Abstract
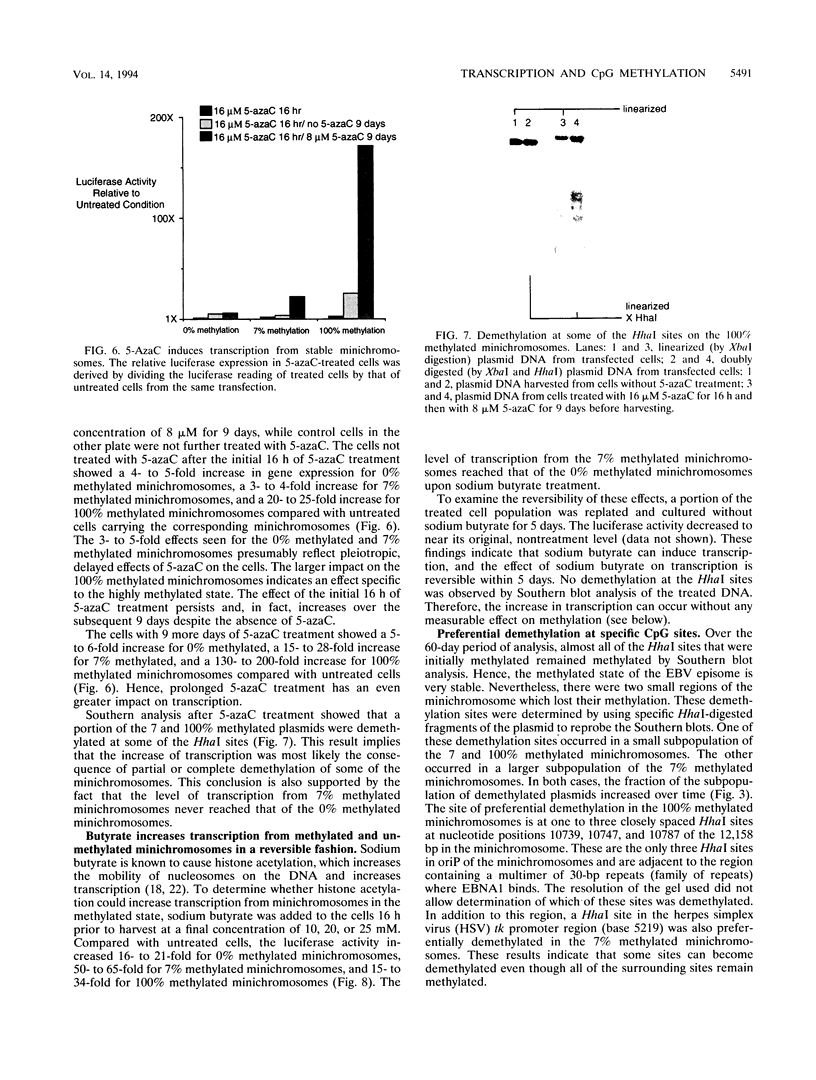
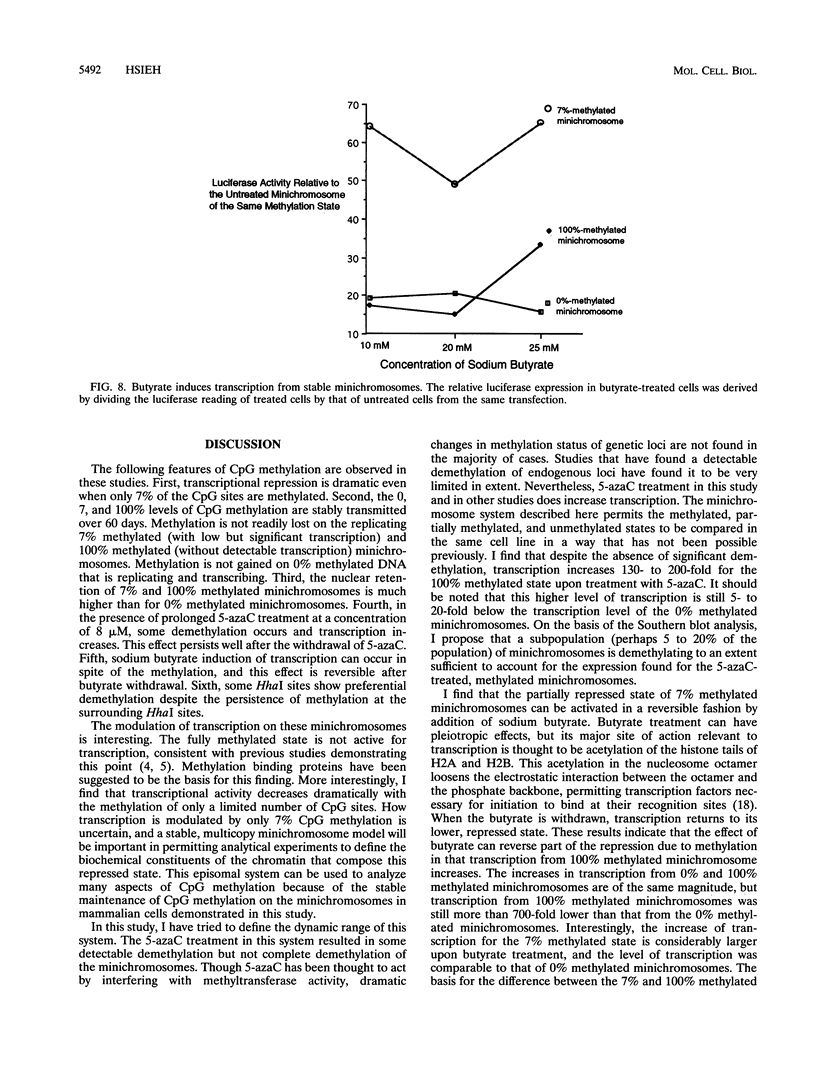
CpG methylation is known to suppress transcription. This repression is generally thought to be related to alterations of chromatin structure that are specified by the methylation. The nature of these chromatin alterations is unknown. Moreover, it has not been clear if the methylation repression occurs in an all-or-none fashion at some critical methylation density, or if intermediate densities of methylation can give intermediate levels of repression. Here I report a stable episomal system which recapitulates many dynamic features of methylation observed in the genome. I have determined the extent of transcriptional repression as a function of four densities of CpG methylation. I find that the repression is a graded but exponential function of the CpG methylation density such that low levels of methylation yield a 67 to 90% inhibition of gene expression. Higher levels of methylation extinguished gene expression completely. Transcription from methylated minichromosomes can be increased by butyrate treatment, suggesting that histone acetylation can reverse some of the repression specified by the methylated state. Sites of preferential demethylation occurred and may have resulted from transcription factor binding or DNA looping.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Antequera F., Macleod D., Bird A. P. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989 Aug 11;58(3):509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992 Jan;11(1):327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBridge R. B., Tang P., Hsia H. C., Leong P. M., Miller J. H., Calos M. P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987 Jan;7(1):379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Claeys K. M., Selker E. U. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993 Dec 10;262(5140):1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- Frappier L., Goldsmith K., Bendell L. Stabilization of the EBNA1 protein on the Epstein-Barr virus latent origin of DNA replication by a DNA looping mechanism. J Biol Chem. 1994 Jan 14;269(2):1057–1062. [PubMed] [Google Scholar]
