Abstract
Background:
Multiparameter flow cytometry is a robust and reliable method for determining tumour DNA content applicable to formalin-fixed paraffin-embedded (FFPE) tissue. This study examined the clinical and pathological associations of DNA content in primary breast cancer using an improved multiparametric technique.
Methods:
The FFPE tissue from 201 primary breast cancers was examined and the cancers categorised according to their DNA content using multiparametric flow cytometry incorporating differential labelling of stromal and tumour cell populations. Mathematical modelling software (ModFit 3.2.1) was used to calculate the DNA index (DI) and percentage S-phase fraction (SPF%) for each tumour. Independent associations with clinical and pathological parameters were sought using backward stepwise Binary Logistic Regression (BLR) and Cox's Regression (CR) analysis.
Results:
Tumours were grouped into four categories based on the DI of the tumour cell population. Low DI tumours (DI=0.76–1.14) associated with progesterone receptor-positive status (P=0.012, BLR), intermediate DI (DI=1.18–1.79) associated with p53 mutant tumours (P=0.001, BLR), high DI (DI⩾1.80) tumours with human epidermal growth factor receptor 2 (HER2)-positive status (P=0.004, BLR) and ‘multiploid tumours' (two or more tumour DNA peaks) did not show any significant associations. Tumours with high SPF% (⩾10%) independently associated with poor overall survival (P=0.027, CR).
Conclusion:
Multiparametric flow analysis of FFPE tissue can accurately assess tumour DNA content. Tumour sub-populations associated with biomarkers of prognosis or likely response to therapy. The alterations in DNA content present the potential for greater understanding of the mechanisms underlying clinically significant biomarker changes in primary breast cancer.
Keywords: breast cancer, multiparametric flow cytometry, DNA content analysis, p53 mutant, HER2
Chromosomal instability (CIN) is a hallmark of cancer, and for the common epithelial tumours complex and clonal karyotypic evolution is the predominant driver of clinical disease. Breast carcinomas show gain and/or loss of either whole chromosomes (aneuploidy) or segments of chromosomes (segmental aneuploidy), which frequently results in abnormal DNA content (Fridlyand et al, 2006). Flow cytometry provides a reliable estimate of the DNA content of tumour populations and provides a global assessment of genetic instability. Flow cytometric and cytogenetic evidence suggests that most common epithelial cancers have a chromosomal constitution in the near diploid or sub-tetraploid range (Cornelisse et al, 1987; Olaharski et al, 2006; Weaver et al, 2006). It is hypothesised that tumours proceed from near diploid to high structural and numerical chromosomal aneuploidy by a process of endoreduplication. This is followed by further non-random gain and/or loss of specific chromosomes or chromosomal regions resulting in an intermediate DNA index (DI) with enhanced malignant potential (Fujiwara et al, 2005; Ganem et al, 2007).
In breast cancer, previous flow cytometry studies have correlated DNA content with tumour grade (Hitchcock et al, 1989), p53 mutation status (Eissa et al, 1997; Friedrich et al, 1997), disease recurrence/relapse (Ewers et al, 1984; Stal et al, 1994a), and survival (Shiao et al, 1997; Utada et al, 1998), suggesting a therapeutic and prognostic clinical relevance. Tumour DNA content, in conjunction with S-phase fraction (SPF), provides a reliable measure of breast cancer aggression (Ferno et al, 1992; Bagwell et al, 2001b). Using Cox's proportional hazard modelling, low DI, node negative, breast cancers were associated with good prognosis (Bagwell et al, 2001a). However, the technical limitations of the single parametric flow cytometry technique, including the inability to differentiate between DNA diploid tumour and normal cell populations (Hedley et al, 1983), does not allow accurate sub-grouping of breast tumours, thus limiting further statistical analysis. Additionally, previous attempts to improve DNA content analysis of formalin-fixed paraffin-embedded (FFPE) samples using the single parameter technique by introducing an external control as a reference were unsuccessful, in particular for the detection of tumours with a DI close to 1.0 (Schutte et al, 1985; Darzynkiewicz, 2010). Further, high DI tumours in the DNA tetraploid range with <25% tumour cells could not be differentiated from low DI tumours with an increased G2/M phase (Bauer, 1993). These limitations of single parametric flow have made DNA content analysis controversial and prevented the technique from achieving clinical utility (Ross et al, 2003).
Dual parameter DNA content flow cytometry of FFPE tissues allows detection of epithelial cells by anti-keratin antibodies (Nylander et al, 1994; Leers et al, 2000). This was further improved by Corver et al (2005), who used anti-keratin and anti-vimentin antibodies to allow specific identification of the tumour epithelial cells along with diploid stromal cells used as an internal reference, thus enabling accurate grouping of tumours based on their DI. This also enabled identification of DNA aneuploid tumour populations with a DI close to 1.0. It has been previously suggested that carcinoma-associated fibroblasts can be genetically abnormal and can have an abnormal DNA content; however, more recent studies have shown that, whatever the gene expression features of carcinoma-associated fibroblasts, the chromosome complement of stromal cells is diploid (Allinen et al, 2004; Hosein et al, 2010; Corver et al, 2011).
Additionally, the multiparametric technique facilitated the identification of high DI tumour (epithelial) cells overlapping the G2/M phase of the diploid stromal cells allowing characterisation of these tumours. These advantages of the multiparametric technique make it a robust platform for DNA content analysis of FFPE tissues and give it the potential to be a clinically useful tool. The aims of this study were to confirm the application of multiparameter flow cytometry on FFPE breast cancer specimens and to examine the clinical and biomarker associations of multiparametric flow analysis in primary breast cancer.
