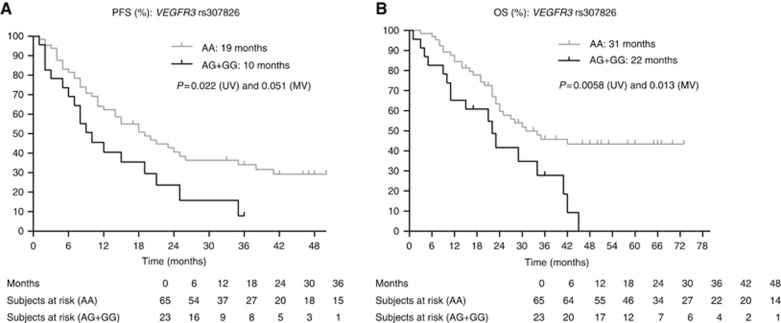
| VEGFR3 rs307826 1480A>G | AG+GG | 23 | 10 | 0.022 | 0.051 | 1.800 | 0.996–3.250 | 6–19 |
| |
AA |
65 |
19 |
|
|
|
|
14–26 |
|
Gene (a) SNP ID |
Polymorphism |
No. of pts |
Median OS (months) |
P-value (UV) |
P-value (MV) |
HR |
95% CI of HR |
95% CI of median OS (months) |
| ABCB1 rs1045642 3435C>T | CC | 25 | 45 | 0.37 | NA | NA | NA | NA |
| CT | 43 | 27 | NA | |||||
| |
TT |
19 |
24 |
|
|
|
|
NA |
| ABCB1 rs1128503 1236C>T | CT+CC | 73 | 34 | 0.055 | 0.025 | 0.415 | 0.193–0.894 | 23–45 |
| |
TT |
15 |
21 |
|
|
|
|
9–30 |
| ABCB1 rs2032582 2677 G>T or G>A | GG | 32 | 35 | 0.49 | NA | NA | NA | NA |
| |
GT/GA |
36 |
34 |
|
|
|
|
NA |
| |
TT/TA |
12 |
24 |
|
|
|
|
NA |
| ABCB1 TCG copy | Present | 16 | 26 | 0.74 | NA | NA | NA | NA |
| |
Absent |
64 |
34 |
|
|
|
|
NA |
| CYP3A5 rs776746 6986G>A | GG | 69 | 30 | 0.92 | NA | NA | NA | NA |
| |
AG |
6 |
NR |
|
|
|
|
NA |
| NR1/2 s3814055 25385C>T | CC+CT | 67 | 30 | 0.46 | NA | NA | NA | NA |
| |
TT |
15 |
31 |
|
|
|
|
NA |
| NR1/2 rs2276707 8055C>T | CC+CT | 78 | 31 | 0.092 | 0.080 | 2.828 | 0.884–9.044 | 24–45 |
| |
TT |
5 |
12 |
|
|
|
|
5–Not reached |
| NR1/3 rs2307424 5719C>T | CC | 45 | 42 | 0.057 | 0.048 | 1.913 | 1.006–3.636 | 25–Not reached |
| |
CT+TT |
43 |
23 |
|
|
|
|
16–34 |
| NR1/3 rs2307418 7738A>C | AA | 61 | 30 | 0.86 | NA | NA | NA | NA |
| |
AC+CC |
27 |
27 |
|
|
|
|
NA |
| NR1/3 rs4073054 7837T>G | TT | 40 | 22 | 0.03 | 0.035 | 1.927 | 1.046–3.549 | 14–34 |
| |
TG+GG |
47 |
35 |
|
|
|
|
28–Not reached |
| NR1/3 CAT copy | Present | 51 | 28 | 0.58 | NA | NA | NA | NA |
| |
Absent |
36 |
30 |
|
|
|
|
NA |
| FGFR2 rs2981582 906C>T | TT | 12 | 23 | 0.97 | NA | NA | NA | NA |
| |
CC |
23 |
25 |
|
|
|
|
NA |
| IL8 rs4073 251T>A | TT | 25 | 23 | 0.68 | NA | NA | NA | NA |
| |
AA |
12 |
31 |
|
|
|
|
NA |
| PDGFRA rs35597368 1580 T>C | TT | 69 | 35 | 0.025 | 0.302 | 1.440 | 0.721–2.875 | 24–Not reached |
| |
TC+CC |
19 |
23 |
|
|
|
|
14–31 |
| VEGFR2 rs1870377 1718T>A | TT | 48 | 24 | 0.63 | NA | NA | NA | NA |
| |
TA+AA |
40 |
30 |
|
|
|
|
NA |
| VEGFR3 (b) rs307821 3971G>T | GT+TT | 24 | 34 | 0.056 | 0.011 | 2.265 | 1.202–4.268 | 11–42 |
| |
GG |
64 |
29 |
|
|
|
|
23–Not reached |
| VEGFR3 rs307826 1480A>G | AG+GG | 23 | 22 | 0.0058 | 0.013 | 2.223 | 1.187–4.163 | 11–34 |
| AA | 65 | 31 | 24–Not reached |
Abbreviations: SNP=single-nucleotide polymorphism; pts=patients; PFS=progression free survival; OS=overall survival; UV=univariate analysis; MV=multivariate analysis; NA=not applicable; HR=hazard ratio; 95% CI=95% confidence interval.
