Abstract
Background:
Several new assays have been developed for high-risk HPV testing of cervical samples; we compare six HPV tests in a screening population.
Methods:
Residual material from liquid-based PreservCyt samples was assayed. Four tests (Hybrid Capture 2, Cobas, Abbott and Becton-Dickinson (BD)) measured HPV DNA while two used RNA (APTIMA and NorChip).
Results:
Positivity rates ranged from 13.4 to 16.3% for the DNA-based tests with a significantly lower positivity rate for the Abbott assay. The Gen-Probe APTIMA assay was positive in 10.3% of women, which was significantly lower than all the DNA tests; the NorChip PreTect HPV-Proofer test was much lower at 5.2%. 40 CIN2+ cases were identified, of which 19 were CIN3+. All CIN3+ cases were HPV positive by all tests except for one, which was negative by the Abbott assay and five which were negative by the NorChip test.
Conclusion:
All HPV tests except NorChip showed high sensitivity for high-grade lesions positive by cytology, suggesting co-testing is unnecessary when using HPV tests. Positivity rates in cytology-negative specimens were similar for the DNA-based tests, but lower for the APTIMA test suggesting this maintains the high sensitivity of DNA tests, but with better specificity.
Keywords: biomarkers, cervix, HPV
There is widespread interest in introducing HPV testing for primary screening and triage. Many studies have shown that HPV DNA testing has higher sensitivity but lower specificity than cytology (Cuzick et al, 2006, 2008a, Ronco et al, 2010). However, there are limited comparisons between the different HPV tests. We have previously carried out studies which have compared a number of adjunctive tests in women with abnormal smears who have been referred to colposcopy (Szarewski et al, 2008, Szarewski et al, 2012). These studies were focused primarily on identifying tests with good sensitivity, and the specificities reflect the typically lower values found in a referral setting. Although this approach does permit a comparison of relative specificity, it is desirable to also compare these tests in a screening setting. A definitive comparison of sensitivity in this setting would require studies of at least 20 000 women, but specificity can be accurately evaluated in a much smaller sample size. Here we compare four DNA-based HPV tests and two RNA-based HPV tests in a screening setting.
Materials and Methods
Residual material was used from the liquid-based cytology PreservCyt samples from 6000 women who attended for a routine 3 or 5 yearly (depending on age) screening smear, and whose samples were sent to the cytology laboratory at St. Mary's Hospital, London. Unsatisfactory cytology samples were excluded; there were no other inclusion/exclusion criteria. Samples were linked to concurrent cytology results and any histology within 6 months of an abnormal smear, and this information was fully anonymised before being transferred for analysis.
Consent was deemed not to be necessary, as the women were not going to be contacted with their result, nor would it be used to influence their management. This also meant that we were unable to access details of any previous screening history. The study was approved by the Imperial NHS Trust Tissue Management Committee and the Multicentre Research Ethics Committee for Wales.
All results are presented based on the local histopathology and the highest grade of abnormality seen in the biopsy or treatment specimen was used. As the study was anonymised, the HPV result was not communicated to the women or the doctor and was not acted upon. This means that women who tested positive for HPV, but had normal cytology would not have been further investigated, and therefore disease ascertainment was not possible in this group. In addition, it was not possible to undertake histology review. However, our experience from previous studies (Szarewski et al, 2012) is that, when pathology review is possible, 5% of biopsies read as CIN2+ are downgraded to<CIN2; conversely, 6% of biopsies reported as<CIN2 are upgraded to CIN2+ following pathology review.
Laboratory methods
In this study the following assays were carried out and scored in strict accordance with the manufacturer's protocol. All tests were carried out in the Centre for Cancer Prevention (by LH & GT), except for PreTect HPV-Proofer, which was carried out at The Doctors Laboratory (SL), and the APTIMA typing test (not the consensus test), which was performed in the Gen-Probe laboratory (see below).
(a) DNA-based detection and genotyping assays:
Hybrid Capture 2 (Qiagen GmbH, Hilden, Germany) detecting 13 HR-HPV genotypes collectively. The Hybrid Capture 2 assay is based on the hybridisation of HPV DNA to a 13 HR HPV RNA probe cocktail. The DNA:RNA hybrid is captured by an antiDNA:RNA antibody and detected by chemiluminescence. Readings over 1 RLU were considered positive.
Cobas 4800 HPV test (Roche Molecular Diagnostics, Pleasanton, CA, USA). The Cobas 4800 HPV test is a qualitative in vitro test for the detection of 14 HR HPV types. The test separately identifies HPV 16 and HPV 18, while concurrently detecting the 12 remaining the high-risk types as a group (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68).