Materials and methods
Patients and tissues
The FFPE tissue blocks from 201 primary, previously untreated operable breast cancers were selected at random from patients diagnosed between 1997 and 2002. The mean age of the patients was 61 years (age range 28–88 years). All patients provided consent; the project received ethical permission from Tayside Tissue Bank under delegated authority from the Local Tayside Research Ethics Committee.
Blocks were reviewed to confirm the presence of >40% tumour cells. Tumours were graded according to the NHS guidelines for pathology reporting of breast disease (Bloom and Richardson, 1957) by a specialist breast pathologist (CAP or LBJ) (Table 1). Human epidermal growth factor receptor 2 (HER2) status was determined using immunohistochemistry (IHC) with confirmation of HER2-positive tumours by fluorescent in situ hybridisation (FISH) (Hsi and Tubbs, 2004; Purdie et al, 2010). The Quickscore (Detre et al, 1995) method was employed to determine oestrogen receptor (ER) and progesterone receptor (PgR) status. P53 mutation status was obtained using the p53 Amplichip test (Roche, Pleasanton, CA, USA) that detects single base pair substitutions and deletions (Baker et al, 2010). Breast cancer-specific overall survival, disease-free survival and recurrence were recorded with a minimum of 5 years follow-up for patients included in the study.
Table 1. Summary of the clinico-histopathological variables for 201 breast cancers studied using flow cytometry for tumour DNA content.
| |
N |
% |
|---|---|---|
| All | 201 | |
|
DNA content | ||
| Low | 79 | 39.3 |
| Intermediate | 42 | 20.8 |
| High | 23 | 11.4 |
| Mixed |
57 |
28.3 |
|
Tumour grade | ||
| 1 | 23 | 11.4 |
| 2 | 79 | 39.3 |
| 3 | 94 | 46.8 |
| Missing |
5 |
2.5 |
|
ER status | ||
| Negative | 48 | 23.9 |
| Positive | 152 | 75.6 |
| Missing |
1 |
0.5 |
|
PgR status | ||
| Negative | 89 | 44.3 |
| Positive | 111 | 55.2 |
| Missing |
1 |
0.5 |
|
HER2 status | ||
| Negative | 170 | 84.6 |
| Positive | 28 | 13.9 |
| Missing |
3 |
1.5 |
|
p53
status | ||
| Wild type | 146 | 72.6 |
| Mutant |
55 |
27.4 |
|
Survival status | ||
| Alive | 163 | 81.1 |
| Dead |
38 |
18.9 |
|
S-phase fraction | ||
| Low | 105 | 52.2 |
| Medium | 41 | 20.4 |
| High | 55 | 27.4 |
Abbreviations: ER, oestrogen receptor; PgR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Tissue processing and staining for flow cytometry
Tissue sections were processed as described by Corver et al (2005). Briefly, 3 × 60 μm thick sections from each block were de-waxed using xylene and rehydrated with decreasing concentrations of alcohol before antigen retrieval using 10 mℳ citric acid buffer (pH 6) for 1 h at 80 °C. Tissue was dissociated with a solution of 0.1% collagenase (Sigma-Aldrich, Dorset, UK) and dispase (Gibco; Invitrogen, Paisley, UK) for 1 h. One million cells were incubated overnight with anti-cytokeratin (2 μg ml−1 MNF116, DAKO (Glostrup, Denmark) and 5 μg ml−1 AE1AE3, Chemicon International (Temecula, CA, USA) MAB3412) and anti-vimentin (diluted 1:5 V9-2b, Antibodies for Research Applications BV, Gouda, The Netherlands; http://www.abforresearch.nl/) primary antibodies. Cells were washed with PBA (500 ml PBS/5% BSA/0.05% Tween-20) and treated with fluorescently conjugated secondary antibodies, FITC-IgG1 (Southern Biotech, Birmingham, AL, USA) for cytokeratin detection and PE-IgG2b (Southern Biotech) for vimentin detection for 30 min. Finally, the cells were resuspended in a solution of 10 μℳ propidium iodide (PI) (Sigma-Aldrich) and 0.1% RNase (Sigma-Aldrich) for 2–3 h or overnight before analysis.
Flow cytometry and analysis
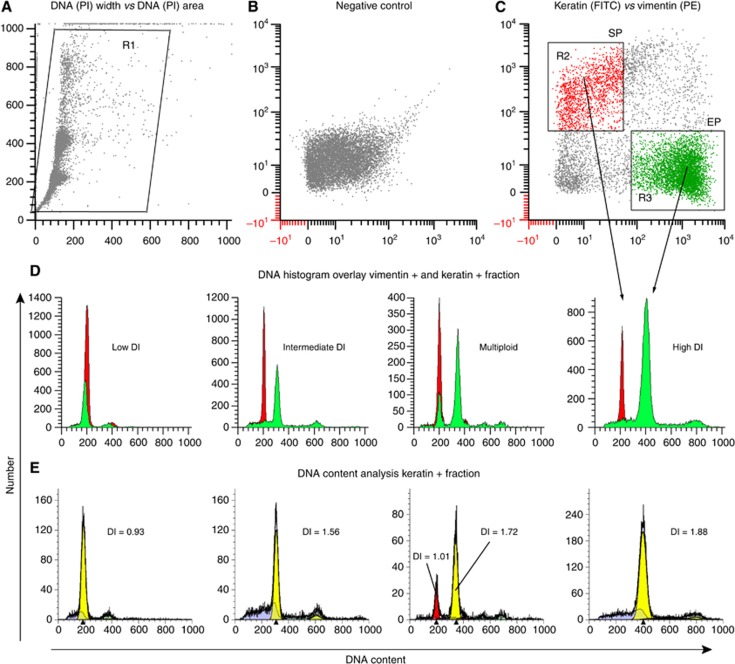
Cell suspensions were analysed using a FACScan (BD Biosciences, San Jose, CA, USA) with CellQuest software (version 3; BD Biosciences). DNA content analysis was performed by acquiring 50 000 events from each sample and simultaneously measuring PI, FITC-labelled epithelial, and PE-labelled stromal cell populations (Corver et al, 1996; Figure 1C). The keratin-positive tumour epithelial cell population detected using flow cytometry ranged from 6% to 90% of the total events analysed. Doublet discrimination was applied to remove cell clumps and enable the analysis of single cell events by using PI area vs PI width signal and noise/debris in front of the first G1 peak was also gated out (Figure 1A) during acquisition. The 530/30 band pass filter was used to detect FITC-labelled epithelial cells in the FL1 channel, PE-labelled stromal cells were detected using the 585/42 band pass filter in the FL2 channel and finally, PI was detected using the 650 long pass filter in the FL3 channel. According to the published guidelines for DNA content analysis from FFPE tissues, samples with a coefficient of variation (CV) of >8% should be excluded from data analysis to ensure robust data generation (Hedley, 1993). All breast cancers included in the present study yielded a CV ranging between 3.05% and 7.84% and the reproducibility of the technique confirmed with 15 randomly selected samples (data not shown).