In the univariate analysis, P-values were calculated by a log-rank test. In the multivariate analysis, P-values were calculated by Cox proportional hazards.
Whenever possible, variants were combined as it was done in the original publications: this was the case for FGFR2, IL8, NR1/2, VEGFR2 and VEGFR3. For ABCB1 and NR1/3, the original publication only reported haplotypes. The haplotypes were tested against PFS and OS in our series and no association with PFS and OS could be shown. Therefore, we checked for each SNP the three subgroups and analysed the PFS and OS curves. For ABCB1, when analysing TT vs TC vs CC in rs1045642 or GG vs GT/GA vs TT/TA in rs2032582, the three curves were overlapping for PFS and OS. Only in ABCB1 rs1128503, when analysing TT vs TC vs CC variants, the CC and CT results were overlapping for PFS and OS and clearly different from the TT results, allowing us to group the results of the CT and CC variants. Concerning NR1/3, for SNP rs2307424, the PFS and OS curves of the CT and TT variants were overlapping, but the curves of the CC variant were clearly distinct. For SNP rs2307418, there were only two CC variant patients: they were grouped with the AC variant patients and tested against the AA variant patients. For SNP rs4073054, the PFS and OS curves of the TG and GG variants were overlapping, but the curves of the TT variant were clearly distinct. This distribution allowed us to test the impact of the CC variant in rs2307424, the AA variant in rs2307418 and the TT variant in rs4073054 vs the combination of the other variants. In case of CYP3A5, there were no AA variants in our series. For PDGFRA, there was only one CC variant: this patient was grouped with the TC variant.
The HR for survival for patients with the GT or TT variants in rs307821 in VEGFR3 vs patients with the GG variant was 2.265, favouring longer survival in patients with the GG genotype. Nevertheless, because of a crossing of the curves, the median OS was longer in the GT and TT variants (see curves).
Figure 1.
(A and B) Kaplan–Meier curves for PFS and OS for SNP rs1128503 in ABCB1. The P-values are indicated for the univariate (UV) and multivariate (MV) analyses.
Figure 2.
Kaplan–Meier curves for PFS for SNP rs2981582 in FGFR2. The P-values are indicated for the univariate (UV) and multivariate (MV) analyses.
Figure 3.
(A and B) Kaplan–Meier curves for PFS and OS for SNP rs2276707 in NR1/2. The P-values are indicated for the univariate (UV) and multivariate (MV) analyses.
Figure 4.
(A and B) Kaplan–Meier curves for PFS and OS for SNP rs4073054 in NR1/3. The P-values are indicated for the univariate (UV) and multivariate (MV) analyses.
Figure 5.
(A and B) Kaplan–Meier curves for PFS and OS for SNP rs35597368 in PDGFRA. The P-values are indicated for the univariate (UV) and multivariate (MV) analyses.
Figure 6.
