Abstract
Background:
The epithelial cell adhesion molecule (EpCAM) is overexpressed on most carcinomas. Dependent on the tumour type, its overexpression is either associated with improved or worse patient survival. For ovarian cancer, however, the role of EpCAM remains unclear.
Methods:
Cell survival of ovarian cancer cell lines was studied after induction or repression of endogenous EpCAM expression using siRNA/cDNA or artificial transcription factors (ATF) consisting of engineered zinc-fingers fused to either a transcriptional activator or repressor domain.
Results:
Two ATFs were selected as the most potent down- and upregulator, showing at least a two-fold alteration of EpCAM protein expression compared with control. Downregulation of EpCAM expression resulted in growth inhibition in breast cancer, but showed no effect on cell growth in ovarian cancer. Induction or further upregulation of EpCAM expression decreased ovarian cancer cell survival.
Conclusion:
The bidirectional ATF-based approach is uniquely suited to study cell-type-specific biological effects of EpCAM expression. Using this approach, the oncogenic function of EpCAM in breast cancer was confirmed. Despite its value as a diagnostic marker and for immunotherapy, EpCAM does not seem to represent a therapeutic target for gene expression silencing in ovarian cancer.
Keywords: EpCAM, ATFs, ovarian cancer, zinc-fingers
The epithelial cell adhesion molecule (EpCAM; CD326) is a transmembrane glycoprotein, overexpressed on the vast majority of carcinomas compared with healthy epithelium (Went et al, 2004). Epithelial cell adhesion molecule is used as a diagnostic marker, has prognostic value for some tumours (Went et al, 2008) and even serves as a therapeutic target in antibody-based therapies (Baeuerle and Gires, 2007). Recently, EpCAM has been identified as a marker for cancer-initiating stem cells (Visvader and Lindeman, 2008) and was shown to be involved in the Wnt-signaling pathway (Maetzel et al, 2009). Upon intramembrane proteolysis of EpCAM, the intracellular domain functions with β-catenin as part of a transcriptional complex inducing c-myc and cyclin A/E expression. In breast cancer, high EpCAM expression is associated with poor prognosis, (Spizzo et al, 2004), which is consistent with the finding that induction of EpCAM in the EpCAM-negative breast cancer cell line Hs578T showed increased proliferation compared with the empty vector control (Gostner et al, 2011). In addition, RNAi-based silencing of EpCAM expression in breast cancer cell lines reduced the oncogenic potential of the treated cells (Osta et al, 2004) and EpCAM expression rescued by cDNA constructs increased cell invasion (Sankpal et al, 2011).
Despite this oncogenic function of EpCAM in breast carcinogenesis, for several tumour types the biological role of EpCAM is far from clear (van der Gun et al, 2010). Clinical observations in ovarian cancer suggest that for this tumour type EpCAM overexpression correlates with decreased overall survival, especially in patients with FIGO stage III/IV (Spizzo et al, 2006), although this could not be confirmed by independent studies (Kim et al, 2003; Heinzelmann-Schwarz et al, 2004). In addition, EpCAM has been described as a marker of human ovarian cancer stem/progenitor cells (Meirelles et al, 2012) and introduction of human EpCAM cDNA in EpCAM-negative mouse ovarian stem-like tumour cells enhanced the tumour-initiating ability (Motohara et al, 2011). Although these studies are far from definitive, they do suggest that downregulation of endogenous EpCAM expression might decrease the oncogenic potential in ovarian cancer.
To our knowledge, so far no data exist on EpCAM expression modulation in human ovarian cell culture systems in contrast to breast and many other tumour types. To investigate the functional role of EpCAM, a method to modulate bidirectionally endogenous expression levels would provide biologically relevant insights. To this end, artificial transcription factors (ATFs) can be constructed by fusing a gene-specific DNA-binding domain to transcriptional modulators (Uil et al, 2003). DNA-binding domains often used are engineered six zinc-fingers targeting an 18-base-pair unique sequence, thereby, enabling specific targeting of virtually any gene (Sera, 2009). Artificial transcription factors are appealing because they allow for both up- (Beltran and Blancafort, 2011) and downregulation (Stolzenburg et al, 2012) of gene expression, by fusing an activator or repressor to the zinc-fingers. Furthermore, compared with siRNA approaches where many mRNA copies have to be targeted, only two gene copies need to be targeted by ATFs. In addition, compared with gene delivery approaches, which often involve the expression of only one cDNA variant, ATFs can induce all splice variants in their natural ratios. Importantly, using this approach, epigenetic effector domains can be targeted to the gene of interest to induce mitotically stable gene expression modulation (De Groote et al, 2012). In this study, bidirectional endogenous EpCAM modulation by ATFs was exploited to investigate the biological role of EpCAM overexpression in ovarian cancer.
