Abstract
In evolution of mammals, some of essential genes for placental development are known to be of retroviral origin, as syncytin-1 derived from an envelope (env) gene of an endogenous retrovirus (ERV) aids in the cell fusion of placenta in humans. Although the placenta serves the same function in all placental mammals, env-derived genes responsible for trophoblast cell fusion and maternal immune tolerance differ among species and remain largely unidentified in the bovine species. To examine env-derived genes playing a role in the bovine placental development comprehensively, we determined the transcriptomic profiles of bovine conceptuses during three crucial windows of implantation periods using a high-throughput sequencer. The sequence reads were mapped into the bovine genome, in which ERV candidates were annotated using RetroTector© (7,624 and 1,542 for ERV-derived and env-derived genes, respectively). The mapped reads showed that approximately 18% (284 genes) of env-derived genes in the genome were expressed during placenta formation, and approximately 4% (63 genes) were detected for all days examined. We verified three env-derived genes that are expressed in trophoblast cells by polymerase chain reaction. Out of these three, the sequence of env-derived gene with the longest open reading frame (named BERV-P env) was found to show high expression levels in trophoblast cell lines and to be similar to those of syncytin-Car1 genes found in dogs and cats, despite their disparate origins. These results suggest that placentation depends on various retrovirus-derived genes that could have replaced endogenous predecessors during evolution.
Keywords: endogenous retrovirus, RNA-seq, syncytin, envelope, cow
Introduction
A certain portion of mammalian genomes corresponds to endogenous retroviruses (ERVs), which are thought to be derived from ancient viral infections of germ cells (Weiss 2006). In fact, sequences of retroviral origins make up 8% and 10% of human and mouse genomes, respectively (Mouse Genome Sequencing Consortium 2002; International Human Genome Sequencing Consortium 2004). An ERV usually consists of 5′- and 3′-long terminal repeats (LTRs), group-specific antigen (gag), protease (pro), polymerase (pol), and envelope (env) genes. Although most ERVs have been inactivated by insertions, deletions, substitutions, and/or epigenetic modifications, a few open reading frames (ORFs) of ERVs are still active and express viral proteins in the hosts. Indeed, some env genes have been found to be essential for placental morphogenesis in humans (Mi et al. 2000; Esnault et al. 2008), mice (Dupressoir et al. 2005), and rabbits (Heidmann et al. 2009). An env gene encodes a protein consisting of two polypeptides; the surface (SU) and the transmembrane (TM) subunits (Ng et al. 1982). It is cleaved by the furin protease, whose recognition sequence is R/K-X-R/K-R (Krysan et al. 1999). The SU subunit is involved in receptor recognition, whereas the TM subunit anchors the entire envelope glycoprotein complex to the membrane and is directly responsible for membrane fusion between cells and virions (Bénit et al. 2001).
The env genes of human ERV-W (HERV-W) and retrovirus-FRD (HERV-FRD), also known as syncytin-1 and syncytin-2, respectively, are expressed in trophoblast cells to induce cell fusion and to avoid rejection by the maternal immune system (Mi et al. 2000; Esnault et al. 2008). In mice, there exist functionally corresponding genes named syncytin-A and syncytin-B, but their origins are different from their human analogs (Dupressoir et al. 2005). Syncytin-Ory1 in rabbit (Heidmann et al. 2009) is another example of similar function but different exogenous viral origin. Recently, Cornelis et al. (2012) reported syncytin-like genes conserved in carnivores (syncytin-Car1). It was the first report of syncytins in the Laurasiatheria clade, but no corresponding genes are found in the genomes of other mammals including noncarnivoran laurasiatherians such as bovines or horses (Cornelis et al. 2012). These observations suggest that during the course of evolution, the incorporation of viral genes into the genome benefited placental development. This can be supported by the differences in morphological diversity observed in mammalian placentas: discoid placenta for humans, mice, and rabbits; zonary placenta for carnivores; cotyledonary placenta for bovines; and diffuse placenta for horses (Wildman et al. 2006; Rawn and Cross 2008). However, the entire picture of functional retroelements during the process of placenta development is still unknown. For the bovine species, in particular, the genes functioning in placental development are unknown despite their importance to the cattle industry. Although artificial insemination technology, widely used for bovine reproduction throughout the world, has contributed in reducing sexually transmitted diseases, the cattle industry has reaped only limited success in improving fertility. In fact, the pregnancy rate has continuously dropped for the last 20 years in Japan (Livestock Improvement Association of Japan, http://liaj.lin.gr.jp/japanese/chosa/index.html). This trend illustrates the need to investigate other factors such as ERVs or retroelements previously regarded as relatively insignificant.