(A and B) Kaplan–Meier curves for PFS and OS for SNP rs307821 in VEGFR3. The P-values are indicated for the univariate (UV) and multivariate (MV) analyses. Note that because of a crossing of the curves, the median OS was longer in the GT and TT variants than in the GG variants of rs307821. Nevertheless, the HR for survival for patients with the GT or TT variants in rs307821 in VEGFR3 vs patients with the GG variant was 2.265 (95% CI 1.202–4.238).
Figure 7.
(A and B) Kaplan–Meier curves for PFS and OS for SNP rs307826 in VEGFR3. The P-values are indicated for the univariate (UV) and multivariate (MV) analyses.
Finally, we also assessed the distribution of various unfavourable SNP genotypes in patients exhibiting a PD vs SD, PR or CR as their best response. On logistic regression, taking into account the MSKCC score, the presence of sarcomatoid dedifferentiation and baseline neutrophil count, the unfavourable genotypes GA/GG in VEGFR3 rs307826 were significantly more frequent in patients experiencing PD as best response when compared with patients experiencing SD, PR or CR as best response (Table 5).
Table 5. Distribution of SNP genotypes in patients exhibiting progressive disease and partial response as the best response.
| Gene (a) | SNP ID | In patients with PD as their best response (n=10) | In patients with SD, PR or CR as their best response (n=78) | P-Value by Fisher's exact | Adjusted P-value by logistic regression | Odds ratio (95% CI) |
|---|---|---|---|---|---|---|
|
Genes involved in pharmacokinetics | ||||||
| ABCB1 | rs1045642 | CC 2/10 (20%) | CC 23/77 (30%) | NS | — | — |
| rs1128503 | TT 3/10 (30%) | TT 12/78 (15%) | NS | — | — | |
| rs2032582 | TT or TA 2/8 (25%) | TT or TA 10/72 (14%) | NS | — | — | |
| TCG copy | Not present 7/7 (100%) | Not present 54/70 (77%) | NS | — | — | |
| CYP3A5 | rs776746 | GG 8/8 (100%) | GG 61/67 (91%) | NS | — | — |
| NR1/2 | rs3814055 | TT 3/8 (38%) | TT 12/74 (16%) | NS | — | — |
| rs2276707 | TT 2/8 (25%) | TT 3/75 (4%) | 0.02 | NS | — | |
| NR1/3 | rs2307424 | CC 6/10 (60%) | CC 36/78 (46%) | NS | — | — |
| rs2307418 | AA 8/10 (80%) | AA 49/78 (63%) | NS | — | — | |
| rs4073054 | TT 7/10 (70%) | TT 31/78 (40%) | 0.08 | NS | — | |
| |
CAT copy |
Present 8/10 (80%) |
Present 40/77 (52%) |
0.09 |
NS |
— |
|
Genes involved in pharmacodynamics | ||||||
| PDGFRA | rs35597368 | TT 8/10 (80%) | TT 61/78 (78%) | NS | — | — |
| VEGFR2 | rs1870377 | TT 7/10 (70%) | TT 41/78 (53%) | NS | — | — |
| VEGFR3 | rs307821 | GT+TT 5/10 (50%) | GT+TT 18/78 (23%) | 0.07 | 0.05 | 5.763 (0.986–33.693) |
| |
rs307826 |
GA+GG 6/10 (60%) |
GA+GG 17/78 (22%) |
0.01 |
0.02 |
7.011 (1.372–42.209) |
|
Genes in alternative proangiogenic factors | ||||||
| FGFR2 | rs2981582 | TT 2/10 (20%) | TT 10/77 (13%) | NS | — | — |
| IL8 | rs4073 | AA 1/9 (11%) | AA 11/70 (16%) | NS | — | — |
Abbreviations: PR=partial response; PD=progressive disease; CR=complete response; SNP=single-nucleotide polymorphism; 95% CI=95% confidence interval; SD=stable disease; NS=nonsignificant.
The logistic regression analysis was adjusted for the presence of sarcomatoid dedifferentiation, the MSKCC score and baseline neutrophils.
