Abstract
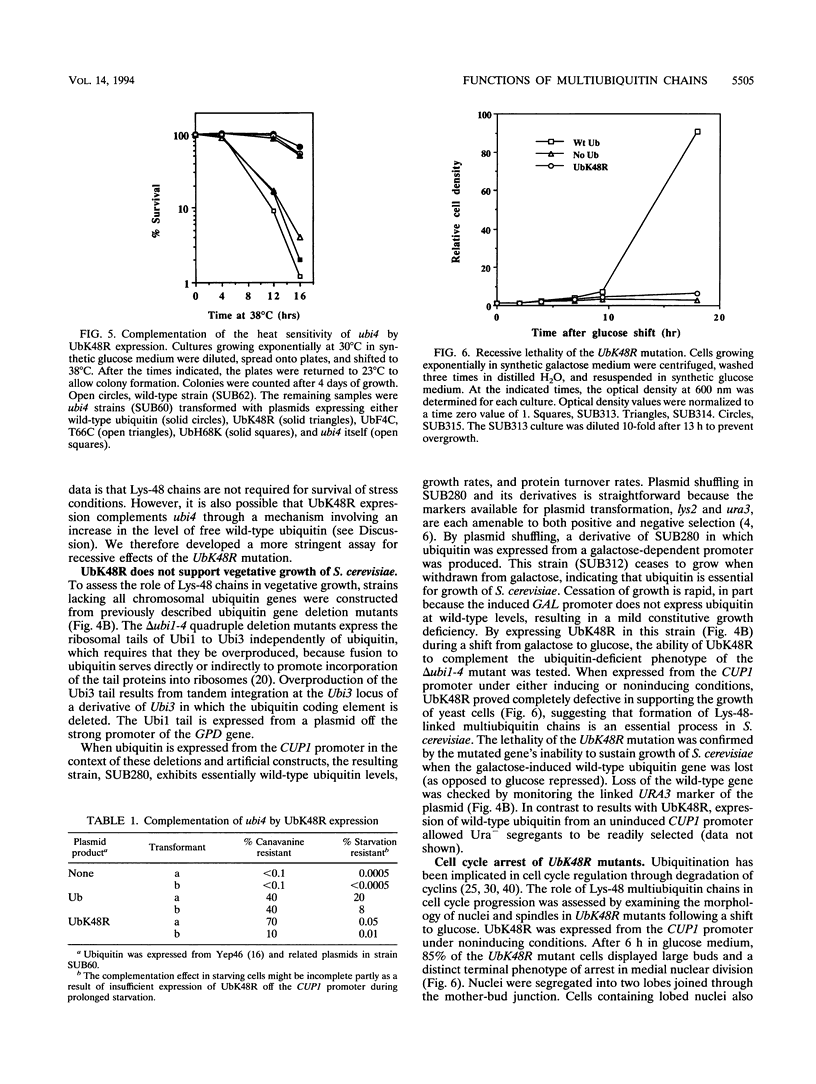
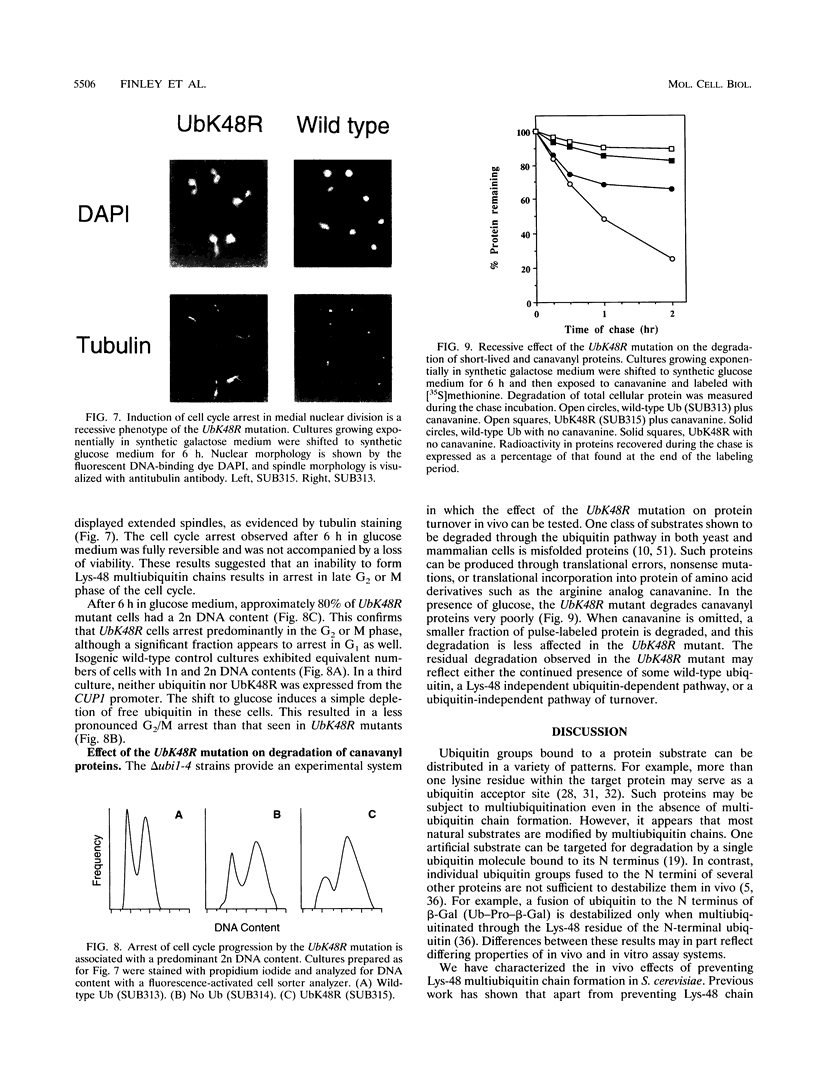
The degradation of many proteins requires their prior attachment to ubiquitin. Proteolytic substrates are characteristically multiubiquitinated through the formation of ubiquitin-ubiquitin linkages. Lys-48 of ubiquitin can serve as a linkage site in the formation of such chains and is required for the degradation of some substrates of this pathway in vitro. We have characterized the recessive and dominant effects of a Lys-48-to-Arg mutant of ubiquitin (UbK48R) in Saccharomyces cerevisiae. Although UbK48R is expected to terminate the growth of Lys-48 multiubiquitin chains and thus to exert a dominant negative effect on protein turnover, overproduction of UbK48R in wild-type cells results in only a weak inhibition of protein turnover, apparently because the mutant ubiquitin can be removed from multiubiquitin chains. Surprisingly, expression of UbK48R complements several phenotypes of polyubiquitin gene (UB14) deletion mutants. However, UbK48R cannot serve as a sole source of ubiquitin in S. cerevisiae, as evidenced by its inability to rescue the growth of ubi1 ubi2 ubi3 ubi4 quadruple mutants. When provided solely with UbK48R, cells undergo cell cycle arrest with a terminal phenotype characterized by replicated DNA, mitotic spindles, and two-lobed nuclei. Under these conditions, degradation of amino acid analog-containing proteins is severely inhibited. Thus, multiubiquitin chains containing Lys-48 linkages play a critical role in protein degradation in vivo.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986 Oct 10;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bartel B., Wünning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990 Oct;9(10):3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Butt T. R., Khan M. I., Marsh J., Ecker D. J., Crooke S. T. Ubiquitin-metallothionein fusion protein expression in yeast. A genetic approach for analysis of ubiquitin functions. J Biol Chem. 1988 Nov 5;263(31):16364–16371. [PubMed] [Google Scholar]
- Chattoo B. B., Sherman F., Azubalis D. A., Fjellstedt T. A., Mehnert D., Ogur M. Selection of lys2 Mutants of the Yeast SACCHAROMYCES CEREVISIAE by the Utilization of alpha-AMINOADIPATE. Genetics. 1979 Sep;93(1):51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989 Mar 24;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., DiGiuseppe J. A., Bercovich B., Orian A., Richter J. D., Schwartz A. L., Brodeur G. M. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Finley D., Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984 May;37(1):57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Cook W. J., Jeffrey L. C., Carson M., Chen Z., Pickart C. M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J Biol Chem. 1992 Aug 15;267(23):16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- Cook W. J., Jeffrey L. C., Kasperek E., Pickart C. M. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J Mol Biol. 1994 Feb 18;236(2):601–609. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]