Abbott RealTime High Risk HPV assay (Abbott Molecular GmbH & Co. KG, Wiesbaden, Germany). This qualitative multiplex real-time test also separately identifies HPV 16 and HPV 18 while concurrently detecting the 12 remaining high-risk types as a group (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68).
BD HPV test (BD Diagnostics, Sparks, MD, USA). This is a real-time PCR (at the time of writing, not yet commercially available), which detects 14 HR HPV types. Type-specific detection is achieved using sequence-specific (non-consensus) E6/E7 DNA amplification. Typing is provided for types 16, 18, 31, 45, 51, 52 and 59. The remaining HPV types are grouped into two pools: (33, 56, 58, 66) and (35, 39, 68).
(b) RNA-based detection assays
PreTect HPV-Proofer (NorChip, Klokkarstua, Norway). PreTect Proofer is a real-time multiplex NASBA assay for isothermal amplification of E6/E7 mRNA expressed by five high-risk HPV types (16, 18, 31, 33 and 45) using proprietary primer sets (Molden et al, 2007).
APTIMA (Gen-Probe Incorp, San Diego, CA, USA). The APTIMA assay is based on target capture, transcription-mediated amplification and hybridisation protection for the detection of E6/E7 mRNA expression of 14 HR HPV types. Specimens which tested positive by APTIMA were then tested with HPV type-specific tests, which detected; Joo et al, 2010). On the basis of the same principle as APTIMA HPV (AHPV) assay for the detection of HPVE6/E7 mRNA from 14 HR HPV genotypes, Gen-Probe has also developed the APTIMA HPV 16 18/45 genotype (AHPV-GT) assay, which specificially detects HPV 16, 18 and 45. The 95% detection limit for these genotypes is 100 copies/reaction. The genotyping accuracy as assessed by agreement with Linear Array genotyping test (Roche Molecular) is 97% for HPV 16 and 93% for HPV18/45.
Statistical analysis
The main outcome measures were specificity and positive predictive value (PPV) in women referred to colposcopy based on cytology findings. Positivity cutpoints were predefined for all tests. Relative sensitivity was also assessed as a secondary end point. Confidence intervals for these were based on binomial statistics. Comparisons between tests were conducted by McNemar's test for matched pairs; odds ratios and 95% confidence intervals for discordant pairs of results are also reported. Additional calculations of relative sensitivity, specificity and PPV were carried out separately for women aged <30 and ⩾30 years. Full details of type-specific results will appear elsewhere, but positivity results for HPV 16, HPV 18 and ‘other high-risk' HPV are reported here.
The worst histology within 6 months of the initial baseline visit was used as the outcome, and histologically confirmed CIN2+ and CIN3+ were taken as the primary end points. Specificity is only reported for <CIN2, as we do not consider the detection of CIN2 to be a ‘false positive'. As this was an anonymised study, no pathology review was possible (see Materials and Methods section above).
All statistical analyses were carried out using Stata 11.2 (StataCorp., College Station, TX, USA).
Results
Residual ThinPrep samples from 6000 women attending for routine (3–5 yearly) screening were assayed. The median age was 37 years (range 20–66, IQR 30–46) and 78% of the women were aged 30 and above. A total of 5682 women (94.7%) had normal cytology and were not referred for colposcopy. An additional 182 (3.0%) had borderline cytology and 35 of these (19%) were referred for colposcopy of which 30 (86%) had a biopsy (Table 1). Most of the women with mild dyskaryosis or worse had colposcopy (108 out of 136, 79.4%) and of these 89 out of 108 (82.4%) had a biopsy.
Table 1. Screening cytology vs worst histology from biopsy or LEEP.
| |
Worst histology |
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cytology result | No biopsy taken | Inadequate/normal | HPV only | Borderline | CIN1 | CIN2 | CIN3/CGIN HG | Invasive cancer | Total |
| Negative |
5682 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
5682 |
| Borderline dyskaryosis |
152 |
2 |
16 |
1 |
7 |
3 |
1 |
0 |
182 |
| Mild dyskaryosis |
42 |
1 |
22 |
1 |
25 |
9 |
5 |
0 |
105 |
| Moderate dyskaryosis |
5 |
0 |
0 |
0 |
4 |
6 |
3 |
0 |
18 |
| Severe dyskaryosis |
0 |
0 |
0 |
0 |
0 |
3 |
9 |
1 |
13 |
| Total | 5881 | 3 | 38 | 2 | 36 | 21 | 18 | 1 | 6000 |
Abbreviations: CIN=cervical intraepithelial neoplasia; HPV=human papillomavirus; LEEP=loop electrosurgical excision procedure.