Variants were combined as follows: ABCB1: a TCG copy was linked to better outcome in van der Veldt et al (2010). Therefore, we analysed the impact of CC in rs1045642, TT in rs1128503 and TT (or TA) in rs2032582; CYP3A5: the GG variant was linked to poor outcome in van der Veldt et al (2010); NR1/2 rs3814055 and rs2276707: the TT variant was linked to poor outcome in van der Veldt et al (2010); NR1/3: a CAT copy was linked to poor outcome in van der Veldt et al (2010). Therefore, we analysed the impact of CC in rs2307424, AA in rs2307418 and TT in rs4073054; FGFR2: the TT variant was linked to poor outcome in Xu et al (2011a, 2011b); IL8: the AA variant was linked to poor outcome in Xu et al (2011a, 2011b); PDGFRA: the TT variant was linked to poor outcome in van der Veldt et al (2010); VEGFR2: the TT variant was linked to poor outcome in van der Veldt et al (2010); VEGFR3 rs307821: the GT/TT variant was linked to poor outcome in Garcia-Donas et al (2011); VEGFR3 rs307826: the GA/GG variant was linked to poor outcome in Garcia-Donas et al (2011).
We could not confirm associations between SNP rs776746 in CYP3A5, rs3814055 in NR1/2, rs11549467 in HIFA, rs1870377 in VEGFR2 and rs4073 in IL8 and outcome.
Discussion
In this retrospective study, we aim to observe the impact of SNPs that have recently been proposed as predictors of outcome to antiangiogenic therapy in metastatic RCC in an independent cohort of patients. We observed significant associations between SNPs in genes involved in sunitinib pharmacokinetics (ABCB1, NR1/2 and NR1/3), sunitinib pharmacodynamics (VEGFR3) and VEGF-independent pro-angiogenic pathways (FGFR2) and the therapeutic outcome of sunitinib in metastatic clear-cell RCC patients. For each of these associated SNPs, we observed similar hazard ratios as reported previously, thereby adding more evidence that these SNPs could be markers associated with outcome on sunitinib. Moreover, for most of these observations, a rationale is available.
As sunitinib was used as a monotherapy and as a first-line treatment, our results were not confounded by concomitant or previous therapies and we could detect significant associations in a series involving only a limited number of patients.
The efflux transporter ABCB1 (ATP binding cassette member B1, formerly known as P-glycoprotein or MDR1) is expressed in the intestine and liver and involved in the oral absorption and biliary secretion of several anticancer drugs (Dietrich et al, 2003). The ABC transporters may also contribute to multidrug resistance in tumours by actively extruding drugs from cancer cells, particularly in RCC cells (Soto-Vega et al, 2009; Walsh et al, 2009). As a consequence, expression levels and functionality of these drug transporters, for instance due to polymorphisms, may have important consequences for the efficacy of sunitinib. The most common functional SNPs in ABCB1 are the synonymous 3435C>T (rs1045642) and 1236C>T (rs1128503) changes and the nonsynonymous 2677G>T change (missense A893S/T rs2032582). Functional studies have shown that the haplotype of these three SNPs (rs1046542, rs1128503 and rs2032582) is a silent mutation and alters the function of the efflux transporter including its substrate specificity. We observed a significant association between the TT variant in rs1128503 1236C>T and shorter PFS and OS. In 89 RCC patients treated with sunitinib, Garcia-Donas et al (2011) observed an association, although not significant, between rs1128503 and PFS (HR 1.42, P=0.089) and OS (HR 1.75, P=0.055), favoring the patients with a C-allele. In 129 RCC patients treated with sunitinib, van der Veldt et al (2010) observed that the presence of a TCG haplotype (rs1045642, rs1128503 and rs2032582) in ABCB1 (and thus the presence of the C-variant in rs1128503) was associated with prolonged PFS (P=0.033) and a tendency for prolonged OS (P=0.078). In 241 patients treated with pazopanib, the wild-type CC variant of rs1128503 was associated with improved OS compared with the wild-type TT genotypes (28 vs 20 months, P=0.009) (Xu et al, 2011b).
Fibroblast growth factor receptor 2 (FGFR2) is a VEGF-independent pro-angiogenic factor. The TT polymorphism in rs2981582 906C>T leads to increased transcription and expression of FGFR2 (Meyer et al, 2008) and thus possibly to increased VEGF-independent angiogenesis. We observed a significant association between the TT variant in rs2981582 and shorter PFS. Data on the impact of rs2981582 in FGFR2 on outcome on TKIs are only available in patients treated with pazopanib. In a series of 380 RCC patients, the TT variant was associated with inferior PFS compared with the CC genotype (P=0.053) (Xu et al, 2011a) and in a group of 241 patients, the TT genotype was associated with inferior OS compared with the CC genotype (median OS 21.4 vs 28.0 months, P=0.02) (Xu et al, 2011b).