In this study, to gain information on active env genes in the bovine genome, high-throughput sequencing technologies with the accurate bovine genome sequence (Bovine Genome Sequencing and Analysis Consortium 2009) and bioinformatics tools were used for genome-wide screening of active ERVs in bovine conceptuses during the implantation period. Recent advances in DNA sequencing technology have enabled us to process increasingly large numbers of nucleotide sequences (Schuster 2008). Previously, we used microarrays to determine genes involved in the process of implantation in mice (Yoshioka et al. 2000), but microarray technology cannot identify expressed ERVs because of the high similarities in their nucleotide sequences. However, next-generation sequencers allow us to distinguish individual ERVs, detecting small nucleotide differences among them. To exploit the expanded capacity of this technology in this study, we collected bovine conceptuses of days 17, 20, and 22 (day 0 = day of estrus) from superovulated Japanese black cattle and sequenced the expressed RNAs using the SOLiD3 System released by Life Technologies (Carlsbad, CA). The sequenced tags were mapped into the bovine genome, which was also verified by quantitative polymerase chain reaction (qPCR). We analyzed the expression of env-derived sequences based on the scanned results of ERV-derived sequences in the bovine genome. Our results indicate that various ERV-derived sequences in the bovine genome are expressed during early pregnancy, suggesting that several genes from env sources may play an active role in placental development of the bovine species.
Materials and Methods
Retro Elements in the Bovine Genome
To identify ERVs in genomes, we used a computer program named RetroTector, which was designed to detect and characterize entire or fragmented ERVs in a given genome sequence (Sperber et al. 2007). Applying this program with default parameters for the Bos taurus genome (unmasked, Bos_taurus.Btau_4.0.55.dna.toplevel.fa) provided by Ensembl (http://www.ensembl.org/), we obtained four types of retrovirus-like sequences in the genome: gag, pro, pol, and env.
Animals and Sampling
All animal procedures in this study were approved by the Committee for Experimental Animals at Zen-noh Embryo Transfer (ET) Center, Hokkaido, and The University of Tokyo, Tokyo, Japan. Estrous synchronization, superovulation, and ET processes were performed as described previously (Ideta et al. 2007). Seven-day embryos (day 0 = day of estrus) were collected from superovulated Japanese black cattle. Twelve embryos derived from the superovulation were transferred nonsurgically into the uterine horn of three Holstein heifers (n = 4 each), ipsilateral to the contralateral on day 7 of the estrous cycle. Elongated conceptuses were collected nonsurgically by uterine flushing on day 17, 20, or 22. These 3 days correspond to a day before the initiation of conceptus attachment to the uterine epithelium, right after attachment, and at the beginning of adhesion-placental formation, respectively. Conceptuses in the uterine flushing media were obtained by centrifugation at 1,000 rpm for 5 min, snap frozen, and transferred to the Laboratory of Animal Breeding at The University of Tokyo.
RNA Extraction
RNA extraction from conceptus tissues (80–100 µg) was performed using Isogen (Nippon gene, Tokyo, Japan) according to the protocol provided by the manufacturer (Nagaoka et al. 2003). For the next-generation sequencer SOLiD analysis, total RNA was depleted of ribosomal RNA (rRNA) molecules using the Ribominus Eukaryote Kit (Life Technologies, Carlsbad, CA). High-throughput sequencing libraries were prepared according to the SOLiD whole-transcriptome library preparation protocol (Ashton-Beaucage et al. 2010). All primary sequencing data can be found in the DDBJ Sequence Read Archive (Kaminuma et al. 2010) under accession number DRA000549.
Mapping Sequence Reads to the Genome
Processed nucleotide sequences from the SOLiD3 for each developmental stage were aligned to the bovine genome. Beforehand, the sequences of SOLiD adapters and barcodes were filtered out. Although the length of each remaining read sequence is 50 nt, four nucleotides from the 3′-terminus were excluded for accuracy, following the SOLiD whole-transcriptome analysis protocol. The Applied Biosystems Whole Transcriptome Analysis Pipeline was used to map the short reads. In this pipeline, each read was divided into two 23-base fragments, and these fragments were mapped to the genome. During this mapping phase, we allowed up to two mismatches and removed reads that aligned to more than 10 locations. In this mapping analysis, we used the reads whose sequence quality scores were 24 or higher, following standard parameters of AB WT Analysis Pipeline. Matching locations were subsequently used to generate counts for identified ERVs and Ensemble-provided coding sequences (Bos_taurus.Btau_4.0.55.gtf.gz).
