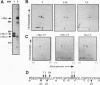
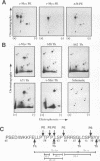
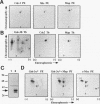
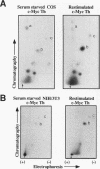
Abstract
The N-terminal domain of the c-Myc protein has been reported to be critical for both the transactivation and biological functions of the c-Myc proteins. Through detailed phosphopeptide mapping analyses, we demonstrate that there is a cluster of four regulated and complex phosphorylation events on the N-terminal domain of Myc proteins, including Thr-58, Ser-62, and Ser-71. An apparent enhancement of Ser-62 phosphorylation occurs on v-Myc proteins having a mutation at Thr-58 which has previously been correlated with increased transforming ability. In contrast, phosphorylation of Thr-58 in cells is dependent on a prior phosphorylation of Ser-62. Hierarchical phosphorylation of c-Myc is also observed in vitro with a specific glycogen synthase kinase 3 alpha, unlike the promiscuous phosphorylation observed with other glycogen synthase kinase 3 alpha and 3 beta preparations. Although both p42 mitogen-activated protein kinase and cdc2 kinase specifically phosphorylate Ser-62 in vitro and cellular phosphorylation of Thr-58/Ser-62 is stimulated by mitogens, other in vivo experiments do not support a role for these kinases in the phosphorylation of Myc proteins. Unexpectedly, both the Thr-58 and Ser-62 phosphorylation events, but not other N-terminal phosphorylation events, can occur in the cytoplasm, suggesting that translocation of the c-Myc proteins to the nucleus is not required for phosphorylation at these sites. In addition, there appears to be an unusual block to the phosphorylation of Ser-62 during mitosis. Finally, although the enhanced transforming properties of Myc proteins correlates with the loss of phosphorylation at Thr-58 and an enhancement of Ser-62 phosphorylation, these phosphorylation events do not alter the ability of c-Myc to transactivate through the CACGTG Myc/Max binding site.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alitalo K., Ramsay G., Bishop J. M., Pfeifer S. O., Colby W. W., Levinson A. D. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature. 1983 Nov 17;306(5940):274–277. doi: 10.1038/306274a0. [DOI] [PubMed] [Google Scholar]
- Alvarez E., Northwood I. C., Gonzalez F. A., Latour D. A., Seth A., Abate C., Curran T., Davis R. J. Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J Biol Chem. 1991 Aug 15;266(23):15277–15285. [PubMed] [Google Scholar]
- Amin C., Wagner A. J., Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993 Jan;13(1):383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Fernandez C., Packham G., Cleveland J. L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich S. J., Cole M. D. Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 1992 Feb;6(2):166–176. doi: 10.1101/gad.6.2.166. [DOI] [PubMed] [Google Scholar]
- Bhatia K., Huppi K., Spangler G., Siwarski D., Iyer R., Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993 Sep;5(1):56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Boulton T. G., Nye S. H., Robbins D. J., Ip N. Y., Radziejewska E., Morgenbesser S. D., DePinho R. A., Panayotatos N., Cobb M. H., Yancopoulos G. D. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 1991 May 17;65(4):663–675. doi: 10.1016/0092-8674(91)90098-j. [DOI] [PubMed] [Google Scholar]
- Boyle W. J., Smeal T., Defize L. H., Angel P., Woodgett J. R., Karin M., Hunter T. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell. 1991 Feb 8;64(3):573–584. doi: 10.1016/0092-8674(91)90241-p. [DOI] [PubMed] [Google Scholar]
- Boyle W. J., van der Geer P., Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Chen R. H., Sarnecki C., Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992 Mar;12(3):915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., Lee W. M. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988 Oct;8(10):4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulić V., Lees E., Reed S. I. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992 Sep 25;257(5078):1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- Fang F., Newport J. W. Evidence that the G1-S and G2-M transitions are controlled by different cdc2 proteins in higher eukaryotes. Cell. 1991 Aug 23;66(4):731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- Farina S. F., Huff J. L., Parsons J. T. Mutations within the 5' half of the avian retrovirus MC29 v-myc gene alter or abolish transformation of chicken embryo fibroblasts and macrophages. J Virol. 1992 May;66(5):2698–2708. doi: 10.1128/jvi.66.5.2698-2708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo E. J., Humphries E. H. An avian retrovirus expressing chicken pp59c-myc possesses weak transforming activity distinct from v-myc that may be modulated by adjacent normal cell neighbors. J Virol. 1991 Dec;65(12):6621–6629. doi: 10.1128/jvi.65.12.6621-6629.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiol C. J., Wang A., Roeske R. W., Roach P. J. Ordered multisite protein phosphorylation. Analysis of glycogen synthase kinase 3 action using model peptide substrates. J Biol Chem. 1990 Apr 15;265(11):6061–6065. [PubMed] [Google Scholar]