The expression of cytochrome P450 CYP3A4, thought to be the key enzyme for the hepatic biotransformation of sunitinib, is regulated by the ligand-activated nuclear receptors NR1I2 (pregnane X receptor) and NR1I3 (constitutive androstane receptor). We observed a significant association between the TT genotype in rs2276707 8055C>T in NR1/2 and a shorter PFS and OS. van der Veldt et al (2010) also found a significant difference in PFS between patients with the CC/CT genotype and patients with the TT genotype, 10.8 vs 6.7 months (P=0.025), but they could not confirm these results on multivariate analysis. Concerning NR1/3, we observed a significant association between the TT variant in SNP rs4073054 and shorter PFS and OS. Prolonged PFS (13.3 vs 8.0 months, P=0.017) was found in 136 patients with absence of a CAT copy in the NR1/3 haplotype (rs2307424, rs2307418 and rs4073054; P=0.021) (van der Veldt et al, 2010). This corresponds with our results, as rs4073054 concerns the T in the CAT haplotype. We also observed a significant association between the CC genotype in rs 2307424 in NR1/3 and better OS, but there is no external validation at this moment for these results and we could not link this finding to the observations of van der Veldt et al (2010).
Platelet-derived growth factor receptor-α is one of the molecular targets of sunitinib. On univariate analysis, we observed a significant association between the TT variant in rs35597368 1580T/C in PDGFRA and longer PFS and OS. The T in rs35597368 corresponds to the T in the GCGT haplotype composed of four SNPs in the gene (rs1800810, rs1800812, rs1800813 and rs35597368). van der Veldt et al (2010) observed on univariate analysis a better OS in patients with a GCGT haplotype in both alleles (GCGT–GCGT), and thus in patients with a TT variant of the SNP, whereas patients with a GCG–other or other–other haplotype had a poorer median OS: 24.2 vs 14.8 months (P=0.002 on univariate analysis but 0.108 on multivariate analysis). We could not confirm the association with PFS and OS on multivariate analysis. The functional impact of this SNP is presently unknown.
The VEGFR3 signalling is involved in embryonic angiogenesis, adult lymphangiogenesis and tumoural angiogenesis (Partanen et al, 1999; Valtola et al, 1999) and is one of the main targets of sunitinib. We observed a significant association between the GT or TT variant in rs307821 3971G>T in VEGFR3 and shorter PFS and OS. Note that because of a crossing of the curves, the median OS was longer in the GT and TT variants than in the GG variants of rs307821. Nevertheless, the HR for survival for patients with the GT or TT variants in rs307821 in VEGFR3 vs patients with the GG variant was 2.265 (95% CI 1.202–4.238). The crossing of the curves is probably because of the limited number of patients in our series. We also observed a significant association between the AG or GG variant in rs307826 1480A>G and shorter OS. In a series of 89 RCC patients treated with sunitinib, TTP for the GT variant of rs307821 of was 6.7 months vs 13.7 months for patients with the GG genotype (P=0.00085) and TTP for the GA variant of rs307826 was 3.6 months vs 13.7 months for patients with the AA genotype (P=0.00049). There was no significant association with OS (Garcia-Donas et al, 2011). In 228 patients treated with pazopanib, OS was 26 months in the AA variant vs 23 months in the AG variant (P=0.04) of rs307826 but, surprisingly, these authors did not find any association between the SNP and PFS (Xu et al, 2011a, 2011b). This matches the observation of van der Veldt et al (2010), who reported no significant effect of rs307826 on PFS after sunitinib treatment.
