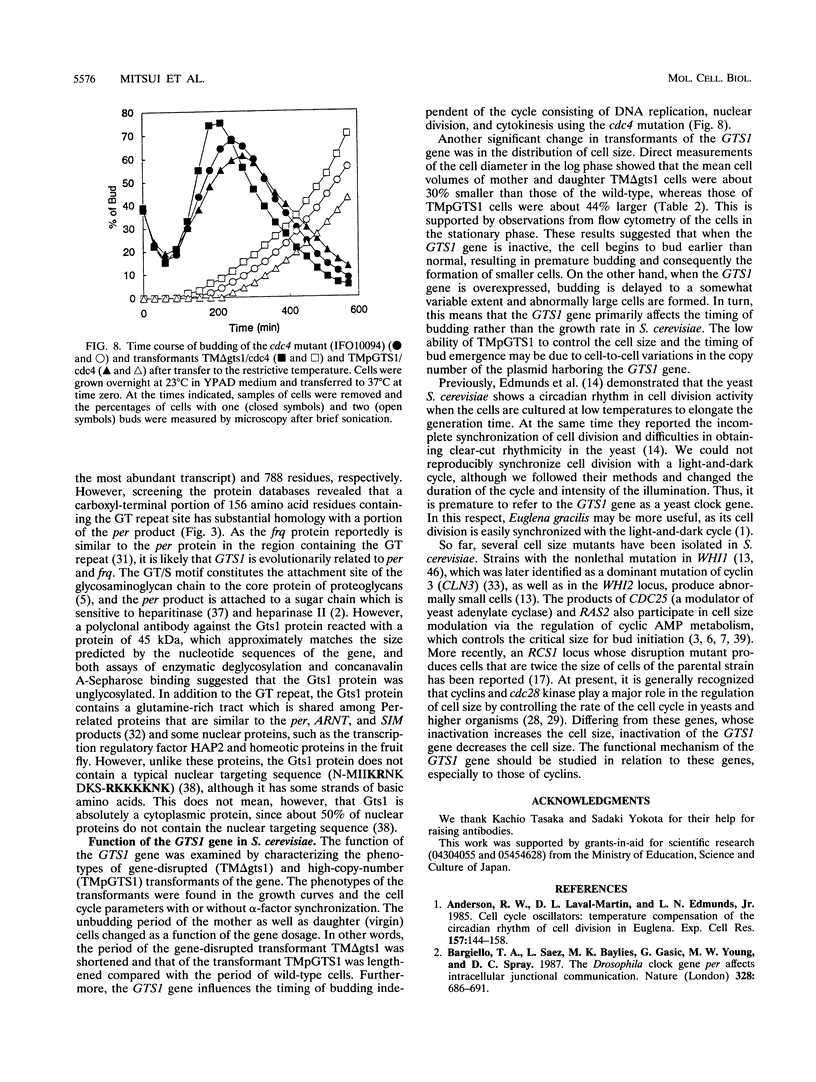
Abstract
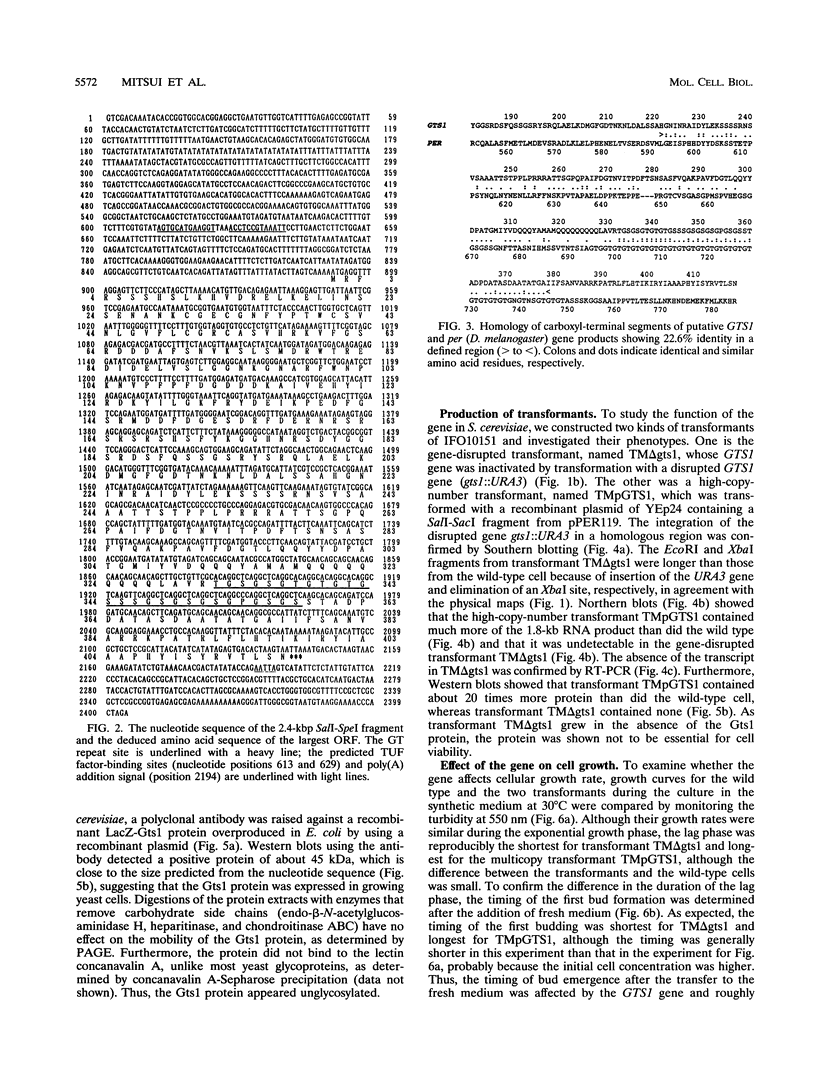
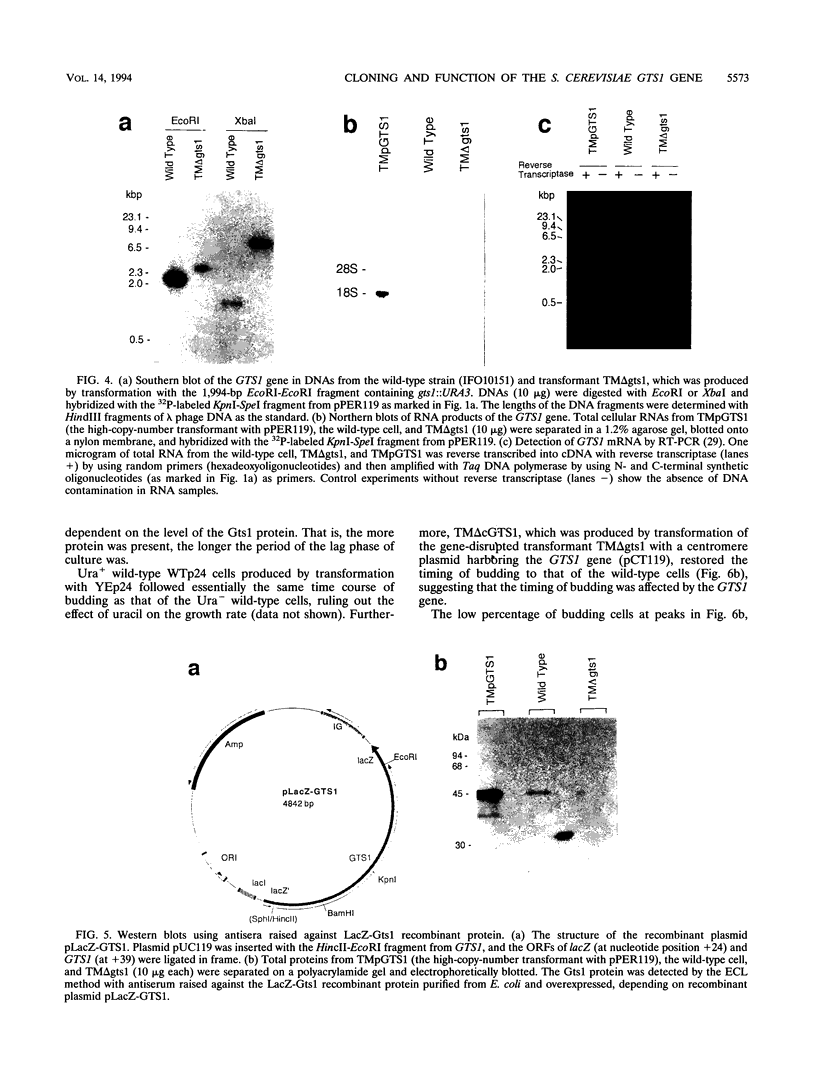
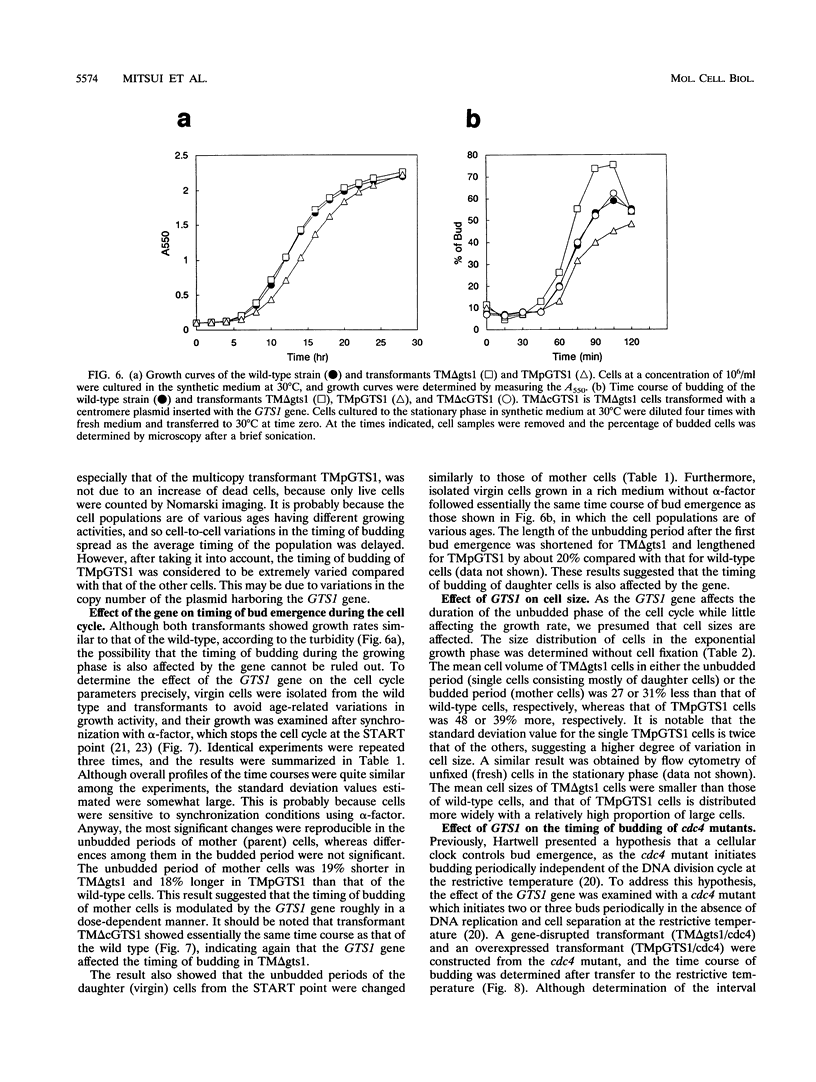
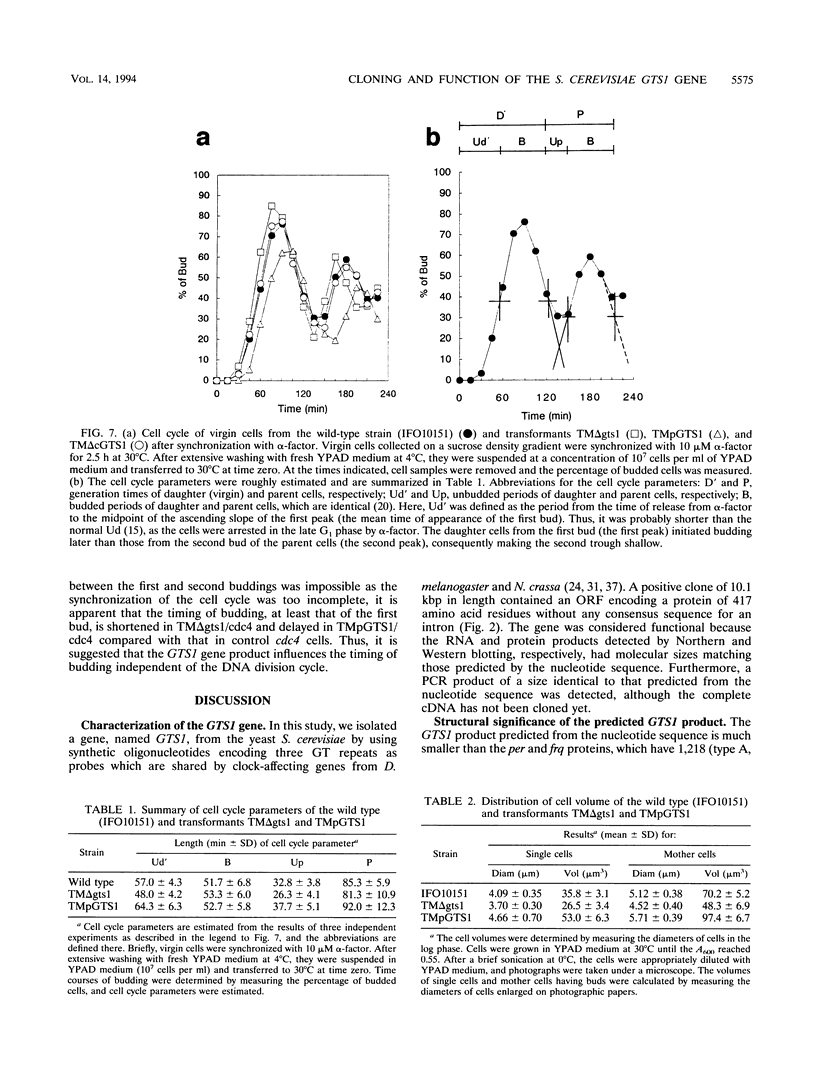
A gene with an open reading frame encoding a protein of 417 amino acid residues with a Gly-Thr repeat was isolated from the yeast Saccharomyces cerevisiae by using synthetic oligonucleotides encoding three Gly-Thr dimers as probes. The deduced amino acid sequence showed partial homology to the clock-affecting gene, per, of Drosophila melanogaster in the regions including the GT repeat. The function of the gene, named GTS1, was examined by characterizing the phenotypes of transformants with different copy numbers of the GTS1 gene produced either by inactivating the GTS1 gene by gene disruption (TM delta gts1) or by transformation with multicopy plasmid pPER119 (TMpGTS1). They grew at similar rates during the exponential growth phase, but the lag phases were shorter for TM delta gts1 and longer for TMpGTS1 cells than that for the wild type. Analyses of their cell cycle parameters using synchronized cells revealed that the unbudding period changed as a function of gene dosage; that is, the periods of TM delta gts1 and TMpGTS1 were about 20% shorter and longer, respectively, than that of the wild-type. Another significant change in the transformants was detected in the distribution of the cell size. The mean cell volume of the TM delta gts1 cells in the unbudded period (single cells) was 27% smaller than that of single wild-type cells, whereas that of single TMpGTS1 cells was 48% larger. Furthermore, in the temperature-sensitive cdc4 mutant, the GTS1 gene affected the timing of budding at the restrictive temperature. Thus, the GTS1 gene product appears to modulate the timing of budding to obtain an appropriate cell size independent of the DNA replication cycle.