- Frykberg L., Graf T., Vennström B. The transforming activity of the chicken c-myc gene can be potentiated by mutations. Oncogene. 1987;1(4):415–422. [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Bhatia K., Magrath I. T., Dang C. V., Dalla-Favera R. Binding and suppression of the Myc transcriptional activation domain by p107. Science. 1994 Apr 8;264(5156):251–254. doi: 10.1126/science.8146655. [DOI] [PubMed] [Google Scholar]
- Gupta S., Seth A., Davis R. J. Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr-58 and Ser-62. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3216–3220. doi: 10.1073/pnas.90.8.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann U., Wenzel A., Frank R., Schwab M. The MYCN protein of human neuroblastoma cells is phosphorylated by casein kinase II in the central region and at serine 367. Oncogene. 1991 Oct;6(10):1745–1751. [PubMed] [Google Scholar]
- Hann S. R., Abrams H. D., Rohrschneider L. R., Eisenman R. N. Proteins encoded by v-myc and c-myc oncogenes: identification and localization in acute leukemia virus transformants and bursal lymphoma cell lines. Cell. 1983 Oct;34(3):789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Eisenman R. N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984 Nov;4(11):2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann S. R., King M. W., Bentley D. L., Anderson C. W., Eisenman R. N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988 Jan 29;52(2):185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Sloan-Brown K., Spotts G. D. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev. 1992 Jul;6(7):1229–1240. doi: 10.1101/gad.6.7.1229. [DOI] [PubMed] [Google Scholar]
- Hateboer G., Timmers H. T., Rustgi A. K., Billaud M., van 't Veer L. J., Bernards R. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson M., Bakardjiev A., Klein G., Lüscher B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene. 1993 Dec;8(12):3199–3209. [PubMed] [Google Scholar]
- Hihara H., Shimizu T., Yamamoto H. Establishment of tumor cell lines cultured from chickens with avian lymphoid leukosis. Natl Inst Anim Health Q (Tokyo) 1974 Winter;14(4):163–173. [PubMed] [Google Scholar]
- Hughes K., Pulverer B. J., Theocharous P., Woodgett J. R. Baculovirus-mediated expression and characterisation of rat glycogen synthase kinase-3 beta, the mammalian homologue of the Drosophila melanogaster zeste-white 3sgg homeotic gene product. Eur J Biochem. 1992 Jan 15;203(1-2):305–311. doi: 10.1111/j.1432-1033.1992.tb19860.x. [DOI] [PubMed] [Google Scholar]
- Hunter T., Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiecik T. E., Shalloway D. Activation and suppression of pp60c-src transforming ability by mutation of its primary sites of tyrosine phosphorylation. Cell. 1987 Apr 10;49(1):65–73. doi: 10.1016/0092-8674(87)90756-2. [DOI] [PubMed] [Google Scholar]
- Koff A., Giordano A., Desai D., Yamashita K., Harper J. W., Elledge S., Nishimoto T., Morgan D. O., Franza B. R., Roberts J. M. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992 Sep 18;257(5077):1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Blackwood E. M., Eisenman R. N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992 Oct 1;359(6394):426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Christenson E., Litchfield D. W., Krebs E. G., Eisenman R. N. Myb DNA binding inhibited by phosphorylation at a site deleted during oncogenic activation. Nature. 1990 Apr 5;344(6266):517–522. doi: 10.1038/344517a0. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. Mitosis-specific phosphorylation of the nuclear oncoproteins Myc and Myb. J Cell Biol. 1992 Aug;118(4):775–784. doi: 10.1083/jcb.118.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Kuenzel E. A., Krebs E. G., Eisenman R. N. Myc oncoproteins are phosphorylated by casein kinase II. EMBO J. 1989 Apr;8(4):1111–1119. doi: 10.1002/j.1460-2075.1989.tb03481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran S., Lee H., Sonenshein G. E. Intracellular association of the protein product of the c-myc oncogene with the TATA-binding protein. Mol Cell Biol. 1994 Feb;14(2):1147–1152. doi: 10.1128/mcb.14.2.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D. W., Street A. J. Nuclear protein phosphorylation and growth control. Biochem J. 1992 Oct 1;287(Pt 1):1–15. doi: 10.1042/bj2870001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M., Enders G. H., Wu C. L., Su L. K., Gorka C., Nelson C., Harlow E., Tsai L. H. A family of human cdc2-related protein kinases. EMBO J. 1992 Aug;11(8):2909–2917. doi: 10.1002/j.1460-2075.1992.tb05360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Norton P. A., Coffin J. M. Bacterial beta-galactosidase as a marker of Rous sarcoma virus gene expression and replication. Mol Cell Biol. 1985 Feb;5(2):281–290. doi: 10.1128/mcb.5.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir R., Dwarki V. J., Rashid D., Verma I. M. Phosphorylation of the C terminus of Fos protein is required for transcriptional transrepression of the c-fos promoter. Nature. 1990 Nov 1;348(6296):80–82. doi: 10.1038/348080a0. [DOI] [PubMed] [Google Scholar]