Materials and methods
Cell culture
HEK293T cells, and the cancer cell lines HeLa (cervix), MCF7, MDA-MB231, SKBR3 (breast) and OVCAR3, CaOV3, SKOV3 and A2780 (ovarian) were cultured in DMEM (BioWhittaker, Walkersville, MD, USA) with 50 μg ml−1 gentamicin sulphate, 2 mℳℒ-glutamine and 10% FBS.
ATF retroviral transduction
Epithelial cell adhesion molecule targeting zinc-finger proteins (ZFPs): one 3-finger (3F-ZFA) and two 6-fingers (referred to as ZFA and ZFB) (Gommans et al, 2007) were subcloned into the retroviral vector pMX-IRES-GFP (pMX) (Beltran and Blancafort, 2011) containing the activator VP64 (a tetrameric repeat of herpes simplex VP16) (Beltran and Blancafort, 2011) or the repressor SKD (Kruppel-associated box) (Stolzenburg et al, 2012). Retroviral particles were produced by co-transfection of pMX with the VSV envelope protein (pMD2.G) and the gag/pol proteins (pMDLg/pRRE) into HEK293T cells using CaPO4. After 2 and 3 days, the viral supernatant was used to transduce the host cell lines in the presence of 5 μg ml−1 polybrene (Sigma, St Louis, MO, USA). Transduction was optimized by, for example, diminishing toxicity to 293 producer cells (lowering the amount of plasmids in the transfection) and by varying the number of host cells seeded. Cells were harvested for analysis, 4 days after the first transduction. As the IRES-GFP present in the pMX vector showed a relative low GFP signal (Beerli et al, 2000), co-transduction with a strong GFP reporter was used to monitor toxicity by downregulation of EpCAM expression in SKBR3 cells.
siRNA or cDNA transfection
siRNA-EpCAM 750 ng (sense, 5′-GGAGAUCACAACGCGUUAUUU and antisense, 5′-AUAACGCGUUGUGAUCUCCUU) or irrelevant-siRNA (irr-siRNA) (1022076; Qiagen, Venlo, Netherlands) or plasmid DNA (generously provided by Giepmans, UMCG and Cirulli (Washington, DC, USA)) (Schnell et al, 2012) was complexed in 200 μl HBS with 20 μl SAINT-2:DOPE (SD; 0.75 mℳ) (Synvolux, Groningen, Netherlands) and pipetted onto the SKOV3 or A2780 cells.
Gene expression analysis
Q-RT-PCR was performed (ABIPrism 7900HT, Applied Biosystems, Nieuwekerk, Netherlands) for EpCAM (Hs00158980_m1, Applied Biosystems) and GAPDH (F, 5′-CCACATCGCTCAGACACCAT-3′ R, 5′-GCGCCCAATACGACCAAAT-3′ probe: 5′-CGTTGACTCCGACCTTCACCTTCCC-3′ (Eurogentec, Maastricht, Netherlands)) in triplicate. Results are shown as relative expression compared with GAPDH levels, using ΔCt method (pMX was set at 1). Epithelial cell adhesion molecule protein was detected by APC-CD326 (BioLegend, Uithoorn, Netherlands). The mean fluorescence intensity (pMX was set at 100%) was measured on a Calibur flow cytometer (Beckton Dickenson Biosciences, San Jose, CA, USA).
Cell growth assays
Transduction was carried out directly in 96-well plates and cell viability MTT assay (M2128; Sigma) was performed at days 4, 5 and 6 after transduction, or transductions were carried out in 6-well plates and host cells were harvested at day 4 after transduction, replated at equal numbers in 96-well plates and MTT assay was performed for the four following days. After 3.5 h of MTT incubation, plates were centrifuged, the medium was aspirated and 200 μl DMSO was added. Absorption was measured at 560 nm (Varioskan microplate spectrophotometer; Thermo Scientific, Breda, Netherlands), and background was subtracted at 670 nm. To estimate cell numbers after treatment, cells were fixed and stained with methanol/acetic acid/water mixture (50%, 20% and 30% respectively), containing 0.01% Coomassie brilliant blue (Sigma) visualizing protein content. In addition, clonogenic survival assay was performed by replating cells in 6-well plates at day 4 after transduction, colonies were allowed to grow for 15–21 days and visualized as described above.