Figure 1.
The four DNA content categories based on multi-parametric analysis. (A) Dot plot showing doublet discrimination to ensure analysis of a single cell population. (B) PI-positive cells as a negative control. (C) Dot plot showing keratin-positive epithelial cell population (EP), and vimentin-positive stromal cell population (SP). (D) DNA content histograms showing an example of a low DI, intermediate DI, high DI, and a multiploid tumour. The epithelial tumour cell population (green) is shown overlapping the normal stromal cell population (red) is shown. Histograms in (E) show the mathematical modelling analysis using the ModFit algorithm. The color reproduction of this figure is available at the British Journal of Cancer journal online.
The DI and percentage SPF (SPF%) were calculated using ModFit 3.2.1 and WinList 6.0 (Verity Software House, Topsham, ME, USA). The two software packages were remotely linked allowing ModFit to use the median of the diploid stromal cells to calculate the DI of the tumour (epithelial) cell population (mean of G0/G1 tumour epithelial peak/mean of G0/G1 reference stromal peak) (Hedley, 1993; Dayal et al., 2008; Corver and ter Haar, 2011). A data file contained all events except those at the lower and higher ends of the linear scale (Figure 1A), to allow the ModFit algorithm to correct for debris, clumps, and aggregates. ModFit analysis also included a gate to select all keratin-positive events in the FSC vs keratin dot plot, thus including doublets, noise and debris, allowing ModFit to handle data correction without manual interference.
Statistical analysis
Flow cytometry data were compared with clinical and histopathological parameters including histological tumour grade, p53 mutation status, HER2 status, ER status, and PgR status. An in-house data analysis tool (INSPIRE), which includes the two tail Fisher's exact test (FET) was used for preliminary statistical analysis (Quinlan et al., 2008). Disease recurrence, disease-free survival, and overall survival were studied using the Kaplan–Meier (KM) log-rank test. Significant results were further analysed to confirm independent associations using the backward stepwise Binary Logistic Regression (BLR) and Cox's Regression (CR) statistical analysis. The strength of the associations was assessed by calculating the β coefficient. The P-values for each association were used to discriminate independent associations between the variables analysed. For all analyses, the null hypothesis was rejected at α level of 5% (P<0.05).
Results
Categorising primary breast tumours
In all, 201 primary breast cancers were grouped into four categories based on the DI of the tumour cell population (Figures 1D and E). Tumours with a DI between 0.76 and1.14 were observed to be in the ‘low DI' range (n=79), ‘intermediate DI' tumours (n=42) had a DI ranging from 1.18 to 1.79, whereas ‘high DI' tumours (n=23) had a DI of ⩾1.80, tumours with two or more peaks from one or more of the above three categories were classed as ‘multiploid' tumours (n=57). DNA index was consistent and no variations were observed when DNA content analysis was repeated for 15 randomly selected tumours ensuring the reliability of the data generated and DI cutoffs were carefully determined based on published recommended standards (Hedley, 1993; Shankey et al, 1993). Figure 2 summarises the DI for all 201 samples used in this study
Figure 2.
A bimodal DNA index distribution curve of 201 breast cancers. The number of samples is represented on the y axis and the DNA index is indicated on the x axis.
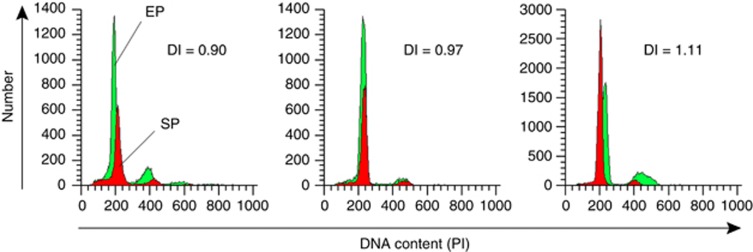
Based on the technical precision of multiparameter flow analysis (Corver et al, 2005), tumours in the low DI range could further be categorised into three groups, (i) group 1 (n=26), with 0.76⩾DI⩽0.95, (ii) group 2 (n=25), with 0.96⩾DI⩽1.05, and (iii) group 3 (n=28), with 1.06⩾DI⩽1.14 (Figure 3).
Figure 3.
Low DI sub-categories. Histograms showing a group 1 low DI tumour with overlapping stromal (SP) and epithelial (EP) cell population and DI=0.90, group 2 low DI=0.97, and group 3 low DI tumour with DI=1.11.