- Dohmen R. J., Madura K., Bartel B., Varshavsky A. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker D. J., Butt T. R., Marsh J., Sternberg E. J., Margolis N., Monia B. P., Jonnalagadda S., Khan M. I., Weber P. L., Mueller L. Gene synthesis, expression, structures, and functional activities of site-specific mutants of ubiquitin. J Biol Chem. 1987 Oct 15;262(29):14213–14221. [PubMed] [Google Scholar]
- Ecker D. J., Butt T. R., Marsh J., Sternberg E., Shatzman A., Dixon J. S., Weber P. L., Crooke S. T. Ubiquitin function studied by disulfide engineering. J Biol Chem. 1989 Jan 25;264(3):1887–1893. [PubMed] [Google Scholar]
- Ecker D. J., Khan M. I., Marsh J., Butt T. R., Crooke S. T. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J Biol Chem. 1987 Mar 15;262(8):3524–3527. [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Ellison M. J., Hochstrasser M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem. 1991 Nov 5;266(31):21150–21157. [PubMed] [Google Scholar]
- Eytan E., Armon T., Heller H., Beck S., Hershko A. Ubiquitin C-terminal hydrolase activity associated with the 26 S protease complex. J Biol Chem. 1993 Mar 5;268(7):4668–4674. [PubMed] [Google Scholar]
- Finley D., Bartel B., Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989 Mar 30;338(6214):394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- Finley D., Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987 Mar 27;48(6):1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Ghiara J. B., Richardson H. E., Sugimoto K., Henze M., Lew D. J., Wittenberg C., Reed S. I. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991 Apr 5;65(1):163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Udvardy A., Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993 Nov 25;366(6453):358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991 Jan 10;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gregori L., Poosch M. S., Cousins G., Chau V. A uniform isopeptide-linked multiubiquitin chain is sufficient to target substrate for degradation in ubiquitin-mediated proteolysis. J Biol Chem. 1990 May 25;265(15):8354–8357. [PubMed] [Google Scholar]
- Haas A. L., Reback P. B., Chau V. Ubiquitin conjugation by the yeast RAD6 and CDC34 gene products. Comparison to their putative rabbit homologs, E2(20K) AND E2(32K). J Biol Chem. 1991 Mar 15;266(8):5104–5112. [PubMed] [Google Scholar]
- Haas A., Reback P. M., Pratt G., Rechsteiner M. Ubiquitin-mediated degradation of histone H3 does not require the substrate-binding ubiquitin protein ligase, E3, or attachment of polyubiquitin chains. J Biol Chem. 1990 Dec 15;265(35):21664–21669. [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ganoth D., Sudakin V., Dahan A., Cohen L. H., Luca F. C., Ruderman J. V., Eytan E. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. J Biol Chem. 1994 Feb 18;269(7):4940–4946. [PubMed] [Google Scholar]
- Hershko A., Heller H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem Biophys Res Commun. 1985 May 16;128(3):1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Ellison M. J., Chau V., Varshavsky A. The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins R. R., Ellison K. S., Ellison M. J. Expression of a ubiquitin derivative that conjugates to protein irreversibly produces phenotypes consistent with a ubiquitin deficiency. J Biol Chem. 1992 May 5;267(13):8807–8812. [PubMed] [Google Scholar]