Results
Bidirectional modulation of EpCAM expression
To identify which ATF could effectively modulate EpCAM expression, ATFs were screened in a panel of carcinoma cell lines by using suboptimal retroviral transductions (Supplementary Figure 1). This screening approach identified ZFASKD and ZFBVP64 as the most potent down- and upregulators of EpCAM expression, respectively. ZFBVP64 showed no effect on Her2neu and ICAM expression (data not shown), whereas an ATF designed to target the Her2neu gene (Beerli et al, 2000) effectively up- (E2C-VP64) or downregulated (E2C-SKD) Her2neu expression in HeLa, but showed no effect on EpCAM expression (Supplementary Figure 1D and E).
Functional validation of downregulation of EpCAM expression by ATFs
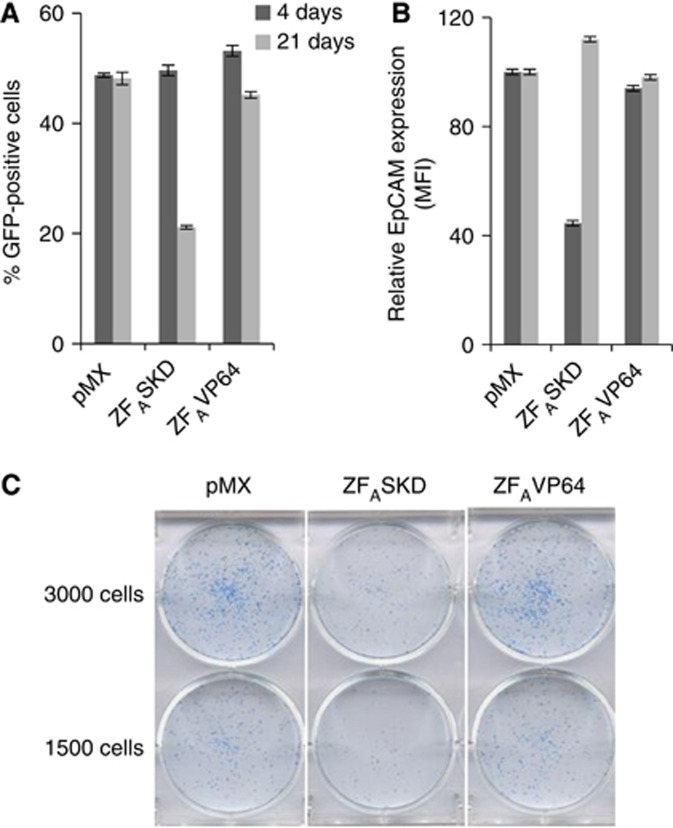
Downregulation of EpCAM expression by RNAi in breast cancer reduces the oncogenic potential (Osta et al, 2004; Sankpal et al, 2011). Therefore, we validated that downregulation of EpCAM expression by ATFs in breast cancer also induced similar phenotypic alterations. SKBR3 cells were co-transduced with ZFASKD and a strong GFP reporter vector to track the percentage of transduced cells. As negative controls, ZFAVP64 and pMX (empty vector control) were used. At 4 days after transduction, all conditions displayed approximately 50% GFP-positive cells (Figure 1A). Compared with pMX, ZFASKD-transduced cells showed a 2.3-fold reduction in EpCAM protein expression, whereas the EpCAM expression of ZFAVP64- and pMX-transduced cells was similar (Figure 1B). After normalizing for cell numbers and subculturing, ZFASKD-transduced cells proliferated less rapidly than the cells transduced with pMX or ZFAVP64, requiring less cell passaging. Indeed, after 17 days of subculturing, the ZFASKD-transduced cell population only contained 20% GFP-positive cells, whereas the cells transduced with pMX or ZFAVP64 still displayed 50% GFP-positive cells (Figure 1A). This finding indicates that the unaffected EpCAM-expressing cell population overgrew the ZFASKD-expressing SKBR3 population, which is also reflected in the restored EpCAM expression of the cell population (Figure 1B). The reduced proliferation displayed by the reduced EpCAM-expressing breast cancer cells was also confirmed by a colony-forming assay. The number of colonies formed by SKBR3 cells expressing ZFASKD was clearly less than the number of colonies formed by the cells transduced with pMX and ZFAVP64 (Figure 1C).
Figure 1.
Downregulation of EpCAM expression by ATFs inhibits cell growth in breast cancer cells. SKBR3 cells were transduced to express the indicated ATFs. At days 4 and 21 after transduction, the percentage of GFP-positive (A) and the relative EpCAM protein expression (B; pMX set at 100) was measured by flow cytometry. (C) Clonogenic survival assay: 4 days after transduction cells were equally replated in 6-well plates and allowed to grow colonies for 16 days when surviving colonies were stained. MFI, mean fluorescence intensity.