Our study has several potential limitations. (1) It was a retrospective analysis of patients treated in several centres without a central protocol dictating schedule and dose modifications or timing of radiological assessments. (2) Because our patients were mainly white, the relevance of these polymorphisms needs to be assessed in other ethnic groups, in whom the described polymorphisms may be less frequent. (3) We failed to genotype SNP rs1126647 in IL-8 because of technical reasons. (4) In case of rs11549467 there was only one heterozygous patient. As a consequence, the impact of this SNP could not be analysed. (5) Concerning SNPs in ABCB1 and NR1/3, in literature, only results of associations with haplotypes were available. (6) Finally, there was a better outcome in our series (PFS 15.0 and OS 29.0 months) compared with the outcome on sunitinib in the pivotal trial (PFS 11.0 and OS 26.0 months; Motzer et al, 2007). This difference is likely because of the patient selection in our series: all the patients had to complete at least one cycle of sunitinib and had to reach at least the first evaluation by CT scan.
Conclusions
We confirmed several associations between polymorphisms in genes linked to pharmacokinetics and pharmacodynamics of sunitinib and therapeutic outcome of patients receiving sunitinib for metastatic RCC. These associations had previously been described in other series of patients treated with sunitinib or pazopanib.
The impact of SNPs in pathways linked to pharmacokinetics and pharmacodynamics of sunitinib shows that besides acquired genetic characteristics of tumour cells, patient's germline genetic variation may also affect the efficacy of anticancer therapy. Moreover, germline DNA is inherited, fixed and relatively insensitive to time and environmental factors, which makes it more reliable than nucleotide and protein biomarkers linked to the tumour.
If the impact of these and other SNPs on outcome on sunitinib could be validated prospectively in independent series, scoring systems based on the combination of several unfavourable or favourable SNPs could be elaborated. When combining these SNPs with clinical and biochemical parameters associated with outcome, we will probably be able to predict more precisely the chance of response to sunitinib and identify primary resistant patients in order to orient them towards other therapies, avoiding unnecessary side effects and costs. Similarly, we will be able to predict more accurately disease progression, which is the time point of secondary resistance to sunitinib. Polymorphisms could also help us to identify those patients whose ideal starting dose of sunitinib could be higher than the usual 50 mg daily, for instance, patients with genotypes and haplotypes leading to lower sunitinib plasma levels.
Acknowledgments
This project is a common project of two kidney tumour banks: the CIT-rein tumour bank (Paris, France) and the University Hospitals Leuven kidney tumour bank (Leuven, Belgium). The CIT-rein project is headed by Professor Stéphane Oudard and Professor Jean-Jacques Patard and was part of the PNES 2007 (Programme National d'Excellence Spécialisée) from the INCa (Institut du Cancer). We sincerely thank the urologists, medical oncologists and pathologists of the next centers, whose biological material was used in the analysis, for their collaboration: Angers: Centre oncologique Paul Papin: Abdel Azzouzi, Rémy Delva, Stéphane Triau, Pierre Bigot; Caen: Centre François Baclesse: Henri Bensadoun, Emmanuel Sevin, François Comoz; Créteil: Hôpital Henri Mondor: Alexandre de la Taille, Bernard Paule, Yves Allory; Suresnes: Hôpital Foch: Thierry Lebret, Christine Théodore, Yves Denoux; Leuven: University Hospitals Leuven: Hendrik Van Poppel, Evelyne Lerut, Joost Berkers, Pascal Wolter, Patrick Schöffski, Robert Paridaens; Limoges: Hôpital Dupuytren: Aurélien Descazeaud, Julien Berger; Lyon: Centre Léon Bérard: Marc Colombel, Sylvie Négrier, Florence Mege-Lechevallier; Marseille: Institut Paoli-Calmettes: Franck Bladou, Gwénaelle Gravis, Myriam Marcy; Nantes: ICO Gauducheau: Olivier Bouchot, Frédéric Rolland, Karine Reanudin; Paris: Hôpital Necker and Saint Joseph: Arnaud Méjean, Virginie Verkarre, Vincent Molinié Poitiers: Jacques Irani, Jean Marc Tourani, Pierre Marie Le Villain; Rennes: Brigitte Laguerre, Jean-Jacques Patard, Nathalie Rioux-Leclercq; Paris: Clinique St-Joseph: Hervé Baumert, Gael Deplanque, Vincent Molinié Strasbourg: CHRU Strasbourg: Didier Jacqmin, Brigitte Duclos, Véronique Lindler; Tours: CHU Tours: Olivier Haillot, Claude Linassier, Franck Fetissof. The tissue collection was coordinated by the Plateforme de Ressources Biologiques de l'Hôpital Européen Georges Pompidou in Paris. We are grateful to Corine Takouchop Teghom, Claudia De Toma and Reza Elaidi for the coordination of the tissue and clinical data collection. Benoit Beuselinck received a grant from the Fondation Martine Midy (Paris, France) (2009–2010). His work is also funded by the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Belgium) (2011–2013). Alexandra Karadimou received a grant from the Hellenic Society of Medical Oncology (Athens, Greece) (2010–2011). Bart Claes is supported by the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT) and by the Emmanuel van der Schueren grant of the Vlaamse Liga tegen Kanker. Diether Lambrechts is supported by the Stichting Tegen Kanker. Evelyne Lerut received fundings from Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Belgium) and Stichting tegen Kanker.