- Hansen R. S., Gartler S. M. 5-Azacytidine-induced reactivation of the human X chromosome-linked PGK1 gene is associated with a large region of cytosine demethylation in the 5' CpG island. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4174–4178. doi: 10.1073/pnas.87.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hsieh C. L., Lieber M. R. CpG methylated minichromosomes become inaccessible for V(D)J recombination after undergoing replication. EMBO J. 1992 Jan;11(1):315–325. doi: 10.1002/j.1460-2075.1992.tb05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. L., McCloskey R. P., Lieber M. R. V(D)J recombination on minichromosomes is not affected by transcription. J Biol Chem. 1992 Aug 5;267(22):15613–15619. [PubMed] [Google Scholar]
- Jahroudi N., Foster R., Price-Haughey J., Beitel G., Gedamu L. Cell-type specific and differential regulation of the human metallothionein genes. Correlation with DNA methylation and chromatin structure. J Biol Chem. 1990 Apr 15;265(11):6506–6511. [PubMed] [Google Scholar]
- Jones C. H., Hayward S. D., Rawlins D. R. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J Virol. 1989 Jan;63(1):101–110. doi: 10.1128/jvi.63.1.101-110.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993 Jan 15;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B., Marsh K., Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985 Feb;5(2):410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen B., Bendixen C., Westergaard O. Histone hyperacetylation is accompanied by changes in DNA topology in vivo. Eur J Biochem. 1991 Oct 1;201(1):107–111. doi: 10.1111/j.1432-1033.1991.tb16262.x. [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R. A., Richards E. J. Arabidopsis thaliana DNA methylation mutants. Science. 1993 Jun 25;260(5116):1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Warren N., Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. 1985 Feb 28-Mar 6Nature. 313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987 Feb;7(2):725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]