The SPF% was also obtained for each tumour using ModFit analysis. Tumours were separated into three categories based on SPF% as described previously (Stal et al, 1994b), low SPF% <5% (n=105), medium SPF%=5–10% (n=41), and high SPF% ⩾10% (n=55) tumours.
Clinical associations
Univariate statistical analysis
Low DI tumours showed significant associations with good prognostic features, including HER2-negative status (P=0.03, FET), ER-positive status (P=0.003, FET), PgR-positive status (P=0.013, FET), p53 wild type (P=0.03, FET), tumour grade 1–2 (P<0.0001, FET), disease-free survival (P=0.0027, KM), and overall survival (P=0.0023, KM). Further, analysis of the low DI cases (into subgroups 1, 2, and 3) indicated that group 2 low DI tumours associated with better prognostic markers when compared with the group 1 low DI tumours. The group 1 low DI tumour category (n=26) associated with poor prognostic features such as poor Nottingham prognostic index (NPI) (Galea et al, 1992) (P=0.03, FET) and tumour grade 3 (P=0.02, FET). Whereas, group 2 low DI tumours (n=25) associated with lymph node-negative status (P=0.02, FET). Group 3 tumours showed no significant associations.
Intermediate DI tumours showed an association with markers of poor prognosis, including ER-negative status (P=0.02, FET), p53 mutant status (P=0.0016, FET), tumour grade 3 (P=0.0014, FET), and disease recurrence (P=0.04, KM). High DI tumours were only associated with HER2-positive status (P=0.006, FET) and multiploid tumours showed no significant associations to any of the clinical parameters examined.
The above tumour categories were further assessed using clinical markers associated with the luminal equivalent (ER, PgR), HER2-positive and triple receptor-negative breast cancers (TNBC). Low DI tumours were significantly associated with clinical markers (ER+ve and/or PgR+ve) that are characteristic of the luminal type cancers (P=0.004, FET), whereas intermediate DI tumours associated with TNBC (P=0.007, FET). The molecular subtyping of these tumours based on microarray expression analysis was outside the scope of this study.
Tumours with high SPF (⩾10%) associated with ER-negative status (P=0.001, FET), PgR-negative status (P=0.001, FET), p53 mutant tumours and intermediate-high DI tumours (P=0.001, FET).
Multivariate statistical analysis
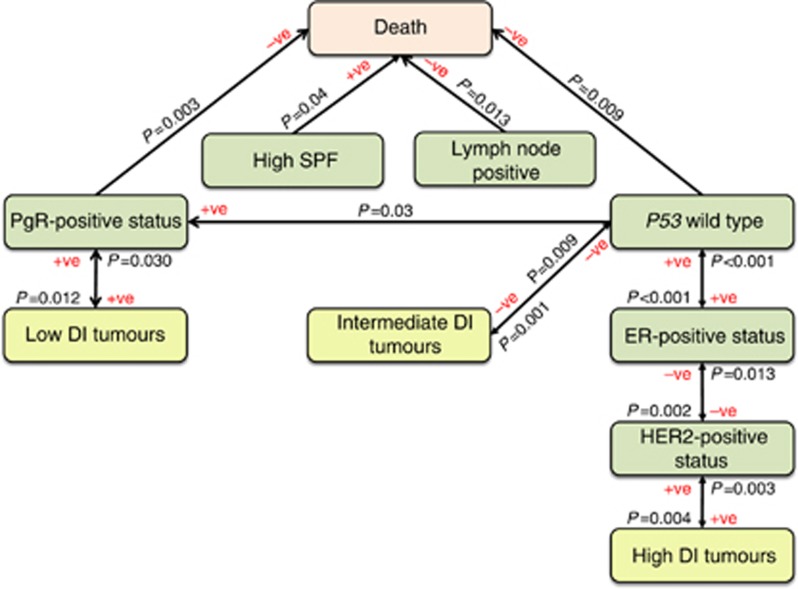
Significant associations obtained from univariate analysis were tested using CR (Table 2) and BLR analyses (Table 3) to determine independent associations. An independent association between tumour DNA content and overall survival was not confirmed by multivariate analysis (Table 3). However, low DI tumours independently associated with PgR-positive status (P=0.012, BLR), intermediate DI tumours associated with p53 mutant status (P=0.001, BLR), and high DI tumours independently associated with HER2-positive status (P=0.004, BLR) (Table 3; Figure 4). High SPF (⩾10%) independently associated (P=0.027, CR) with low patient survival (Table 2; Figure 4).
Table 2. Cox's regression (CR) analysis.
| Response variable | Predictor variables | Hazards ratio | 95% Low | 95% High | P-value |
|---|---|---|---|---|---|
| Death |
PgR positive |
0.32 |
0.149 |
0.686 |
0.003 |
| |
p53 mutant |
2.61 |
1.266 |
5.383 |
0.009 |
| |
Lymph node positive |
2.782 |
1.239 |
6.249 |
0.013 |
| High SPF (⩾10) | 2.218 | 1.094 | 4.499 | 0.027 |
Abbreviations: PgR=progesterone receptor; SPF=S-phase fraction.
All variables were analysed by backwards step-wise Cox's proportional hazards regression model to find independent associations with survival. Independent associations were confirmed with PgR-positive status, p53 mutation, positive lymph nodes, and high S-phase fraction (SPF3).