Positivity rates were in the range of 13.4–16.3% for the DNA-based tests (Table 2). Positivity rates were generally higher among women aged <30 years compared with women ⩾30 years. For the DNA-based tests, positivity ranged from 25.0 to 29.0% among younger women (aged <30 years). Among women 30 years or older, positivity rates for the DNA-based tests ranged from 10.0 to 12.6% (Supplementary Appendix Table A2). Results separately for normal and abnormal cytology are also given in Supplementary Appendix Table A2 by age.
Table 2. High-risk (HR) HPV positivity overall and for types 16 and 18 for different tests (ordered by overall high-risk HPV positivity).
| Test | Number tested | Number adequate result | Positive (%) | HPV 16 positive (%) | HPV 18 positive (%) | Other HR HPV positive (%) |
|---|---|---|---|---|---|---|
| BD HPV |
5998 |
5965 |
970 (16.3) |
168 (2.8) |
71 (1.2) |
834 (14.0) |
| Roche Cobas |
5986 |
5979 |
959 (16.0) |
211 (3.5) |
86 (1.4) |
801 (13.4) |
| Qiagen Hybrid Capture 2 |
5984 |
5984 |
906 (15.1) |
— |
— |
— |
| Abbott RealTime High Risk HPV |
5994 |
5990 |
801 (13.4) |
180 (3.0) |
70 (1.2) |
634 (10.6) |
| Gen-Probe APTIMA |
6000 |
5995 |
620 (10.3) |
128 (2.1) |
63 (1.1)a |
— |
| NorChip PreTect HPV-Proofer | 5969 | 5747b | 297 (5.2) | 116 (2.0) c | 51 (0.9) c | 139 (2.4)d |
Abbreviations: HPV=human papillomavirus; HR=high risk.
Tested HPV 18/45 combined.
n=76 U1A mRNA not detected, n=132 testing failed for technical reasons, n=10 failed quality control, n=4 low U1A mRNA (indeterminate result).
Samples were excluded if the overall result was inadequate.
Includes only types 31, 33 and 45.
The Gen-Probe APTIMA assay was positive in 10.3% of all women and this was significantly lower than all the DNA tests (P<0.0001 in all cases). The NorChip PreTect HPV-Proofer had a significantly and substantially lower positivity rate than all other tests, being 5.2%, which was equal to the positivity rate for borderline or worse cytology.
Relative positivity rates for HPV 16 mirrored the findings for all high-risk types (Table 2), being 2.8–3.5% for the three DNA tests that provided typing and 2.1% for each of the RNA tests. HPV 18 positivity was lower, ranging from 0.9 to 1.4% and showed less difference between tests, where the only significant difference was Roche Cobas being higher than the RNA-based tests. Positivity for only non 16/18 high-risk types showed similar patterns as for all high-risk types, with the exception of NorChip PreTect HPV-Proofer, which was much lower due, in part, to the restricted number of HPV types tested (only 31, 33 and 45).
There was a high concordance between the Roche Cobas, the Abbott RealTime High Risk HPV assay and the BD assay, with kappa values in excess of 0.8 for all pairwise comparisons (Table 3). The Roche Cobas and BD tests had very similar performance, but both were significantly more often positive than the Abbott RealTime High Risk HPV assay, which was significantly less often positive than all the other DNA tests. Kappa values for the HPV DNA tests and Gen-Probe APTIMA were also high for type 16, although Roche Cobas was more often positive than the other tests for this type. Compared with all other tests, there was poorer correlation with NorChip PreTect Proofer, which was less often positive (Supplementary Appendix Table A3).
Table 3. Summary of discordant results between different tests.