Estimation of Differential Expression
To evaluate gene expression level independent of variance in gene lengths and the number of reads among samples, we applied the widely recognized quantification measurement, reads per kilobase of exon model per million mapped reads (RPKM, Mortazavi et al. 2008) as follows:
where C and N are the number of reads that are mapped into the gene and the whole genome, respectively, and L is the length of the gene.
In Silico Evaluation of env-Derived Sequence and Primer-Binding Site
We performed a hidden Markov model (HMM) search to the candidate env-derived sequences using a computer program hmmsearch in HMMER3 (Eddy 2011) with TLV coat motif (PF00429), which was constructed by viral envelope proteins including a syncytin-1 protein in humans in the Pfam database (Finn et al. 2010). For these sequences, the number of TM regions in a sequence was estimated by the TMHMM program (Krogh et al. 2001) with default parameters. The primer-binding site (PBS) was predicted using the Genomic tRNA Database (Chan and Lowe 2009).
PCR and qPCR
SOLiD data were validated by comparing RNA quantification with eight genes including DLX3, GADD45A, IFNT, TP53, CDH1, MMP2, EGFR, and ITGA4, which are expressed during conceptus attachment and early placental development periods in the bovine species. For PCR and real-time PCR analyses of conceptus RNA, isolated RNA (1,000 ng) was reverse transcribed to cDNA using ReverTra Ace qPCR RT kit (TOYOBO, Osaka, Japan) in a 10 µl reaction volume, and the resulting cDNA (RT template) was stored at 4 °C until use. For PCR analysis of conceptus mRNA, poly(A)+ RNA was initially prepared from isolated RNA (10 µg) through the use of GenElute mRNA Miniprep Kit (Sigma-Aldrich, Tokyo, Japan), which was reverse transcribed to cDNA with ReverTra Ace in a 20 µl reaction volume. In both cases, the cDNA reaction mixture was diluted 1:10 using DNase, RNase-free molecular biology grade water, and 3 µl was taken for each amplification reaction. RT template (cDNA) was subjected to PCR or real-time PCR amplification using primers presented in supplementary table S1, Supplementary Material online. After 30 cycles, amplification products were separated on 1.5% (w/v) agarose gels. In addition, PCR products were subcloned and verified by DNA sequencing. qPCR reactions were performed using the SYBR Green kit (Takara Biomedicals, Tokyo, Japan) and the Applied Biosystems thermal cycle system (7900HT, Applied Biosystems, Tokyo, Japan) as described previously (Sakurai et al. 2009). Average threshold (Ct) values for all mRNAs examined were calculated and normalized to Ct values for ACTB mRNA. These procedures also applied to the expression analysis of functional env-derived genes.
We used the following bovine cell lines: CT-1 and F3 from trophoblasts; EPI and STR from uteri; oCG from ovarian cumulus-granulosa cells; EF from ear-fibroblast cells; bIE from intestinal epithelial cells; MDBK and CKT-1 from kidney cells; 23CLN from lymph nodes; and BoMAC from macrophage cells. The details are summarized in the supplementary materials and methods, Supplementary Material online.
To estimate the age of BERV-P insertion, genomic DNA was extracted from a B. taurus (Holstein) kidney, B. javanicus (Bali cattle), Bubalus bubalis (water buffalo), Tragelaphus spekii (sitatunga), and Ovis aries (sheep) peripheral blood mononuclear cells using lysis buffer (1 M Tris pH 7.4, 0.5 M ethylenediaminetetraacetic acid, 5 M NaCl, and 10% SDS) and purified using phenol:chloroform:isoamyl alcohol (25:24:1 v/v) and following isopropanol precipitation. Genomic PCR was performed using KOD-Plus-Neo (TOYOBO, Osaka, Japan), 10 ng genomic DNA/10 µl PCR mixture, and 0.3 µM of primer pairs described in supplementary table S2, Supplementary Material online. PCR thermal cycling condition was as follows: 1 cycle at 94 °C for 2 min, 30 cycles at 98 °C for 10 s, and 68 °C for 1 min, followed by 1 cycle at 68 °C for 7 min. PCR amplicons were electrophoresed in 0.8 or 2% agarose gels and visualized by ethidium bromide staining.