Table 3. Backward stepwise Binary Logistic Regression (BLR) statistical analysis.
| Response variable | Predictor variables | β | P-value | OR (95% CI) |
|---|---|---|---|---|
| PgR |
p53 wild type |
0.72 |
0.030 |
2.05 (1.07–3.93) |
| |
Low DI tumours |
−0.67 |
0.030 |
0.51 (0.28–0.94) |
|
p53 wild type |
ER |
1.69 |
<0.001 |
5.40 (2.62–11.13) |
| |
Intermediate DI tumours |
−1.02 |
0.009 |
0.36 (0.17–0.78) |
| ER |
p53 wild type |
1.68 |
<0.001 |
5.37 (2.38–12.14) |
| |
HER2 |
1.20 |
0.013 |
3.34 (1.29–8.63) |
| HER2 |
ER |
1.38 |
0.002 |
3.96 (1.65–9.46) |
| |
High DI tumours |
−1.55 |
0.003 |
0.21 (0.08–0.59) |
| Low DI tumours |
PgR |
−0.76 |
0.012 |
0.47 (0.26–0.84) |
| Intermediate DI tumours |
p53 wild type |
−1.17 |
0.001 |
0.31 (0.15–0.63) |
| High DI tumours | HER2 | −1.42 | 0.004 | 0.24 (0.09–0.64) |
Abbreviations: CI=confidence interval; DI=DNA index; ER=oestrogen receptor; HER2=human epidermal growth factor receptor 2; OR=odds ratio; PgR=progesterone receptor.
Variables were analysed by backwards step-wise BLR. All significant associations between the predictor and response variables are shown and non-significant associations are omitted for brevity and clarity.
Figure 4.
Independent associations of DNA content categories with prognostic markers. Independent associations between DNA content categories and molecular markers associated with breast cancer prognosis using BLR and CR analysis. The sense of association (negative or positive) and the corresponding level of confidence (P-value) are also indicated in the figure. The direction of the arrows indicates the direction of association found by BLR and CR. DI, DNA index; SPF%, S-phase fraction.
Discussion
This study has, for the first time, applied the multiparametric flow technique to FFPE tissue from 201 primary breast cancers. The technique allows assessment of tumour DNA content by differential staining of keratin expressing epithelial tumour cells (FITC labelled) from vimentin expressing normal stromal cells (PE labelled) (Corver et al, 2005). The ability of this technique to utilise routinely processed FFPE clinical material of mixed tumour and stromal tissues is beneficial compared with other methods of DNA analysis, such as deep sequencing, that requires a minimum of 50–70% tumour cells from frozen tissue and relies on other labour intensive techniques. Reassuringly, the DI values obtained in this study from FFPE sections show a similar distribution (Figure 2) to previously published data on fresh/frozen tissue material (Kimmig et al, 2001), underpinning the reliability of this multiparameter flow technique for DNA content analysis of FFPE tissues.
The association of low DI tumours with good prognostic indicators in breast cancer confirms previous findings (Cunningham et al, 1994; Spiethoff et al, 2000). Furthermore, reliable subgrouping of low DI FFPE tumours based on the DI, while not possible using the single parametric technique, demonstrated an association of group 2 low DI tumours with lymph node-negative status (P=0.02, FET), not previously reported using FFPE tissue. This suggests that the technical deficiencies of single parameter flow cytometry may have masked clinically relevant associations between lymph-node status and tumour DNA content (Beerman et al, 1990; Bergers et al, 1996; Spiethoff et al, 2000).
Tumours belonging to the intermediate DI, high DI, and multiploid categories (Figure 1D and E) could not be appropriately grouped in previous studies due to the technical limitations of the single parameter technique to differentiate between epithelial and stromal cell populations. As a result, multiploid tumours may have been incorrectly allocated to either the intermediate or high DI tumour groups. A reliable separation of intermediate DI and high DI tumours could only be achieved as exemplified here with the multiparametric flow method. The association of DNA content categories with different markers of prognosis (Figure 4) in the present study indicates that these tumours clinically behave differently. This highlights the clinical potential of multiparametric flow analysis of breast cancer to determine sub-groups of cancers within tumour categories previously identified by DNA content using single parametric flow analysis.
Biomarker associations
The PgR expression and tumour p53 mutation have been postulated as independent markers of prognosis in breast cancer (Overgaard et al, 2000; Liu et al, 2009) as confirmed here (Table 3; Figure 4). An association of low DI tumours with ER-positive and PgR-positive status has been previously reported (Leers and Nap, 2001); however, for the first time we demonstrate multivariate analysis showing an independent association of low DI tumours with PgR, but not ER, positive IHC.
Tumours with aneuploid DNA have been associated with poor prognostic features (Friedrich et al, 1997; Leers and Nap, 2001). On further interrogation these tumours revealed two distinct subgroups, (i) intermediate DI tumours, which associated with poor prognostic features including p53 mutation and triple negative status; and (ii) high DI tumours associated with HER2 positivity but not with any other markers of prognosis. Multivariate statistical analysis confirmed these associations suggesting the presence of distinct molecular categories within the aneuploid tumours of potential clinical significance.
High SPF% independently associated with poor survival using CR analysis. Previous studies have failed to identify SPF% as an independent prognostic marker using single parameter DNA content analysis in lymph node-positive breast cancer patients (Witzig et al, 1991, 1993) but have found an association with overall survival in lymph node-negative tumours (Cunningham et al, 1994) and, recently, in both early onset and advanced stage breast cancer (Vielh et al, 2005). In the present study, SPF% was not found to be significantly associated with lymph-node status; however, both lymph-node status and SPF% were independently associated with survival (Table 2; Figure 4). The SPF% data presented here are inherently different from previously reported studies as the tumour (epithelial) cell fraction SPF% has been specifically calculated, not the total SPF%. Interestingly, high SPF% also associated with intermediate and high DI tumours (P=0.001, FET), the tumour categories in this study associated with poor prognostic markers.