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. W., Laval-Martin D. L., Edmunds L. N., Jr Cell cycle oscillators. Temperature compensation of the circadian rhythm of cell division in Euglena. Exp Cell Res. 1985 Mar;157(1):144–158. doi: 10.1016/0014-4827(85)90158-2. [DOI] [PubMed] [Google Scholar]
- Bargiello T. A., Saez L., Baylies M. K., Gasic G., Young M. W., Spray D. C. The Drosophila clock gene per affects intercellular junctional communication. Nature. 1987 Aug 20;328(6132):686–691. doi: 10.1038/328686a0. [DOI] [PubMed] [Google Scholar]
- Baroni M. D., Martegani E., Monti P., Alberghina L. Cell size modulation by CDC25 and RAS2 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Jun;9(6):2715–2723. doi: 10.1128/mcb.9.6.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylies M. K., Bargiello T. A., Jackson F. R., Young M. W. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. 1987 Mar 26-Apr 1Nature. 326(6111):390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- Bourdon M. A., Oldberg A., Pierschbacher M., Ruoslahti E. Molecular cloning and sequence analysis of a chondroitin sulfate proteoglycan cDNA. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1321–1325. doi: 10.1073/pnas.82.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Camonis J. H., Kalékine M., Gondré B., Garreau H., Boy-Marcotte E., Jacquet M. Characterization, cloning and sequence analysis of the CDC25 gene which controls the cyclic AMP level of Saccharomyces cerevisiae. EMBO J. 1986 Feb;5(2):375–380. doi: 10.1002/j.1460-2075.1986.tb04222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheifetz S., Andres J. L., Massagué J. The transforming growth factor-beta receptor type III is a membrane proteoglycan. Domain