| BD HPV | Roche Cobas | Qiagen Hybrid Capture 2 | Abbott RealTime High Risk HPV | Gen-Probe APTIMA | NorChip PreTect HPV-Proofer | |
|---|---|---|---|---|---|---|
| BD HPV |
— |
0.83 |
0.78 |
0.81 |
0.65 |
0.33 |
| Roche Cobas |
(145, 134)a |
— |
0.75 |
0.85 |
0.64 |
0.35 |
| |
1.1 (0.8, 1.4) |
|
|
|
|
|
| Qiagen Hybrid Capture 2 |
(204, 141) |
(225, 174) |
— |
0.78 |
0.68 |
0.35 |
| |
1.4 (1.2, 1.8) |
1.3 (1.1, 1.6) |
|
|
|
|
| Abbott RealTime High Risk HPV |
(227, 57) |
(194, 35) |
(216, 108) |
— |
0.70 |
0.39 |
| |
4.0 (3.0, 5.4) |
5.5 (3.9, 8.2) |
2.0 (1.6, 2.5) |
|
|
|
| Gen-Probe APTIMA |
(421, 70) |
(414, 77) |
(353, 68) |
(279, 97) |
— |
0.45 |
| |
6.0 (4.7, 7.9) |
5.4 (4.2, 6.9) |
5.2 (4.0, 6.8) |
2.9 (2.3, 3.7) |
|
|
| NorChip PreTect HPV-Proofer |
(684, 61) |
(670, 54) |
(625, 63) |
(532, 65) |
(375, 82) |
— |
| 11.2 (8.6, 14.8) | 12.4 (9.4, 16.7) | 9.9 (7.6, 13.1) | 8.2 (6.3, 10.8) | 4.6 (3.6, 5.9) |
Number in top half of Table represent kappa values. Numbers in bottom half of Table give discordant pairs (column positive/row negative vs column negative/row positive) and OR (95% CI).
145 Tested positive for BD HPV and negative for Roche Cobas, and 134 tested negative for BD HPV and positive for Roche Cobas.
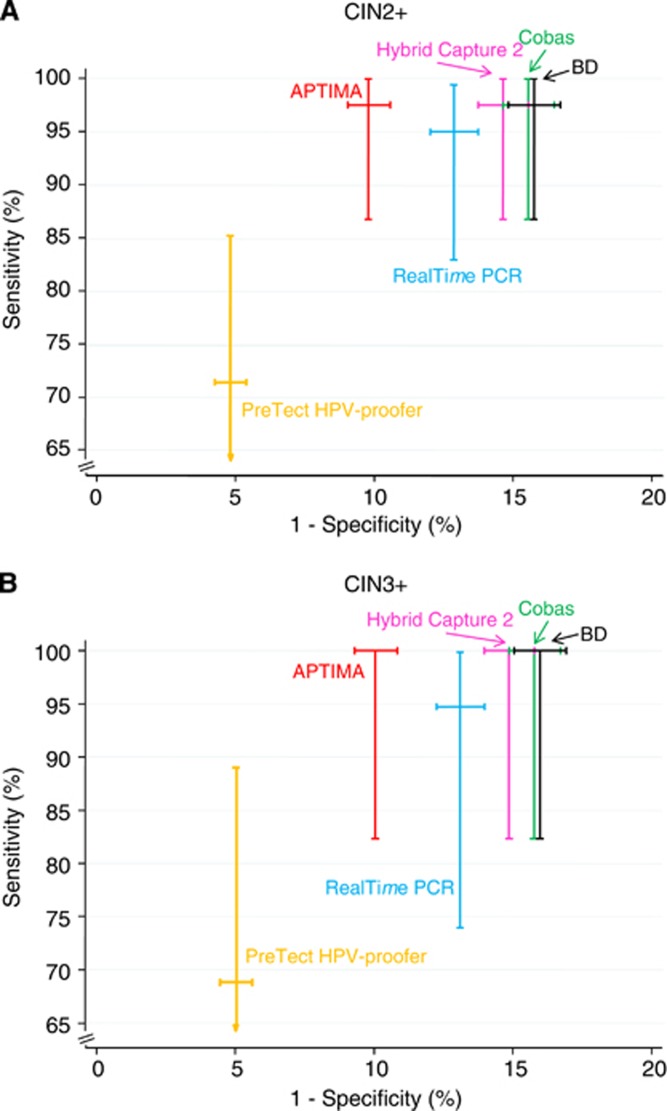
A total of 40 CIN2+ cases were identified, of which 19 were CIN3+. Of these, 5 of the CIN2 and 11 of the CIN3+ cases were aged 30 or above. All of the CIN3+ cases were HPV positive by all tests, except for the NorChip assay and one case, from a woman aged 29 years, which was missed by the Abbott RealTime High Risk HPV assay. Five CIN3+ cases were negative by the NorChip Test, and three cases had an inadequate result for technical reasons (Table 4 and Figure 1). Of the five cases with a negative result by NorChip test, two were under the age of 30.