Stéphane Oudard received honorarium from Novartis, Pfizer, Roche, Sanofi-Aventis, Keocyt, Amgen and Bayer. Patrick Schöffski received honoraria from Pfizer and GSK for participation in educational activities in the context of renal cell carcinoma. Pierre Bigot received honorarium from Novartis. Jean Jacques Patard is a consultant and principal investigator in Pfizer trials. The other authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Beuselinck B, Oudard S, Rixe O, Wolter P, Blesius A, Ayllon J, Elaidi R, Schöffski P, Barrascout E, Morel A, Escudier B, Lang H, Zucman-Rossi J, Medioni J. Negative impact of bone metastasis on outcome in clear cell renal cell carcinoma treated with Sunitinib. Ann Oncol. 2011;22:794–800. doi: 10.1093/annonc/mdq554. [DOI] [PubMed] [Google Scholar]
- Dietrich CG, Geier A, Oude Elferink RPJ. ABC of oral bioavailability: transporters as gatekeepers in the gut. Gut. 2003;52:1788–1795. doi: 10.1136/gut.52.12.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Donas J, Esteban E, Leandro-García LJ, Castellano DE, Del Alba AG, Climent MA, Arranz JA, Gallardo E, Puente J, Bellmunt J, Mellado B, Martínez E, Moreno F, Font A, Robledo M, Rodríguez-Antona C. Single nucleotide polymorphism associations with response and toxic effects in patients with advanced renal-cell carcinoma treated with first-line sunitinib: a multicentre, observational, prospective study. Lancet Oncol. 2011;12:1143–1150. doi: 10.1016/S1470-2045(11)70266-2. [DOI] [PubMed] [Google Scholar]
- Golshayan AR, George S, Heng DY, Elson P, Wood LS, Mekhail TM, Garcia JA, Aydin H, Zhou M, Bukowski RM, Rini BI. Metastatic sarcomatoid renal cell carcinoma treated with vascular endothelial growth factor-targeted therapy. J Clin Oncol. 2009;27:235–241. doi: 10.1200/JCO.2008.18.0000. [DOI] [PubMed] [Google Scholar]
- Heng DY, Xie W, Regan MM, Warren MA, Golshayan AR, Sahi C, Eigl BJ, Ruether JD, Cheng T, North S, Venner P, Knox JJ, Chi KN, Kollmannsberger C, McDermott DF, Oh WK, Atkins MB, Bukowski RM, Rini BI, Choueiri TK. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: results from a large, multicenter study. J Clin Oncol. 2009;27:5794–5799. doi: 10.1200/JCO.2008.21.4809. [DOI] [PubMed] [Google Scholar]
- Houk BE, Bello CL, Poland B, Rosen LS, Demetri GD, Motzer RJ. Relationship between exposure to sunitinib and efficacy and tolerability endpoints in patients with cancer: results of a pharmacokinetic/pharmacodynamic meta-analysis. Cancer Chemother Pharmacol. 2010;66:357–371. doi: 10.1007/s00280-009-1170-y. [DOI] [PubMed] [Google Scholar]
- Meyer KB, Maia AT, O'Reilly M, Teschendorff AE, Chin SF, Caldas C, Ponder BA. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;5:e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer R, Bacik J, Mazumbar M. Prognostic factors for survival of patients with stage IV renal cell carcinoma: Memorial Sloan Kettering Cancer Center experience. Clin Cancer Res. 2004;10:6202S–6203S. doi: 10.1158/1078-0432.CCR-040031. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Oudard S, Negrier S, Szczylik C, Pili R, Bjarnason GA, Garcia-del-Muro X, Sosman JA, Solska E, Wilding G, Thompson JA, Kim ST, Chen I, Huang X, Figlin RA. Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol. 2009;27:3584–3590. doi: 10.1200/JCO.2008.20.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST, Chen I, Bycott PW, Baum CM, Figlin RA. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- Partanen TA, Alitalo K, Miettinen M. Lack of lymphatic vascular specificity of vascular endothelial growth factor 3 in 185 vascular tumors. Cancer. 1999;86:2406–2412. [PubMed] [Google Scholar]