structure of the receptor. J Biol Chem. 1988 Nov 15;263(32):16984–16991. [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Hall J. C., Rosbash M. Interspecific comparison of the period gene of Drosophila reveals large blocks of non-conserved coding DNA. EMBO J. 1988 Dec 1;7(12):3929–3937. doi: 10.1002/j.1460-2075.1988.tb03279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R., Peixoto A. A., Thackeray J. R., Dalgleish R., Kyriacou C. P. Length polymorphism in the threonine-glycine-encoding repeat region of the period gene in Drosophila. J Mol Evol. 1991 Mar;32(3):238–246. doi: 10.1007/BF02342746. [DOI] [PubMed] [Google Scholar]
- Coté G. G., Brody S. Circadian rhythms in Drosophila melanogaster: analysis of period as a function of gene dosage at the per (period) locus. J Theor Biol. 1986 Aug 21;121(4):487–503. doi: 10.1016/s0022-5193(86)80104-7. [DOI] [PubMed] [Google Scholar]
- Cross F. R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds L. N., Jr, Apter R. I., Rosenthal P. J., Shen W. K., Woodward J. R. Light effects in yeast: persisting oscillations in cell division activity and amino acid transport in cultures of Saccharomyces cerevisiae entrained by light-dark cycles. Photochem Photobiol. 1979 Nov;30(5):595–601. doi: 10.1111/j.1751-1097.1979.tb07186.x. [DOI] [PubMed] [Google Scholar]
- Ewer J., Hamblen-Coyle M., Rosbash M., Hall J. C. Requirement for period gene expression in the adult and not during development for locomotor activity rhythms of imaginal Drosophila melanogaster. J Neurogenet. 1990 Nov;7(1):31–73. doi: 10.3109/01677069009084151. [DOI] [PubMed] [Google Scholar]
- Gil R., Zueco J., Sentandreu R., Herrero E. RCS1, a gene involved in controlling cell size in Saccharomyces cerevisiae. Yeast. 1991 Jan;7(1):1–14. doi: 10.1002/yea.320070102. [DOI] [PubMed] [Google Scholar]