- Palmieri S., Kahn P., Graf T. Quail embryo fibroblasts transformed by four v-myc-containing virus isolates show enhanced proliferation but are non tumorigenic. EMBO J. 1983;2(12):2385–2389. doi: 10.1002/j.1460-2075.1983.tb01750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papas T. S., Lautenberger J. A. Sequence curiosity in v-myc oncogene. Nature. 1985 Nov 21;318(6043):237–237. doi: 10.1038/318237a0. [DOI] [PubMed] [Google Scholar]
- Pearson R. B., Kemp B. E. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- Pulverer B. J., Fisher C., Vousden K., Littlewood T., Evan G., Woodgett J. R. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994 Jan;9(1):59–70. [PubMed] [Google Scholar]
- Ramsay G. M., Moscovici G., Moscovici C., Bishop J. M. Neoplastic transformation and tumorigenesis by the human protooncogene MYC. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2102–2106. doi: 10.1073/pnas.87.6.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay G., Hayman M. J., Bister K. Phosphorylation of specific sites in the gag-myc polyproteins encoded by MC29-type viruses correlates with their transforming ability. EMBO J. 1982;1(9):1111–1116. doi: 10.1002/j.1460-2075.1982.tb01305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resar L. M., Dolde C., Barrett J. F., Dang C. V. B-myc inhibits neoplastic transformation and transcriptional activation by c-myc. Mol Cell Biol. 1993 Feb;13(2):1130–1136. doi: 10.1128/mcb.13.2.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. B., Vila J., Lansing T. J., Potts W. M., Weber M. J., Parsons J. T. Activation of the oncogenic potential of the avian cellular src protein by specific structural alteration of the carboxy terminus. EMBO J. 1987 Aug;6(8):2359–2364. doi: 10.1002/j.1460-2075.1987.tb02512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach P. J. Multisite and hierarchal protein phosphorylation. J Biol Chem. 1991 Aug 5;266(22):14139–14142. [PubMed] [Google Scholar]
- Rossomando A. J., Payne D. M., Weber M. J., Sturgill T. W. Evidence that pp42, a major tyrosine kinase target protein, is a mitogen-activated serine/threonine protein kinase. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6940–6943. doi: 10.1073/pnas.86.18.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A. L., Carruthers C., Gutjahr T., Roeder R. G. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature. 1993 Sep 23;365(6444):359–361. doi: 10.1038/365359a0. [DOI] [PubMed] [Google Scholar]
- Saksela K., Mäkelä T. P., Hughes K., Woodgett J. R., Alitalo K. Activation of protein kinase C increases phosphorylation of the L-myc trans-activator domain at a GSK-3 target site. Oncogene. 1992 Feb;7(2):347–353. [PubMed] [Google Scholar]
- Sarid J., Halazonetis T. D., Murphy W., Leder P. Evolutionarily conserved regions of the human c-myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci U S A. 1987 Jan;84(1):170–173. doi: 10.1073/pnas.84.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A., Gonzalez F. A., Gupta S., Raden D. L., Davis R. J. Signal transduction within the nucleus by mitogen-activated protein kinase. J Biol Chem. 1992 Dec 5;267(34):24796–24804. [PubMed] [Google Scholar]
- Shrivastava A., Saleque S., Kalpana G. V., Artandi S., Goff S. P., Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993 Dec 17;262(5141):1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Vennstrom B., Hayman M. J., Enrietto P. J. Nucleotide sequence of HBI, a novel recombinant MC29 derivative with altered pathogenic properties. J Virol. 1985 Dec;56(3):969–977. doi: 10.1128/jvi.56.3.969-977.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Spotts G. D., Hann S. R. Enhanced translation and increased turnover of c-myc proteins occur during differentiation of murine erythroleukemia cells. Mol Cell Biol. 1990 Aug;10(8):3952–3964. doi: 10.1128/mcb.10.8.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street A. J., Blackwood E., Lüscher B., Eisenman R. N. Mutational analysis of the carboxy-terminal casein kinase II phosphorylation site in human c-myc. Curr Top Microbiol Immunol. 1990;166:251–258. doi: 10.1007/978-3-642-75889-8_31. [DOI] [PubMed] [Google Scholar]
- Symonds G., Hartshorn A., Kennewell A., O'Mara M. A., Bruskin A., Bishop J. M. Transformation of murine myelomonocytic cells by myc: point mutations in v-myc contribute synergistically to transforming potential. Oncogene. 1989 Mar;4(3):285–294. [PubMed] [Google Scholar]
- Tikhonenko A. T., Hartman A. R., Linial M. L. Overproduction of v-Myc in the nucleus and its excess over Max are not required for avian fibroblast transformation. Mol Cell Biol. 1993 Jun;13(6):3623–3631. doi: 10.1128/mcb.13.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- Woodgett J. R. A common denominator linking glycogen metabolism, nuclear oncogenes and development. Trends Biochem Sci. 1991 May;16(5):177–181. doi: 10.1016/0968-0004(91)90071-3. [DOI] [PubMed] [Google Scholar]
- de Vries-Smits A. M., Burgering B. M., Leevers S. J., Marshall C. J., Bos J. L. Involvement of p21ras in activation of extracellular signal-regulated kinase 2. Nature. 1992 Jun 18;357(6379):602–604. doi: 10.1038/357602a0. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S., Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993 Dec 24;262(5142):2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]