Biological role of EpCAM expression in ovarian cancer
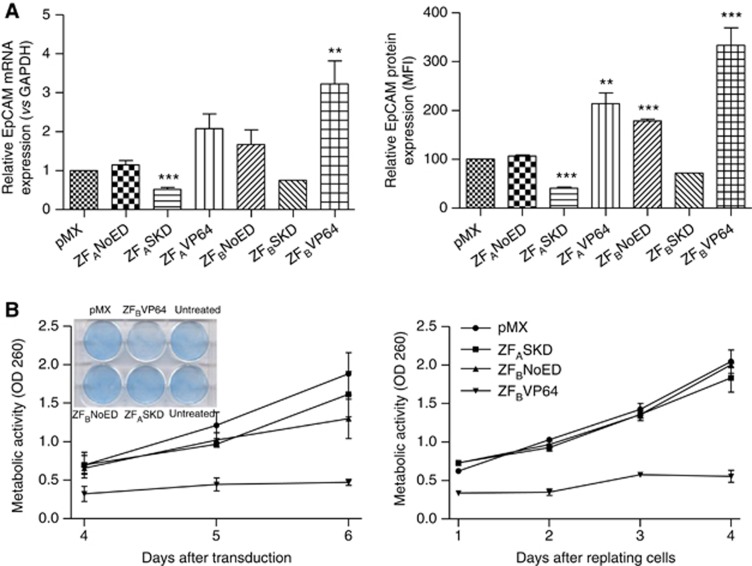
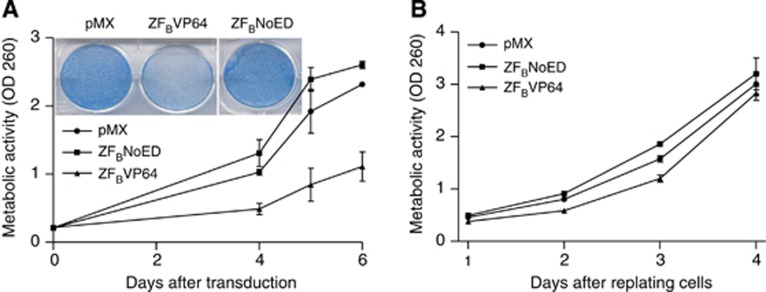
As the biological role of EpCAM in ovarian cancer is unclear (van der Gun et al, 2010), we set out to provide such insights by bidirectionally modulating EpCAM expression in intermediate EpCAM-expressing ovarian cancer cells (SKOV3). The transduction protocol was improved to yield at least 80% of GFP-positive ovarian cancer cells expressing ZFASKD or ZFBVP64. Compared with cells transduced with pMX, ZFASKD-expressing cells displayed a two-fold reduction of EpCAM expression on RNA (0.5±0.1; P<0.001) and on protein level (41±2% expression left; P<0.001) (Figure 2A). The ZFAVP64- and ZFBVP64-transduced cells demonstrated increased EpCAM protein expression (213±22% P<0.01 and 340±33% P<0.001), respectively). Cells transduced to express ZFBNoED (NoED=no effector domain) without an effector domain also exhibited upregulation (178±4% P<0.001), which was not observed for ZFANoED (Figure 2A).
Figure 2.
Phenotypical effect of up- and downregulation of EpCAM expression in ovarian cancer. SKOV3 cells were transduced to express the indicated ATFs in 6- and 96-well plates. (A) At day 4 after transduction, cells were harvested and analysed for EpCAM mRNA (left; pMX set at 1) and protein (right; pMX set at 100) expression (mean±s.e.m., n⩾5; t-test **P<0.01, ***P<0.001). (B) At day 4 after transduction, cells were fixed, stained (inset) and 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was performed at days 4, 5 and 6 after transduction (left) or cells were replated at equal cell numbers and MTT assay was performed for the four following days (right). GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MFI, mean fluorescence intensity; OD, optical density.
Because ZFASKD and ZFBVP64 modulated EpCAM expression most effectively, these ATFs were further selected to investigate the phenotypical change due to EpCAM gene downregulation vs overexpression. Unexpectedly, at day 4 after transduction, not ZFASKD- but ZFBVP64-transduced cells showed less cell survival as shown by staining (Figure 2B, left inset) compared with the cells transduced with pMX (empty vector control). Cells transduced with pMX, ZFBNoED or ZFASKD showed comparable survival. This observation was confirmed by cell viability MTT assays performed on days 4, 5 and 6 after transduction: less cell survival was observed after transduction with ZFBVP64, whereas pMX, ZFASKD and ZFBNoED displayed the same cell growth rate (Figure 2B, left). Also, after harvesting the different transduced cell populations at day 4 after transduction, MTT assays performed on the equal numbers of replated cells showed exactly the same trend: ZFBVP64-transduced cells did poorly attach to the wells and were hardly capable to proliferate, whereas ZFASKD and ZFBNoED showed the same growth rate as pMX (Figure 2B, right). Furthermore, enforced ectopic cDNA expression of EpCAM did not result in growth induction of ovarian cells (Supplementary Figure 2A). To exclude that the expression of VP64 itself was toxic for the cells, transductions were performed with an irrelevant ZFP fused to VP64 (ZFirrVP64), with ZFAVP64 and the controls ZFANoED and pMX. Again, ZFBVP64-transduced cells clearly showed less cell survival (Supplementary Figure 2B), whereas all other constructs displayed comparable cell numbers with pMX.