5′- and 3′-Rapid Amplification of cDNA Ends
5′- and 3′-rapid amplification of cDNA ends (RACE) was performed with the SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA) according to the manufacturer’s instructions. Prime STAR HS (TaKaRa) was used in PCR reactions. Each PCR product was subcloned into a pGEM T-Easy vector (Promega, Madison, WI), which was then subjected to sequence analysis using an ABI PRISM 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA).
Phylogenetic Analysis of Expressed env Sequences
To characterize the origin and evolution of expressed env-derived genes identified in the bovine genome, we conducted a molecular phylogenetic analysis as follows. We used amino acid sequences coded by the following endogenous envelope genes expressed during conceptus attachment and early placental development periods: syncytin-1 (NP_055405.3, Mi et al. 2000), syncytin-2 (NP_997465.1, Esnault et al. 2008), and HERV-K Env (AAF88168.1, Andersson et al. 2002) in humans; syncytin-A and syncytin-B (NM_001013751 and NM_173420.3, respectively, Dupressoir et al. 2005) in mice; syncytin-Ory1 in rabbits (ACZ58381.1, Heidmann et al. 2009); and syncytin-Car1 in dogs and cats (JN587092.1 and JN587095.1, respectively, Cornelis et al. 2012). We also used the two Env genes of the bovine endogenous viruses, BERV-K1 (BAJ72717.1) and K2 (BAJ72718.1), which were recently found to be transcribed in the bovine placenta (Baba et al. 2011). In addition, we added the following nine Env sequences of exogenous viruses: bovine leukemia virus (AAO21338.2), bovine immunodeficiency virus (AAA91274.1), Jembrana disease virus (ABB72001.1), ovine enzootic nasal tumor virus (ACX93982.1), reticuloendotheliosis virus (ACJ65654.1), simian retrovirus 4 (ADC52789.1), Mason–Pfizer monkey virus (AAA47712.1), avian leukosis virus (AAU06813.1), and Jaagsiekte sheep retrovirus (AAD45228.2). For each sequence, a furin cleavage site (consensus R/K-X-R/K-R) that divides the sequence into two subunits, the SU and the TM subunits, was identified. Because the TM subunit is known to be conserved in many retroviral elements (Bénit et al. 2001), we used the TM subunits for our subsequent phylogenetic analysis. The multiple alignments of the sequences were computed by the computer program L-INS-i in MAFFT (Katoh et al. 2005). The gapped regions of the multiple alignments were removed by trimAI with a “gappyout” option (Capella-Gutiérrez et al. 2009). For the multiple alignment, we applied a computational tool, PROTTEST 3 (Darriba 2011), that calculates an Akaike's Information Criterion (AIC) score for each amino acid replacement model with two parameters: a discrete gamma distribution to account for heterogeneity in evolutionary rates among sites (α) and an estimation of the proportion of invariant sites (P). Using the AIC scores, we selected the LG model (Le and Gascuel 2008) with α = 3.27 and P = 0.01. The phylogenetic tree was constructed using the maximum-likelihood method (Felsenstein 1981) as implemented in the RAxML v7.2.6 (Stamatakis 2006). The robustness of the phylogenetic tree was evaluated by fast bootstrapping (Stamatakis et al. 2008) with 1,000 pseudoreplicate data sets.
Results
Predicted Genomic ERV Elements
We identified 7,624 ERV-derived elements (gag-, pro-, pol-, and env-derived sequences) in the bovine genome using RetroTector (see Materials and Methods). The summary of all ERV-derived elements obtained is presented in table 1 (supplementary table S3, Supplementary Material online, for each chromosome). Of the seven genera, gammaretrovirus-derived genes represent most in our analysis (5,131 sequences). The most abundant retroelement is pol derived (3,250 sequences), followed by env, pro, and gag in the genome, a pattern also noted in the same order as in humans, chimpanzees, rhesus macaques, and dogs (Sperber et al. 2007). We sought the longest ORF for each env-derived sequence, and their lengths range from 45 to 2,085 nucleic acids (15–695 amino acids). Using amino acid sequences of these ORFs, we conducted motif searches using hmmsearch in HMMER3 (Eddy 2011) with the HMM of the PF00429 profile (TLV coat) in the Pfam database (Finn et al. 2010) that was identified in human syncytin-1. When an amino acid sequence of an env-derived sequence is similar to that of syncytin-1, the E value of the HMM search is close to 0. Because the TM domain is known to be essential for fusogenic activity (Chang et al. 2004), it is reasonable to expect that other proteins functioning in conceptus attachment have TM domains. Indeed, the TM domain is found in syncytin proteins in humans, mice, rabbits, dogs, and cats. Therefore, env-derived sequences were analyzed using a computer program TMHMM, which is widely used for the prediction of TM domains in a given amino acid sequence (Krogh et al. 2001). Results of these env-derived sequences examined are summarized with given IDs in supplementary table S4, Supplementary Material online.