- Hall J. C. Genetics of circadian rhythms. Annu Rev Genet. 1990;24:659–697. doi: 10.1146/annurev.ge.24.120190.003303. [DOI] [PubMed] [Google Scholar]
- Hamblen-Coyle M., Konopka R. J., Zwiebel L. J., Colot H. V., Dowse H. B., Rosbash M., Hall J. C. A new mutation at the period locus of Drosophila melanogaster with some novel effects on circadian rhythms. J Neurogenet. 1989 Aug;5(4):229–256. doi: 10.3109/01677068909066210. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Culotti J., Pringle J. R., Reid B. J. Genetic control of the cell division cycle in yeast. Science. 1974 Jan 11;183(4120):46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Genetic control of the cell division cycle in yeast. II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971 Jul 14;59(1):183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Unger M. W. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol. 1977 Nov;75(2 Pt 1):422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H., Yahara I. Specific early-G1 blocks accompanied with stringent response in Saccharomyces cerevisiae lead to growth arrest in resting state similar to the G0 of higher eucaryotes. J Cell Biol. 1984 Apr;98(4):1185–1193. doi: 10.1083/jcb.98.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson F. R., Bargiello T. A., Yun S. H., Young M. W. Product of per locus of Drosophila shares homology with proteoglycans. Nature. 1986 Mar 13;320(6058):185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- Kyriacou C. P., Hall J. C. Circadian rhythm mutations in Drosophila melanogaster affect short-term fluctuations in the male's courtship song. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6729–6733. doi: 10.1073/pnas.77.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Marini N. J., Reed S. I. Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells of the budding yeast S. cerevisiae. Cell. 1992 Apr 17;69(2):317–327. doi: 10.1016/0092-8674(92)90412-6. [DOI] [PubMed] [Google Scholar]