siRNA-based silencing of EpCAM expression in ovarian cancer has no effect on cell growth
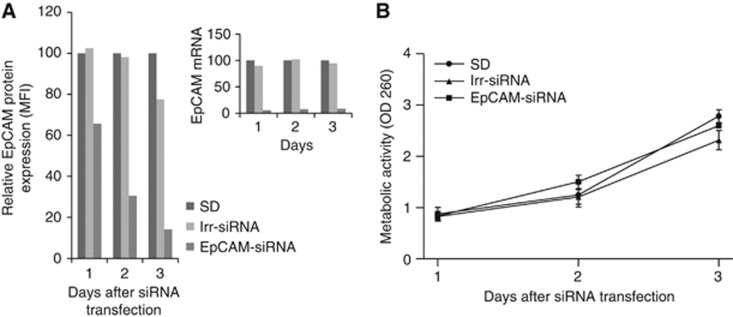
The biological effect of increased EpCAM expression was in contrast with our initial hypothesis that reduced EpCAM expression in ovarian cancer would result in growth inhibition. To confirm that indeed downregulation of EpCAM expression had no effect on cell proliferation, we validated our finding with the conventional approach of silencing EpCAM expression by siRNA. After transfection of EpCAM-siRNA into SKOV3 cells, EpCAM RNA expression was almost completely silenced and protein expression was reduced by 95% (Figure 3A), whereas irr-siRNA had no effect on EpCAM expression. Despite the efficient downregulation of EpCAM, again equal growth rates were observed for EpCAM-siRNA- and irr-siRNA-treated cells (Figure 3B).
Figure 3.
SiRNA-based silencing of EpCAM expression in ovarian cancer has no effect on cell growth. SKOV3 cells were transfected with irr-siRNA or EpCAM-siRNA. At days 1 to 3 after transfection, EpCAM mRNA, protein expression (A) and 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay (B) were analysed (SD set at 100=delivery agent). MFI, mean fluorescence intensity; OD, optical density.
Increased EpCAM expression levels correlated with cell growth inhibition
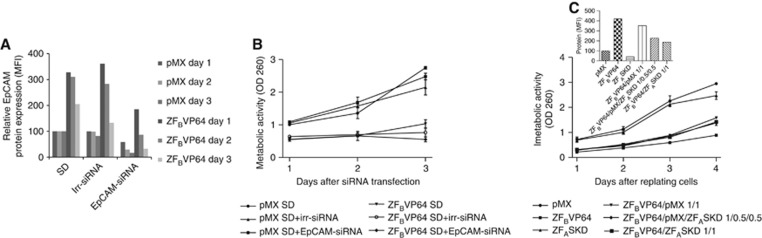
To investigate whether the growth inhibition associated with induced EpCAM overexpression could be phenotypically rescued, ZFBVP64-transduced SKOV3 cells were transfected with EpCAM-siRNA. At day 1 after siRNA transfection, the ZFBVP64-induced 3.3-fold upregulation of EpCAM protein expression was reduced by the combination with siRNA to only 1.8-fold upregulation compared with ZFBVP64 and irr-siRNA treatment (Figure 4A). At day 3, only 16% EpCAM expression compared with mock-treated cells was left. Despite the efficient downregulation of EpCAM expression at day 3, this reduction could not rescue the growth inhibition caused by the initial ATF-induced EpCAM overexpression (Figure 4B).
Figure 4.
Increased EpCAM expression levels correlate with cell growth inhibition. SKOV3 cells were transduced with pMX and ZFBVP64. At day 4 after transduction, cells were equally replated. Next day, siRNA transfection was performed, and subsequently at 3 days, EpCAM protein (A) and 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay (B) were analysed. (C) SKOV3 cells were transduced with a constant amount of ZFBVP64 virus supernatant mixed with pMX and/or ZFASKD. At day 4 after transduction, EpCAM protein expression was measured (inset) and transduced cells were replated at equal cell numbers, and MTT assay was performed for the four following days.