- Holloway S. L., Glotzer M., King R. W., Murray A. W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993 Jul 2;73(7):1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Johnson E. S., Bartel B., Seufert W., Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992 Feb;11(2):497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J., Reins H. A., Schobert C., Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993 Jan 28;361(6410):369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Solomon M. J., Kirschner M. W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989 May 25;339(6222):280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Okazaki K., Furuno N., Watanabe N., Sagata N. The 'second-codon rule' and autophosphorylation govern the stability and activity of Mos during the meiotic cell cycle in Xenopus oocytes. EMBO J. 1992 Jul;11(7):2433–2446. doi: 10.1002/j.1460-2075.1992.tb05308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkaynak E., Finley D., Solomon M. J., Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987 May;6(5):1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parag H. A., Raboy B., Kulka R. G. Effect of heat shock on protein degradation in mammalian cells: involvement of the ubiquitin system. EMBO J. 1987 Jan;6(1):55–61. doi: 10.1002/j.1460-2075.1987.tb04718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Kasperek E. M., Beal R., Kim A. Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1). J Biol Chem. 1994 Mar 11;269(10):7115–7123. [PubMed] [Google Scholar]
- Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993 Mar 25;268(9):6065–6068. [PubMed] [Google Scholar]
- Reiss Y., Heller H., Hershko A. Binding sites of ubiquitin-protein ligase. Binding of ubiquitin-protein conjugates and of ubiquitin-carrier protein. J Biol Chem. 1989 Jun 25;264(18):10378–10383. [PubMed] [Google Scholar]
- Rose I. A., Warms J. V. A specific endpoint assay for ubiquitin. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1477–1481. doi: 10.1073/pnas.84.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993 Nov 5;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Seufert W., Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990 Feb;9(2):543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Berleth E., Pickart C., Prakash S., Prakash L. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 1991 Aug;10(8):2187–2193. doi: 10.1002/j.1460-2075.1991.tb07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992 May 29;69(5):725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C. E., Cook W. J. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987 Apr 5;194(3):531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C. E., Wilkinson K. D., Vierstra R. D., Hatfield P. M., Cook W. J. Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J Biol Chem. 1987 May 5;262(13):6396–6399. [PubMed] [Google Scholar]
- Wiebel F. F., Kunau W. H. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature. 1992 Sep 3;359(6390):73–76. doi: 10.1038/359073a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson K. D., Cox M. J., O'Connor L. B., Shapira R. Structure and activities of a variant ubiquitin sequence from bakers' yeast. Biochemistry. 1986 Sep 9;25(18):4999–5004. doi: 10.1021/bi00366a005. [DOI] [PubMed] [Google Scholar]