- Linskens M., Tyers M., Futcher B. CLN3 functions in both daughter and mother cells of S. cerevisiae. Cell. 1993 Feb 26;72(4):487–488. doi: 10.1016/0092-8674(93)90067-z. [DOI] [PubMed] [Google Scholar]
- McClung C. R., Fox B. A., Dunlap J. C. The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature. 1989 Jun 15;339(6225):558–562. doi: 10.1038/339558a0. [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Lewis J. O., Wharton K. A., Jr, Crews S. T. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991 Dec 20;67(6):1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Nash R., Tokiwa G., Anand S., Erickson K., Futcher A. B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988 Dec 20;7(13):4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto A. A., Campesan S., Costa R., Kyriacou C. P. Molecular evolution of a repetitive region within the per gene of Drosophila. Mol Biol Evol. 1993 Jan;10(1):127–139. doi: 10.1093/oxfordjournals.molbev.a039993. [DOI] [PubMed] [Google Scholar]
- Peixoto A. A., Costa R., Wheeler D. A., Hall J. C., Kyriacou C. P. Evolution of the threonine-glycine repeat region of the period gene in the melanogaster species subgroup of Drosophila. J Mol Evol. 1992 Nov;35(5):411–419. doi: 10.1007/BF00171819. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Hardwick K. G., Lewis M. J. Sorting of soluble ER proteins in yeast. EMBO J. 1988 Jun;7(6):1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P., Jacquier A. C., Abovich N., Petersen G., Rosbash M. The period clock locus of D. melanogaster codes for a proteoglycan. Cell. 1986 Jul 4;46(1):53–61. doi: 10.1016/0092-8674(86)90859-7. [DOI] [PubMed] [Google Scholar]
- Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991 Feb 8;64(3):615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Robinson L. C., Gibbs J. B., Marshall M. S., Sigal I. S., Tatchell K. CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science. 1987 Mar 6;235(4793):1218–1221. doi: 10.1126/science.3547648. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. S., Bargiello T. A., Clark B. T., Jackson F. R., Young M. W. An unusual coding sequence from a Drosophila clock gene is conserved in vertebrates. Nature. 1985 Oct 3;317(6036):445–448. doi: 10.1038/317445a0. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P. E., Goodey A. R., Carter B. L. Genes which control cell proliferation in the yeast Saccharomyces cerevisiae. Nature. 1980 Nov 27;288(5789):401–404. doi: 10.1038/288401a0. [DOI] [PubMed] [Google Scholar]
- Sweet D. J., Pelham H. R. The Saccharomyces cerevisiae SEC20 gene encodes a membrane glycoprotein which is sorted by the HDEL retrieval system. EMBO J. 1992 Feb;11(2):423–432. doi: 10.1002/j.1460-2075.1992.tb05071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais M. L., Huet J., Buhler J. M., Sentenac A. Contacts between the factor TUF and RPG sequences. J Biol Chem. 1990 Aug 25;265(24):14669–14674. [PubMed] [Google Scholar]
- Yu Q., Colot H. V., Kyriacou C. P., Hall J. C., Rosbash M. Behaviour modification by in vitro mutagenesis of a variable region within the period gene of Drosophila. Nature. 1987 Apr 23;326(6115):765–769. doi: 10.1038/326765a0. [DOI] [PubMed] [Google Scholar]
- Zehring W. A., Wheeler D. A., Reddy P., Konopka R. J., Kyriacou C. P., Rosbash M., Hall J. C. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell. 1984 Dec;39(2 Pt 1):369–376. doi: 10.1016/0092-8674(84)90015-1. [DOI] [PubMed] [Google Scholar]