Table 1.
Distribution of ERV-Derived Elements in the Bovine Genome
| α | β | γ | δ | ε | G | s | Sum | |
|---|---|---|---|---|---|---|---|---|
| gag | 3 | 303 | 927 | 77 | 0 | 23 | 7 | 1,340 |
| pro | 5 | 359 | 1,014 | 65 | 0 | 10 | 39 | 1,492 |
| pol | 25 | 868 | 2,125 | 106 | 0 | 7 | 119 | 3,250 |
| env | 11 | 311 | 1,065 | 56 | 1 | 2 | 96 | 1,542 |
| Sum | 44 | 1,841 | 5,131 | 304 | 1 | 42 | 261 | 7,624 |
Note.—The symbols represent the following genera: α, alpharetroviruses; β, betaretroviruses; γ, gammaretroviruses; δ, deltaretroviruses; ε, epsilonretroviruses; G, Gypsy elements; and s, spumaviruses.
Mapping Short Reads and Its Validation
We mapped short reads from SOLiD sequencer into the bovine genome for the early pregnancy period (days 17, 20, and 22). The detailed information of the short reads obtained from SOLiD sequencer and mapped into the genome is summarized in table 2. The relative frequency of the sequence quality scores of mapped short reads for each day is shown in supplementary figure S1, Supplementary Material online. We then examined the expression level of various gene types annotated by Ensembl: protein coding, rRNA, tRNA, micro RNA (miRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), other functional nuclear RNA, retrotransposed element, pseudogene, mitochondrial rRNA, and mitochondrial tRNA. As the numbers of mapped reads differ in different samples and the lengths of different genes vary, we calculated RPKM values for all genes examined to compare gene expression levels in this study (see Materials and Methods). The RPKM values were validated using qPCR assays as summarized in supplementary figure S2, Supplementary Material online.
Table 2.
The Number of Mapped SOLiD Reads
| Sample | Reads Processed from the Sequencer | Reads Mapped | Reads Used in This Study | Reads Filtered | Reads with More Than 10 Mappings | Read Counts Allowing Multiple Hits within 10 Times |
|---|---|---|---|---|---|---|
| Day 17 | 91,084,477 | 68,482,322 (75.2%) | 56,508,808 | 11,973,514 | 8,115,964 | 172,435,337 |
| Day 20 | 85,022,587 | 65,176,129 (76.7%) | 53,530,938 | 11,645,191 | 7,927,851 | 142,294,526 |
| Day 22 | 82,230,027 | 61,700,453 (75.0%) | 50,664,880 | 11,035,573 | 7,026,966 | 139,083,864 |
RNA-Seq Analysis
Various snoRNAs, 60–150 nt long noncoding RNAs that guide modifications of selected nucleotides in rRNAs or spliceosomal RNAs (Kiss 2001, 2002; Bachellerie et al. 2002), were highly expressed in each day. Analyzing RPKM values of Ensembl annotations, we found snoRNA accounted for 82, 79, and 91 of the top 100 most highly expressed genes for days 17, 20, and 22 as presented in supplementary tables S5, S6, and S7, Supplementary Material online, respectively, despite snoRNA making up only 1.9% of all Ensembl-annotated features in the cattle genome (586 of 30,273). For example, SNORD100 (HBII-429) showed the highest RPKM values among snoRNAs for each day (supplementary tables S5–S7 and fig. S3, Supplementary Material online). snoRNAs are 60–150 nt long noncoding RNAs that guide modifications of selected nucleotides in rRNAs or spliceosomal RNAs (Kiss 2001, 2002; Bachellerie et al. 2002). A large amount of protein products during conceptus development period could be provided in part by snoRNA expression. Although rRNAs are usually the most abundant in cells, we excluded rRNA from our sample preparation (see Materials and Method). The data also showed that other noncoding RNAs such as snRNAs and miRNAs were abundant in the genome. These results indicated that various noncoding RNAs may be involved in developmental processes of bovine conceptuses.