- Patil S, Figlin R, Hutson T, Michaelson M, Négrier S, Kim T, Huang X, Motzer R. Prognostic factors for progression free survival and overall survival with sunitinib targeted therapy and with cytokine as first-line therapy in patients with metastatic renal cell carcinoma. Ann Oncol. 2011;22:295–300. doi: 10.1093/annonc/mdq342. [DOI] [PubMed] [Google Scholar]
- Reumers J, De Rijk P, Zhao H, Liekens A, Smeets D, Cleary J, Van Loo P, Van Den Bossche M, Catthoor K, Sabbe B, Despierre E, Vergote I, Hilbush B, Lambrechts D, Del-Favero J. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat Biotechnol. 2011;30:61–68. doi: 10.1038/nbt.2053. [DOI] [PubMed] [Google Scholar]
- Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- Soto-Vega E, Arroyo C, Richaud-Patin Y, Garcia-Carrasco M, Vazquez-Lavista LG, Llorente L. P-glycoprotein activity in renal clear cell carcinoma. Urol Oncol. 2009;27:363–366. doi: 10.1016/j.urolonc.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Valtola R, Salven P, Heikkil P, Taipale J, Joensuu H, Rehn M, Pihlajaniemi T, Weich H, deWaal R, Alitalo K. VEGFR-3 and its ligand VEGF-C are associated with angiogenesis in breast cancer. Am J Pathol. 1999;154:1381–1390. doi: 10.1016/S0002-9440(10)65392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veldt AA, Eechoute K, Gelderblom H, Gietema J, Guchelaar HJ, van Erp NP, van den Eertwegh AJ, Haanen JB, Mathijssen RH, Wessels JA. Genetic polymorphisms associated with a prolonged progression-free survival in patients with metastatic renal cell cancer treated with sunitinib. Clin Cancer Res. 2010;17:620–629. doi: 10.1158/1078-0432.CCR-10-1828. [DOI] [PubMed] [Google Scholar]
- Walsh N, Larkin A, Kennedy S, Connolly L, Ballot J, Ooi W, Gullo G, Crown J, Clynes M, O'Driscoll L. Expression of multidrug resistance markers ABCB1 (MDR-1/P-gp) and ABCC1 (MRP-1) in renal cell carcinoma. BMC Urol. 2009;9:6. doi: 10.1186/1471-2490-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CF, Bing NX, Ball HA, Rajagopalan D, Sternberg CN, Hutson TE, de Souza P, Xue ZG, McCann L, King KS, Ragone LJ, Whittaker JC, Spraggs CF, Cardon LR, Mooser VE, Pandite LN. Pazopanib efficacy in renal cell carcinoma: evidence for predictive genetic markers in angiogenesis-related and exposure-related genes. J Clin Oncol. 2011a;29:2557–2564. doi: 10.1200/JCO.2010.32.9110. [DOI] [PubMed] [Google Scholar]
- Xu CF, Ball HA, Bing N, Sternberg C, Xue Z, McCann L, King K, Spraggs C, Mooser V, Pandite LN. Association of genetic markers in angiogenesis- or exposure-related genes with overall survival in pazopanib-treated patients with advanced renal cell carcinoma. J Clin Oncol. 2011b;29 (Suppl 7:abstr 303. [Google Scholar]