Table 4. Relative sensitivity, specificity and PPV of different tests for the detection of high-grade disease based on 19 CIN3+ cases and 40 CIN2+ cases.
| Test | Sensitivity (95% CI) | Specificity (95% CI) | PPV (%,all) (95% CI) | PPV (%,only for those referred for colposcopy) (95% CI) |
|---|---|---|---|---|
|
BD HPV | ||||
| CIN3+ |
100.0 (82.4–100.0) |
|
2.0 (1.2–3.0) |
16.5 (10.3–24.6) |
| CIN2+ |
97.5 (86.8–99.9) |
84.3 (83.3–85.2) |
4.0 (2.9–5.5) |
33.9 (25.3–43.3) |
|
Roche Cobas | ||||
| CIN3+ |
100.0 (82.4–100.0) |
|
2.0 (1.2–3.1) |
16.7 (10.3–24.8) |
| CIN2+ |
97.5 (86.8–99.9) |
84.5 (83.6–85.4) |
4.1 (2.9–5.5) |
34.2 (25.6–43.7) |
|
Qiagen Hybrid Capture 2 | ||||
| CIN3+ |
100.0 (82.4–100.0) |
|
2.1 (1.3–3.3) |
16.0 (9.9–23.8) |
| CIN2+ |
97.5 (86.8–99.9) |
85.4 (84.5–86.3) |
4.3 (3.1–5.8) |
32.8 (24.4–42.0) |
|
Abbott RealTime High Risk HPV | ||||
| CIN3+ |
94.7 (74.0–99.9) |
|
2.2 (1.3–3.5) |
16.1 (9.8–24.2) |
| CIN2+ |
95.0 (83.1–99.4) |
87.2 (86.3–88.0) |
4.7 (3.4–6.5) |
33.9 (25.3–43.5) |
|
Gen-Probe APTIMA | ||||
| CIN3+ |
100.0 (82.4–100.0) |
|
3.1 (1.9–4.7) |
17.4 (10.8–25.9) |
| CIN2+ |
97.5 (86.8–99.9) |
90.2 (89.5–91.0) |
6.3 (4.5–8.5) |
35.8 (26.8–45.5) |
|
NorChip PreTect HPV-Proofera | ||||
| CIN3+ |
68.8 (41.3–89.0) |
|
3.7 (1.9–6.5) |
20.0 (10.4–33.0) |
| CIN2+ | 71.4 (53.7–85.4) | 95.2 (94.7–95.8) | 8.4 (5.5–12.2) | 45.5 (32.0–59.4) |
Abbreviations: CI=confidence interval; HPV=human papillomavirus.
Based on 16 cases of CIN3+ and 35 cases of CIN2+.
Figure 1.
Summary graph of the sensitivity and specificity results for the detection of (A) CIN2+ and (B) CIN3+ (with 95% CI).
The CIN3+ case, in a woman aged 29, missed by the Abbott assay was positive by Hybrid Capture 2 with an RLU of 85, and was additionally typed by Linear Array (courtesy of Dr C Wheeler), and showed strong bands for types 53, 66, 67, 70, 81, 82 and 84. For the other tests it was generally a low-positive result for non 16/18 HPV types, and the BD assay was positive for the (33, 56, 58, 66) pool. Sensitivity for CIN2+ was again lower for the NorChip test, but of the 21 cases of CIN2, all but one were positive by all other tests, and the same case was negative throughout, suggesting that the histology may have been inaccurate in this case.
As specificity for CIN3+ would treat CIN2 as a false positive, which is inappropriate, we only report specificity for less than CIN2. As seen from Table 4 and Figure 1, the tests with lower positivity rates have higher specificity, with a 95.0% value for the NorChip test 90.2% for Gen-Probe, 87.2% for the Abbott assay and the three other DNA tests ranging from 84.3 to 85.4%. The sensitivity and specificity are generally higher for women over 30 years compared with women <30 years, although the relative performance of the tests was similar (Supplementary Appendix Table A1).
PPVs for CIN2+ and CIN3+ are also shown in Table 4, both for all women and only those referred for colposcopy. The former are artificially low due to non-referral of all negative and most borderline smears, while the latter is an accurate measure only for women with mild dyskaryosis or high-grade cytologic abnormalities. The relative order of the performance of the tests was similar for PPV and specificity, with high PPVs seen for tests with high specificity.
Discussion
The main strength of this study is a head- to-head comparison of six HPV tests in a screening population in which all women were evaluated by all tests. No other such comparison exists and we are only aware of studies comparing two or at most three different tests, usually against Hybrid Capture 2, that is, vs Gen-Probe APTIMA (Monsenego et al, 2011, Wu et al, 2010), the Abbott RealTime High Risk HPV assay (Carozzi et al, 2011, Poljak et al, 2011) and Roche Cobas/Linear Array (Castle et al, 2011, Gage et al, 2012, Wright et al, 2012).