The identification of clinically distinct DNA content sub-groups in the present study suggests that a reliable DNA content analysis platform is required and achievable to investigate the complex biological mechanisms that drive cancer. The SPF% and DNA content analysis using multiparameter flow analysis associate significantly with prognosis and warrant further investigation. Moreover, an understanding of the basic biology responsible for tetraploidy and sub-tetraploidy in breast cancer is likely to have direct relevance to similar events in other common epithelial tumours.
Conclusion
Multiparameter DNA content analysis of FFPE primary breast cancers identifies categories of breast cancer, based on DI, associated with conventional clinical and pathological parameters of prognostic and therapeutic significance. Investigating the mechanisms involved in tumour cell DNA content may provide future therapeutic opportunities in breast cancer.
Ethical approval
Ethical approval was obtained from the Tayside Tissue Bank under delegated authority from the Tayside Local Research Ethics Committee for all patient samples included in this study.
Acknowledgments
We would like to thank the Tayside Tissue Bank facility for providing the tissue samples for the study. The clinical comparisons in the study would not be possible without the contributions of Miss Alison Ashfield and Miss Lea-Jane Birse-Stewart-Bell. This study was funded by MRC DHPA (Medical Research Council Dorothy Hodgkin Post Graduate Award), Anonymous Trust Grant, Ninewells Cancer Campaign Bear Necessities funding and Breast Cancer Research (Scotland).
Authors contribution
JHSD conducted all experiments in the laboratory and drafted the manuscript. MJS provided advice on intellectual content of the manuscript and data interpretation. CAP and LBJ were responsible for specialist advice on tissue material used for the study and for all of the pathological data. LB and PQ conducted statistical analysis. WEC and NTT provided training for multiparametric flow analysis in Leiden. WEC performed WinList/ModFit flow cytometric data analysis. AMT and NRP designed and supervised the project and critically revised the manuscript.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang HY, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6 (1:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Bagwell CB, Clark GM, Spyratos F, Chassevent A, Bendahl PO, Stal O, Killander D, Jourdan ML, Romain S, Hunsberger B, Baldetorp B. Optimizing flow cytometric DNA ploidy and S-phase fraction as independent prognostic markers for node-negative breast cancer specimens. Cytometry. 2001a;46 (3:121–135. doi: 10.1002/cyto.1097. [DOI] [PubMed] [Google Scholar]
- Bagwell CB, Clark GM, Spyratos F, Chassevent A, Bendahl PO, Stal O, Killander D, Jourdan ML, Romain S, Hunsberger B, Wright S, Baldetorp B.2001bDNA and cell cycle analysis as prognostic indicators in breast tumors revisited Clin Lab Med 21(4875–895.x [PubMed] [Google Scholar]
- Baker L, Quinlan PR, Patten N, Ashfield A, Birse-Stewart-Bell LJ, McCowan C, Bourdon JC, Purdie CA, Jordan LB, Dewar JA, Wu L, Thompson AM. p53 mutation, deprivation and poor prognosis in primary breast cancer. Br J Cancer. 2010;102 (4:719–726. doi: 10.1038/sj.bjc.6605540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer KD. Quality control issues in DNA content flow cytometry. Ann NY Acad Sci. 1993;677:59–77. doi: 10.1111/j.1749-6632.1993.tb38765.x. [DOI] [PubMed] [Google Scholar]
- Beerman H, Kluin PM, Hermans J, van de Velde CJ, Cornelisse CJ. Prognostic significance of DNA-ploidy in a series of 690 primary breast cancer patients. Int J Cancer. 1990;45 (1:34–39. doi: 10.1002/ijc.2910450108. [DOI] [PubMed] [Google Scholar]
- Bergers E, van Diest PJ, Baak JP. Cell cycle analysis of 932 flow cytometric DNA histograms of fresh frozen breast carcinoma material. Correlations between flow cytometric, clinical, and pathologic variables. MMMCP Collaborative Group. Multicenter Morphometric Mammary Carcinoma Project Collaborative Group. Cancer. 1996;77 (11:2258–2266. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2258::AID-CNCR12>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Bloom HJ, Richardson WW. Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer. 1957;11 (3:359–377. doi: 10.1038/bjc.1957.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelisse CJ, van de Velde CJ, Caspers RJ, Moolenaar AJ, Hermans J. DNA ploidy and survival in breast cancer patients. Cytometry. 1987;8 (2:225–234. doi: 10.1002/cyto.990080217. [DOI] [PubMed] [Google Scholar]
- Corver WE, Bonsing BA, Abeln EC, Vlak-Theil PM, Cornelisse CJ, Fleuren GJ. One-tube triple staining method for flow cytometric analysis of DNA ploidy and phenotypic heterogeneity of human solid tumors using single laser excitation. Cytometry. 1996;25 (4:358–366. doi: 10.1002/(SICI)1097-0320(19961201)25:4<358::AID-CYTO7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Corver WE, ter Haar NT.2011High-resolution multiparameter DNA flow cytometry for the detection and sorting of tumor and stromal subpopulations from paraffin-embedded tissues Curr Protoc CytomChapter 7: Unit 7 37 [DOI] [PubMed]