As downregulation of EpCAM expression after upregulation of EpCAM expression had no effect on cell survival, we investigated whether we could prevent the induced upregulation by simultaneous downregulation of EpCAM expression. SKOV3 cells were transduced with a constant amount of ZFBVP64 virus supernatant mixed with pMX and or ZFASKD. Although the pMX–ATF mixtures induced a gradual decrease of EpCAM upregulation (Figure 4C, inset), the lowest induced upregulation was still 1.9-fold and only a slight recovery from growth inhibition was observed (Figure 4C). Subsequent induction of different levels of endogenous EpCAM expression (e.g. by titration of pMX-6-ZF-B-VP64 with ZFASKD) confirmed that exceeding a threshold of EpCAM expression resulted in irreversible growth inhibition (Supplementary Figure 3).
Induction of EpCAM expression in EpCAM-negative ovarian cancer cell line
To further validate our unexpected finding that up-regulation of EpCAM expression resulted in growth inhibition in ovarian cancer, we investigated the phenotypical changes due to induction of EpCAM in the EpCAM-negative ovarian cancer cell line A2780. Indeed, ZFBVP64-transduced A2780 cells clearly displayed a lower number of surviving cells than pMX-transduced cells and again growth inhibition was observed (Figure 5A). Transfection of an EpCAM cDNA-expressing plasmid also induced growth inhibition (Supplementary Figure 2). When surviving infected cells were harvested at day 4 and replated at equal densities, the same growth rate was obtained for cells transduced with pMX, ZFBNoED or ZFBVP64 (Figure 5B). The difference in growth capacity of direct treated cells (Figure 5A) compared with the replated surviving cell population (Figure 5B) can be explained by the cytotoxic effect of the induced EpCAM expression. At day 4, only 35±3% of the A2780 cells was still GFP-positive compared with 85±2% and 80±10% for ZFBNoED and ZFASKD, respectively. In contrast, ZFBVP64-transduced SKOV3 cells displayed at day 4 after transduction 84±5% GFP-positive cells compared with 96±1% and 93±6% for ZFBNoED and ZFASKD, respectively.
Figure 5.
Induction of EpCAM expression in EpCAM-negative ovarian cancer cell line. A2780 cells were transduced to express the indicated ATFs in 6- and 96-wellsplates. (A) At day 4, transduced cells were fixed and stained, or 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT assay was performed at days 4, 5 and 6 after transduction. (B) Transduced cells were harvested at day 4 after transduction, replated at equal cell numbers in 96-well plates and MTT assay was performed for the four following days. OD, optical density.
Discussion
Dependent on the tumour type, EpCAM overexpression has been associated with either decreased or increased overall survival of patients (van der Gun et al, 2010). Although this dual role is also reflected by EpCAM modulation studies, for human ovarian cancer no such siRNA/cDNA studies have been reported and the biological role of EpCAM here remains unclear. This study is the first to report that downregulation of EpCAM expression in human ovarian cancer does not decrease growth rate, not when using a repressive ATF nor after siRNA-mediated downregulation. Importantly, ATF-induced EpCAM downregulation did result in cell growth inhibition for breast cancer, confirming previous RNA interference studies for this carcinoma type (Osta et al, 2004; Sankpal et al, 2011).
As our findings suggest the absence of an oncogenic role for EpCAM in human ovarian cancer, we set out to induce or further upregulate EpCAM expression in EpCAM-negative or low expressing ovarian cancer cells. As has also been reported for renal cell (Klatte et al, 2009) and thyroid carcinomas (Ralhan et al, 2010), we observed an inhibition in cell growth by inducing EpCAM expression. As ablation of p53 expression has been associated with an increase in EpCAM expression (Sankpal et al, 2009), the commonly observed EpCAM overexpression in ovarian cancer might be a consequence of p53 dysfunction, ubiquitously observed for ovarian cancers (Ahmed et al, 2010). Indeed, although EpCAM expression enhanced the tumour-initiating ability of ovarian stem-like cells, a significant outgrowth of tumour mass was only observed for EpCAM-positive cells when p53 was downregulated (Motohara et al, 2011). In addition, a growth-promoting role of EpCAM via Wnt signalling, an important pathway in cancers, might not be of general importance in the development of cancer. Whereas EpCAM overexpression in MDA-MB231 could activate Wnt signalling, this was not observed in Hs578T (Gostner et al, 2011). Alternatively, the high EpCAM expression commonly observed on ovarian cancer cells could be explained by a recent postulation stating that high-grade serous ovarian cancer arises from the fallopian tube instead of the ovary (Reitsma et al, 2012). The relative increase of EpCAM protein expression in ovarian cancer (2.8 on a scale of 0–3) compared with healthy fallopian tube epithelium (1.8) is only 1.5-fold, whereas the increase of EpCAM protein expression compared with ovarian surface epithelium (0.2) is 14-fold (Emmanuel et al, 2011).