Our estimates of sensitivity refer only to the ‘relative sensitivity' in women whose cytology results indicated a need for referral. In most cases this was mild dyskaryosis or worse, so that any disease in women with negative or borderline cytologic changes (regardless of their HPV status) would not be detected. Only in the case where different HPV tests had different sensitivities in women testing negative (or borderline) for cytology, which was not reflected in those with positive cytology result, could this distort the relative performance in terms of sensitivity.
As the vast majority of cytologically negative (or borderline) smears will not be associated with high-grade disease pathologically, the effect on not having full ascertainment in this group will only have a small impact on specificity. Again, unless the detection of the disease differs substantially by test (which is unlikely given the high sensitivity of all tests when cytology was positive) this will have a very small effect on the ‘relative' specificities of the different tests.
The study confirms the relative ordering of the specificities for these six tests seen previously in a referral population (Szarewski et al, 2008, 2012), but now also in a screening context. In this study, more than 75% of the women were aged 30 or above (Supplementary Appendix Table A1), a group in which HPV testing is currently recommended by many groups. The use of HPV testing in younger women in currently debated and our results are relevant to both groups. In addition, all of the tests except the NorChip PreTect HPV-Proofer showed a very high sensitivity for CIN2+ in the women referred for colposcopy due to abnormal cytology. Due to the design of the study, we were not able to assess sensitivity in those with negative cytology or borderline changes. Ideally, outcomes would be evaluated at the next screening round for those women testing HPV positive.
The NorChip test showed lower sensitivity and because of this it is more suited to be a triage test to reduce the referral rate than as a primary screening test, where those who are negative would not be subject to short-term follow-up.
Of the highly sensitive tests, the Gen-Probe APTIMA assay was the most specific, with ∼5% fewer ‘false positives'. On a relative basis, this comes to about 15% fewer false positives than seen for the other highly sensitive DNA tests. Again, specificity has to be regarded as ‘relative', as some of those HPV-positive women may well have harboured a high-grade lesion, which was not detected due to non-referral of women with negative or borderline cytology. However, the impact of this on specificity is likely to be small, and the impact on ‘relative specificity' is even smaller, as most of these cases are likely to be positive on all the sensitive HPV tests.
Meijer et al (2009) have provided criteria for validating new HPV tests. Their work focussed on using a single population, while our approach has been to establish sensitivity primarily in a referral population and specificity primarily in a screening population. In the referral population (Szarewski et al, 2012) we have established that four tests (Roche Cobas, Gen-Probe APTIMA, Abbot RealTime High Risk HPV assay and BD HPV assay) achieved the required sensitivity and ‘specificity' compared with Hybrid Capture 2. Here we report further evidence that specificity is achieved in a screening population, and sensitivity was virtually 100% in all cases for these four tests. A specificity that was 98% of that of Hybrid Capture 2 in this setting (thus meeting the guidelines) would require a specificity of 87.0% or higher, which all tests satisfy. However, we acknowledge that there were not enough cases of CIN2+ in the screening population to validate that the lower 95% CI for sensitivity was greater than 95% of the sensitivity achieved for Hybrid Capture 2 in this study—97.5%, (see Table 4).
Reproducibility of these tests has previously been established in a range of other studies (Carozzi et al, 2005, Dockter et al, 2009, LeBar et al, 2009, Carozzi et al, 2011, Heideman et al, 2011, Poljak et al, 2012) but was not formally performed in this study.
Conclusions
In this evaluation of six HPV tests from residual liquid-based screening cytology specimens, all tests except for NorChip showed high sensitivity for high-grade lesions that were positive by cytology, suggesting that they are suitable for primary screening and that dual co-testing with cytology as well is unnecessary. Positivity rates in cytology-negative specimens were similar for the DNA-based tests, but were lower for the APTIMA test, suggesting it can maintain the high sensitivity of the DNA tests, but with a better specificity, so that fewer women would need triage tests or short-term follow-up. However, a long-term low-risk period after a negative test has yet to be demonstrated for APTIMA or any RNA-based test, as has been shown for some of the DNA-based tests, especially Hybrid Capture 2 (Dillner et al, 2008, Cuzick et al, 2008b, Mesher et al, 2010, Rijkaart et al, 2012). Direct demonstration of this is desirable to support its use in primary screening, The NorChip test had lower sensitivity but higher specificity, suggesting its role may be more in triage than primary screening.