- Corver WE, Ter Haar NT, Dreef EJ, Miranda NF, Prins FA, Jordanova ES, Cornelisse CJ, Fleuren GJ. High-resolution multi-parameter DNA flow cytometry enables detection of tumour and stromal cell subpopulations in paraffin-embedded tissues. J Pathol. 2005;206 (2:233–241. doi: 10.1002/path.1765. [DOI] [PubMed] [Google Scholar]
- Corver WE, Ter Haar NT, Fleuren GJ, Oosting J. Cervical carcinoma-associated fibroblasts are DNA diploid and do not show evidence for somatic genetic alterations. Cell Oncol (Dordr) 2011;34 (6:553–563. doi: 10.1007/s13402-011-0061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Ingle JN, Jung SH, Cha SS, Wold LE, Farr G, Witzig TE, Krook JE, Wieand HS, Kovach JS. p53 gene expression in node-positive breast cancer: relationship to DNA ploidy and prognosis. J Natl Cancer Inst. 1994;86 (24:1871–1873. doi: 10.1093/jnci/86.24.1871. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z.2010Critical aspects in analysis of cellular DNA content Curr Protoc CytomChapter 7: Unit7 2 [DOI] [PMC free article] [PubMed]
- Dayal JHS, Sales M, Purdie C, Hislop RG, Thompson AM, Pratt NR.2008Flow analysis of aneuploidy and the chromosome instability phenotype J Med Genet vol 45(supplement 1) pp:s66British Human Genetics Conference [Google Scholar]
- Detre S, Saclani Jotti G, Dowsett M. A "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48 (9:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa S, Khalifa A, el-Gharib A, Salah N, Mohamed MK. Multivariate analysis of DNA ploidy, p53, c-erbB-2 proteins, EGFR, and steroid hormone receptors for prediction of poor short term prognosis in breast cancer. Anticancer Res. 1997;17 (2B:1417–1423. [PubMed] [Google Scholar]
- Ewers SB, Langstrom E, Baldetorp B, Killander D. Flow-cytometric DNA analysis in primary breast carcinomas and clinicopathological correlations. Cytometry. 1984;5 (4:408–419. doi: 10.1002/cyto.990050419. [DOI] [PubMed] [Google Scholar]
- Ferno M, Baldetorp B, Borg A, Olsson H, Sigurdsson H, Killander D. Flow cytometric DNA index and S-phase fraction in breast cancer in relation to other prognostic variables and to clinical outcome. Acta Oncol. 1992;31 (2:157–165. doi: 10.3109/02841869209088897. [DOI] [PubMed] [Google Scholar]
- Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, McLennan J, Ziegler J, Chin K, Devries S, Feiler H, Gray JW, Waldman F, Pinkel D, Albertson DG. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich K, Dimmer V, Haroske G, Meyer W, Theissig F, Kunze KD. Correlation between p53 status, DNA ploidy, proliferation rate and nuclear morphology in breast cancer. An image cytometric study. Anal Cell Pathol. 1997;15 (2:85–97. doi: 10.1155/1997/719876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437 (7061:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat. 1992;22 (3:207–219. doi: 10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17 (2:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Hedley DW. DNA Cytometry Consensus Conference. DNA flow cytometry and breast cancer. Breast Cancer Res Treat. 1993;28 (1:51–53. doi: 10.1007/BF00666356. [DOI] [PubMed] [Google Scholar]
- Hedley DW, Friedlander ML, Taylor IW, Rugg CA, Musgrove EA. Method for analysis of cellular DNA content of paraffin-embedded pathological material using flow cytometry. J Histochem Cytochem. 1983;31 (11:1333–1335. doi: 10.1177/31.11.6619538. [DOI] [PubMed] [Google Scholar]
- Hitchcock A, Ellis IO, Robertson JF, Gilmour A, Bell J, Elston CW, Blamey RW. An observation of DNA ploidy, histological grade, and immunoreactivity for tumour-related antigens in primary and metastatic breast carcinoma. J Pathol. 1989;159 (2:129–134. doi: 10.1002/path.1711590207. [DOI] [PubMed] [Google Scholar]
- Hosein AN, Wu M, Arcand SL, Lavallee S, Hebert J, Tonin PN, Basik M. Breast Carcinoma-Associated Fibroblasts Rarely Contain p53 Mutations or Chromosomal Aberrations. Cancer Res. 2010;70 (14:5770–5777. doi: 10.1158/0008-5472.CAN-10-0673. [DOI] [PubMed] [Google Scholar]
- Hsi ED, Tubbs RR. Guidelines for HER2 testing in the UK. J Clin Pathol. 2004;57 (3:241–242. doi: 10.1136/jcp.2003.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmig R, Wimberger P, Kapsner T, Hillemanns P. Flow cytometric DNA analysis using cytokeratin labeling for identification of tumor cells in carcinomas of the breast and the female genital tract. Anal Cell Pathol. 2001;22 (3:165–178. doi: 10.1155/2001/746827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leers MP, Nap M. Steroid receptor heterogeneity in relation to DNA index in breast cancer: a multiparameter flow cytometric approach on paraffin-embedded tumor samples. Breast J. 2001;7 (4:249–259. doi: 10.1046/j.1524-4741.2001.20108.x. [DOI] [PubMed] [Google Scholar]
- Leers MP, Schutte B, Theunissen PH, Ramaekers FC, Nap M. A novel flow cytometric steroid hormone receptor assay for paraffin-embedded breast carcinomas: an objective quantification of the steroid hormone receptors and direct correlation to ploidy status and proliferative capacity in a single-tube assay. Hum Pathol. 2000;31 (5:584–592. doi: 10.1053/hp.2000.6700. [DOI] [PubMed] [Google Scholar]