As EpCAM is frequently overexpressed in ovarian cancer, anti-EpCAM antibodies are successfully exploited to recruit immune effector cells, resulting in ovarian cancer cell lysis (Richter et al, 2010). Despite this important role of EpCAM in ovarian cancer therapy, we show here that inhibition of EpCAM expression does not offer a promising therapeutic approach for ovarian cancer.
Acknowledgments
We thank Drs V Cirulli (University of Washington), BNG Giepmans (UMCG) for providing EpCAM cDNA and C Barbas (Scripps, La Jolla, US) for E2C constructs. This work was financed by the National Dutch Scientific Research Organisation (Grant: NWO-ZonMW/VIDI/91786373) and EU FP7 (EuroTransBio: ProTuMa ETB09008).
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, Stewart C, Fereday S, Caldas C, Defazio A, Bowtell D, Brenton JD. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle PA, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007;96:417–423. doi: 10.1038/sj.bjc.6603494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Dreier B, Barbas CF. Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran AS, Blancafort P. Reactivation of MASPIN in non-small cell lung carcinoma (NSCLC) cells by artificial transcription factors (ATFs) Epigenetics. 2011;7:350–360. doi: 10.4161/epi.6.2.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groote ML, Verschure PJ, Rots MG. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012;40:10596–10613. doi: 10.1093/nar/gks863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel C, Gava N, Kennedy C, Balleine RL, Sharma R, Wain G, Brand A, Hogg R, Etemadmoghadam D, George J, Australian Ovarian Cancer Study Group, Birrer MJ, Clarke CL, Chenevix-Trench G, Bowtell DD, Harnett PR, deFazio A. Comparison of expression profiles in ovarian epithelium in vivo and ovarian cancer identifies novel candidate genes involved in disease pathogenesis. PLoS One. 2011;6:e17617. doi: 10.1371/journal.pone.0017617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommans WM, McLaughlin PM, Lindhout BI, Segal DJ, Wiegman DJ, Haisma HJ, van der Zaal BJ, Rots MG. Engineering zinc finger protein transcription factors to downregulate the epithelial glycoprotein-2 promoter as a novel anti-cancer treatment. Mol Carcinogen. 2007;46:391–401. doi: 10.1002/mc.20289. [DOI] [PubMed] [Google Scholar]
- Gostner JM, Fong D, Wrulich OA, Lehne F, Zitt M, Hermann M, Krobitsch S, Martowicz A, Gastl G, Spizzo G. Effects of EpCAM overexpression on human breast cancer cell lines. BMC Cancer. 2011;11:45. doi: 10.1186/1471-2407-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzelmann-Schwarz VA, Gardiner-Garden M, Henshall SM, Scurry J, Scolyer RA, Davies MJ, Heinzelmann M, Kalish LH, Bali A, Kench JG, Edwards LS, Vanden Bergh PM, Hacker NF, Sutherland RL, O'Brien PM. Overexpression of the cell adhesion molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin Cancer Res. 2004;10:4427–4436. doi: 10.1158/1078-0432.CCR-04-0073. [DOI] [PubMed] [Google Scholar]
- Kim JH, Herlyn D, Wong KK, Park DC, Schorge JO, Lu KH, Skates SJ, Cramer DW, Berkowitz RS, Mok SC. Identification of epithelial cell adhesion molecule autoantibody in patients with ovarian cancer. Clin Cancer Res. 2003;9:4782–4791. [PubMed] [Google Scholar]
- Klatte T, Pantuck AJ, Said JW, Seligson DB, Rao NP, LaRochelle JC, Shuch B, Zisman A, Kabbinavar FF, Belldegrun AS. Cytogenetic and molecular tumor profiling for type 1 and type 2 papillary renal cell carcinoma. Clin Cancer Res. 2009;15:1162–1169. doi: 10.1158/1078-0432.CCR-08-1229. [DOI] [PubMed] [Google Scholar]
- Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- Meirelles K, Benedict LA, Dombkowski D, Pepin D, Preffer FI, Teixeira J, Tanwar PS, Young RH, Maclaughlin DT, Donahoe PK, Wei X. Human ovarian cancer stem/progenitor cells are stimulated by doxorubicin but inhibited by Mullerian inhibiting substance. Proc Natl Acad Sci USA. 2012;109:2358–2363. doi: 10.1073/pnas.1120733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohara T, Masuko S, Ishimoto T, Yae T, Onishi N, Muraguchi T, Hirao A, Matsuzaki Y, Tashiro H, Katabuchi H, Saya H, Nagano O. Transient depletion of p53 followed by transduction of c-Myc and K-Ras converts ovarian stem-like cells into tumor-initiating cells. Carcinogenesis. 2011;32:1597–1606. doi: 10.1093/carcin/bgr183. [DOI] [PubMed] [Google Scholar]
- Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, Cole DJ, Gillanders WE. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004;64:5818–5824. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]
- Ralhan R, Cao J, Lim T, MacMillan C, Freeman J, Walfish P. EpCAM nuclear localization identifies aggressive thyroid cancer and is a marker for poor prognosis. BMC Cancer. 2010;10:331. doi: 10.1186/1471-2407-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitsma W, de Bock GH, Oosterwijk JC, Bart J, Hollema H, Mourits MJ. Support of the ‘fallopian tube hypothesis' in a prospective series of risk-reducing salpingo-oophorectomy specimens. Eur J Cancer. 2012;49:132–141. doi: 10.1016/j.ejca.2012.07.021. [DOI] [PubMed] [Google Scholar]
- Richter CE, Cocco E, Bellone S, Silasi DA, Ruttinger D, Azodi M, Schwartz PE, Rutherford TJ, Pecorelli S, Santin AD. High-grade, chemotherapy-resistant ovarian carcinomas overexpress epithelial cell adhesion molecule (EpCAM) and are highly sensitive to immunotherapy with MT201, a fully human monoclonal anti-EpCAM antibody. Am J Obstet Gynecol. 2010;203:582.e1–582.e7. doi: 10.1016/j.ajog.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankpal NV, Mayfield JD, Willman MW, Fleming TP, Gillanders WE. Activator protein 1 (AP-1) contributes to EpCAM-dependent breast cancer invasion. Breast Cancer Res. 2011;13:R124. doi: 10.1186/bcr3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankpal NV, Willman MW, Fleming TP, Mayfield JD, Gillanders WE. Transcriptional repression of epithelial cell adhesion molecule contributes to p53 control of breast cancer invasion. Cancer Res. 2009;69:753–757. doi: 10.1158/0008-5472.CAN-08-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell U, Dijk F, Sjollema KA, Giepmans BN. Immunolabeling artifacts and the need for live-cell imaging. Nat Methods. 2012;9:152–158. doi: 10.1038/nmeth.1855. [DOI] [PubMed] [Google Scholar]
- Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev. 2009;61:513–526. doi: 10.1016/j.addr.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Spizzo G, Went P, Dirnhofer S, Obrist P, Moch H, Baeuerle PA, Mueller-Holzner E, Marth C, Gastl G, Zeimet AG. Overexpression of epithelial cell adhesion molecule (Ep-CAM) is an independent prognostic marker for reduced survival of patients with epithelial ovarian cancer. Gynecol Oncol. 2006;103:483–488. doi: 10.1016/j.ygyno.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Spizzo G, Went P, Dirnhofer S, Obrist P, Simon R, Spichtin H, Maurer R, Metzger U, von Castelberg B, Bart R, Stopatschinskaya S, Kochli OR, Haas P, Mross F, Zuber M, Dietrich H, Bischoff S, Mirlacher M, Sauter G, Gastl G. High Ep-CAM expression is associated with poor prognosis in node-positive breast cancer. Breast Cancer Res Treat. 2004;86:207–213. doi: 10.1023/B:BREA.0000036787.59816.01. [DOI] [PubMed] [Google Scholar]
- Stolzenburg S, Rots MG, Beltran AS, Rivenbark AG, Yuan X, Qian H, Strahl BD, Blancafort P. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res. 2012;40:6725–6740. doi: 10.1093/nar/gks360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uil TG, Haisma HJ, Rots MG. Therapeutic modulation of endogenous gene function by agents with designed DNA-sequence specificities. Nucleic Acids Res. 2003;31:6064–6078. doi: 10.1093/nar/gkg815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Gun BT, Melchers LJ, Ruiters MH, de Leij LF, McLaughlin PM, Rots MG. EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis. 2010;31:1913–1921. doi: 10.1093/carcin/bgq187. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Went P, Dirnhofer S, D S, Moch H, Spizzo G. Expression and prognostic significance of EpCAM. J Cancer Mol. 2008;3:169–174. [Google Scholar]
- Went PTH, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Frequent EpCAM protein expression in human carcinomas. Hum Pathol. 2004;35:122–128. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.