Acknowledgments
We thank the staff of the cytopathology department at the St. Mary's hospital for their help and co-operation, in particular Rezzeline Mendiola, Deputy Laboratory Manager. We also thank Martha Bicket and Josephine Salverda for their assistance with this study. This study was supported by Cancer Research UK Programme Grants (C569/A10404, and C8162/A10406), and supplemented with financial contributions and/or assay kits from each of the manufacturers whose tests were involved (Qiagen, Gen-Probe Incorporated, Abbott Molecular, BD Diagnostics, NorChip and Roche Diagnostics).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Carozzi FM, Burroni E, Bisanzi S, Puliti D, Confortini M, Giorgi Rossi P, Sani C, Scalisi A, Chini F. Comparison of clinical performance of Abbott RealTime High Risk HPV test with that of hybrid capture 2 assay in a screening setting. J Clin Microbiol. 2011;49 (4:1446–1451. doi: 10.1128/JCM.02311-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carozzi FM, Del Mistro A, Confortini M, Sani C, Puliti D, Trevisan R, De Marco L, Tos AG, Girlando S, Palma PD, Pellegrini A, Schiboni ML, Crucitti P, Pierotti P, Vignato A, Ronco G. Reproducibility of HPV DNA testing by hybrid capture 2 in a screening setting. Am J Clin Pathol. 2005;124 (5:716–721. doi: 10.1309/84E5-WHJQ-HK83-BGQD. [DOI] [PubMed] [Google Scholar]
- Castle PE, Stoler MH, Wright TC, Sharma A, Wright TL, Behrens CM. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol. 2011;12 (9:880–890. doi: 10.1016/S1470-2045(11)70188-7. [DOI] [PubMed] [Google Scholar]
- Cuzick J, Clavel C, Petry KU, Meijer CJ, Hoyer H, Ratnam S, Szarewski A, Birembaut P, Kulasingam S, Sasieni P, Iftner T. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int J Cancer. 2006;119:1095–1101. doi: 10.1002/ijc.21955. [DOI] [PubMed] [Google Scholar]
- Cuzick J, Szarewski A, Mesher D, Cadman L, Austin J, Perryman K, Ho L, Terry G, Sasieni P, Dina R, Soutter WP. Long-term follow-up of cervical abnormalities among women screened by HPV testing and cytology-results from the Hammersmith study. Int J Cancer. 2008b;122 (10:2294–2300. doi: 10.1002/ijc.23339. [DOI] [PubMed] [Google Scholar]
- Cuzick J, Arbyn M, Sankaranarayanan R, Tsu V, Ronco G, Mayrand MH, Dillner J, Meijer CJ. Overview of human papillomavirus based and other novel options for cervical cancer screening in developed and developing countries. Vaccine. 2008a;26 (Suppl 10:K29–K41. doi: 10.1016/j.vaccine.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Dillner J, Rebolj M, Birembaut P, Petry U, Szarewski A, Munk C, de Sanjose S, Naucler P, Lloveras B, Kjaer S, Cuzick J, van Ballegooijen M, Clavel C, Iftner T. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ. 2008;337:a1754. doi: 10.1136/bmj.a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockter J, Schroder A, Eaton B, Wang A, Sikhamsay N, Morales L, Giachetti C. Analytical characterization of the APTIMA HPV assay. J Clin Virol. 2009;45 (Suppl 1:S39–S47. doi: 10.1016/S1386-6532(09)70007-1. [DOI] [PubMed] [Google Scholar]
- Gage JC, Sadorra M, Lamere BJ, Kail R, Aldrich C, Kinney W, Fetterman B, Lorey T, Schiffman M, Castle PE, PaP Cohort Study Group et al. Comparison of the cobas human papillomavirus (HPV) test with the hybrid capture 2 and linear array HPV DNA tests. J Clin Microbiol. 2012;50 (1:61–65. doi: 10.1128/JCM.05989-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideman DA, Hesselink AT, Berkhof J, van Kemenade F, Melchers WJ, Daalmeijer NF, Verkuijten M, Meijer CJ, Snijders PJ. Clinical validation of the cobas 4800 HPV test for cervical screening purposes. J Clin Microbiol. 2011;49 (11:3983–3985. doi: 10.1128/JCM.05552-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S, Salva N, Nater T, Dockter J.2010Preliminary evaluation of the APTIMA HPV 16 18/45 genotype assay for the detection of human papillomavirus types 16, 18 and 45 in liquid Pap specimensPoster 252 IPV Montreal (poster)