- Liu S, Chia SK, Mehl E, Leung S, Rajput A, Cheang MC, Nielsen TO. Progesterone receptor is a significant factor associated with clinical outcomes and effect of adjuvant tamoxifen therapy in breast cancer patients. Breast Cancer Res Treat. 2009;119 (1:53–61. doi: 10.1007/s10549-009-0318-0. [DOI] [PubMed] [Google Scholar]
- Nylander K, Stenling R, Gustafsson H, Roos G. Application of dual parameter analysis in flow cytometric DNA measurements of paraffin-embedded samples. J Oral Pathol Med. 1994;23 (4:190–192. doi: 10.1111/j.1600-0714.1994.tb01111.x. [DOI] [PubMed] [Google Scholar]
- Olaharski AJ, Sotelo R, Solorza-Luna G, Gonsebatt ME, Guzman P, Mohar A, Eastmond DA. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis. 2006;27 (2:337–343. doi: 10.1093/carcin/bgi218. [DOI] [PubMed] [Google Scholar]
- Overgaard J, Yilmaz M, Guldberg P, Hansen LL, Alsner J. TP53 mutation is an independent prognostic marker for poor outcome in both node-negative and node-positive breast cancer. Acta Oncol. 2000;39 (3:327–333. doi: 10.1080/028418600750013096. [DOI] [PubMed] [Google Scholar]
- Purdie CA, Jordan LB, McCullough JB, Edwards SL, Cunningham J, Walsh M, Grant A, Pratt N, Thompson AM. HER2 assessment on core biopsy specimens using monoclonal antibody CB11 accurately determines HER2 status in breast carcinoma. Histopathology. 2010;56 (6:702–707. doi: 10.1111/j.1365-2559.2010.03533.x. [DOI] [PubMed] [Google Scholar]
- Quinlan PR, Reed C, Baker L, Thompson AM.2008INSPIRE: An Integrated Agent Based System for Hypothesis Generation within Cancer Datasets WI-IAT vol. 3587–590.IEEE/WIC/ACM International Conference on Web Intelligence and Intelligent Agent Technology, 2008 [Google Scholar]
- Ross JS, Linette GP, Stec J, Ross MS, Anwar S, Boguniewicz A. DNA ploidy and cell cycle analysis in breast cancer. Am J Clin Pathol. 2003;120 (Suppl:S72–S84. doi: 10.1309/QD096UGF70T5H46G. [DOI] [PubMed] [Google Scholar]
- Schutte B, Reynders MM, Bosman FT, Blijham GH. Flow cytometric determination of DNA ploidy level in nuclei isolated from paraffin-embedded tissue. Cytometry. 1985;6 (1:26–30. doi: 10.1002/cyto.990060106. [DOI] [PubMed] [Google Scholar]
- Shankey TV, Rabinovitch PS, Bagwell B, Bauer KD, Duque RE, Hedley DW, Mayall BH, Wheeless L, Cox C. Guidelines for implementation of clinical DNA cytometry. International Society for Analytical Cytology. Cytometry. 1993;14 (5:472–477. doi: 10.1002/cyto.990140503. [DOI] [PubMed] [Google Scholar]
- Shiao YH, Chen VW, Lehmann HP, Wu XC, Correa P. Patterns of DNA ploidy and S-phase fraction associated with breast cancer survival in blacks and whites. Clin Cancer Res. 1997;3 (4:587–592. [PubMed] [Google Scholar]
- Spiethoff A, Schenck A, Bohrer M. Relationship of DNA ploidy to hormone receptor status and proliferation in invasive breast carcinoma. J Cancer Res Clin Oncol. 2000;126 (12:707–710. doi: 10.1007/s004320000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stal O, Carstensen JM, Wingren S, Rutqvist LE, Skoog L, Klintenberg C, Nordenskjold B. Relationship of DNA ploidy and S-phase fraction to survival after first recurrence of breast cancer. Acta Oncol. 1994a;33 (4:423–429. doi: 10.3109/02841869409098439. [DOI] [PubMed] [Google Scholar]
- Stal O, Skoog L, Rutqvist LE, Carstensen JM, Wingren S, Sullivan S, Andersson AC, Dufmats M, Nordenskjold B. S-phase fraction and survival benefit from adjuvant chemotherapy or radiotherapy of breast cancer. Br J Cancer. 1994b;70 (6:1258–1262. doi: 10.1038/bjc.1994.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utada Y, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G. [Relationship between DNA ploidy and survival in breast cancer] Gan To Kagaku Ryoho. 1998;25 (Suppl 3:431–435. [PubMed] [Google Scholar]
- Vielh P, Carton M, Padoy E, de Rycke Y, Klijanienko J, El-Naggar AK, Asselain B. S-phase fraction as an independent prognostic factor of long-term overall survival in patients with early-stage or locally advanced invasive breast carcinoma. Cancer. 2005;105 (6:476–482. doi: 10.1002/cncr.21348. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Cleveland DW.2006Cell biology: nondisjunction, aneuploidy and tetraploidy Nature 442(7104E9–E10.discussion E10 [DOI] [PubMed] [Google Scholar]
- Witzig TE, Gonchoroff NJ, Therneau T, Gilbertson DT, Wold LE, Grant C, Grande J, Katzmann JA, Ahmann DL, Ingle JN. DNA content flow cytometry as a prognostic factor for node-positive breast cancer. The role of multiparameter ploidy analysis and specimen sonication. Cancer. 1991;68 (8:1781–1788. doi: 10.1002/1097-0142(19911015)68:8<1781::aid-cncr2820680822>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Witzig TE, Ingle JN, Schaid DJ, Wold LE, Barlow JF, Gonchoroff NJ, Gerstner JB, Krook JE, Grant CS, Katzmann JA. DNA ploidy and percent S-phase as prognostic factors in node-positive breast cancer: results from patients enrolled in two prospective randomized trials. J Clin Oncol. 1993;11 (2:351–359. doi: 10.1200/JCO.1993.11.2.351. [DOI] [PubMed] [Google Scholar]