- LeBar W, Taylor S, Hook EW.2009Reproducibility of the BD Viper System with XTR Technology (Extracted Mode) with PreservCyt Liquid Based Cytology MediumPresented at 25th Clinical Virology Symposium: Daytona Beach, FL
- Meijer CJ, Berkhof J, Castle PE, Hesselink AT, Franco EL, Ronco G, Arbyn M, Bosch FX, Cuzick J, Dillner J, Heideman DA, Snijders PJ. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int J Cancer. 2009;124 (3:516–520. doi: 10.1002/ijc.24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesher D, Szarewski A, Cadman L, Cubie H, Kitchener H, Luesley D, Menon U, Hulman G, Desai M, Ho L, Terry G, Williams A, Sasieni P, Cuzick J. Long-term follow-up of cervical disease in women screened by cytology and HPV testing: results from the HART study. Br J Cancer. 2010;102 (9:1405–1410. doi: 10.1038/sj.bjc.6605619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molden T, Kraus I, Skomedal H, Nordstrom T, Karlsen F. PreTect HPV-Proofer: real-time detection and typing of E6/E7 mRNA from carcinogenic human papillomaviruses. J Virol Methods. 2007;142 (1–2:204–212. doi: 10.1016/j.jviromet.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Monsonego J, Hudgens MG, Zerat L, Zerat JC, Syrjänen K, Halfon P, Ruiz F, Smith JS. Evaluation of oncogenic human papillomavirus RNA and DNA tests with liquid-based cytology in primary cervical cancer screening: the FASE study. Int J Cancer. 2011;129 (3:691–701. doi: 10.1002/ijc.25726. [DOI] [PubMed] [Google Scholar]
- Poljak M, Cuzick J, Kocjan BJ, Iftner T, Dillner J, Arbyn M. Nucleic Acid tests for the detection of alpha human papillomaviruses. Vaccine. 2012;30 (Suppl 5:F100–F106. doi: 10.1016/j.vaccine.2012.04.105. [DOI] [PubMed] [Google Scholar]
- Poljak M, Ostrbenk A, Seme K, Ucakar V, Hillemanns P, Vrtacnik Bokal E, Jancar N, Klavs I. Comparison of clinical and analytical performance of the Abbott RealTime High Risk HPV test and Hybrid Capture 2 in population-based cervical cancer screening. J Clin Microbiol. 2011;49 (5:1721–1729. doi: 10.1128/JCM.00012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkaart DC, Berkhof J, Rozendaal L, van Kemenade FJ, Bulkmans NW, Heideman DA, Kenter GG, Cuzick J, Snijders PJ, Meijer CJ. Human papillomavirus testing for the detection of high-grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol. 2012;13 (1:78–88. doi: 10.1016/S1470-2045(11)70296-0. [DOI] [PubMed] [Google Scholar]
- Ronco G, Giorgi-Rossi P, Carozzi F, Confortini M, Dalla Palma P, Del Mistro A, Ghiringhello B, Girlando S, Gillio-Tos A, De Marco L, Naldoni C, Pierotti P, Rizzolo R, Schincaglia R, Zorzi P, ZappaM M, Segnan N, Cuzick J. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol. 2010;11:249–257. doi: 10.1016/S1470-2045(09)70360-2. [DOI] [PubMed] [Google Scholar]
- Szarewski A, Ambroisine L, Cadman L, Austin J, Ho L, Terry G, Liddle S, Dina R, McCarthy J, Buckley H, Bergeron C, Soutter WP, Lyons D, Cuzick J. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol Biomarkers Prev. 2008;11:3033–3042. doi: 10.1158/1055-9965.EPI-08-0508. [DOI] [PubMed] [Google Scholar]
- Szarewski A, Mesher D, Cadman L, Austin J, Ashdown-Barr L, Ho L, Terry G, Liddle S, Young M, Stoler M, McCarthy J, Wright C, Bergeron C, Soutter WP, Lyons D, Cuzick J. The Predictors 2 Study: comparison of seven tests for high grade cervical intraepithelial neoplasia in women with abnormal smears: the Predictors 2 Study. J Clin Microbiol. 2012;50 (6:1867–1873. doi: 10.1128/JCM.00181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TC, Stoler MH, Behrens CM, Apple R, Derion T, Wright TL. The ATHENA human papillomavirus study: design, methods, and baseline results. Am J Obstet Gynecol. 2012;206 (1:46–e1-46.e11. doi: 10.1016/j.ajog.2011.07.024. [DOI] [PubMed] [Google Scholar]
- Wu R, Belinson SE, Du H, Na W, Qu X, Wu R, Liu Y, Wang C, Zhou Y, Zhang L, Belinson JL. Human papillomavirus messenger RNA assay for cervical cancer screening: the Shenzhen Cervical Cancer Screening Trial I. Int J Gynecol Cancer. 2010;20 (8:1411–1414. doi: 10.1111/IGC.0b013e3181f29547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.