Next, we examined the expression levels of env-derived sequences for each day of trophoblast development. It was found that the numbers of env-derived sequences with one or more mapped reads (i.e., possibly expressed env-derived sequences) were 178, 150, and 157 for days 17, 20, and 22, respectively (supplementary table S4, Supplementary Material online), although most of the env-derived sequences exhibited small RPKM values (supplementary fig. S4, Supplementary Material online). Among 1,542 env-derived elements predicted by RetroTector in the bovine genome (table 1), 146 sequences were detected on one day, whereas 75 were detected on 2 days, and 63 were detected on all 3 days (fig. 1). In addition, we analyzed the expressed ERV elements of each day (fig. 2A). Comparing the proportion of ERVs expressed with those in the genome (fig. 2B), we found that the proportion of ERVs differs, depending on the types of ERVs (for each day, P < 0.01, Pearson's χ2 test). The proportion of expressed env- and pol-derived sequences exceeded that of detected ERVs for each day, whereas the proportions of expressed pro-derived sequences were smaller than those found in the genome. These results indicate that some pol-derived sequences and env-derived ones might be involved in trophectoderm and placental development (see Discussion).
Fig. 1.—
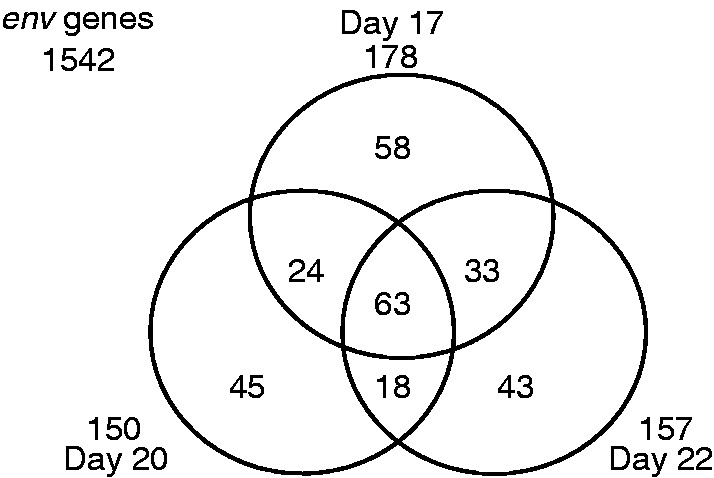
Summary of env-derived sequences detected by the SOLiD sequencing. This Venn diagram exclusively shows the number of shared env-derived sequences detected by the SOLiD sequencing for day 17 (top middle), day 20 (bottom left), or day 22 (bottom right).
Fig. 2.—
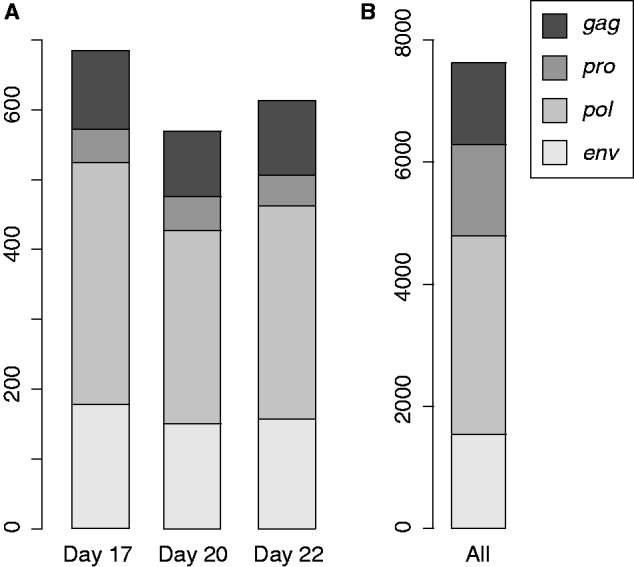
The frequency of possibly expressed ERV elements for each day and ERV elements in the genome. The frequencies of each ERV element with SOLiD reads for each day (A) and predicted in the genome (B) are shown. Colored bars as defined in the key indicate the numbers of endogenous elements without overlapping.
Searching for env-Derived Elements Expressed during Placentation
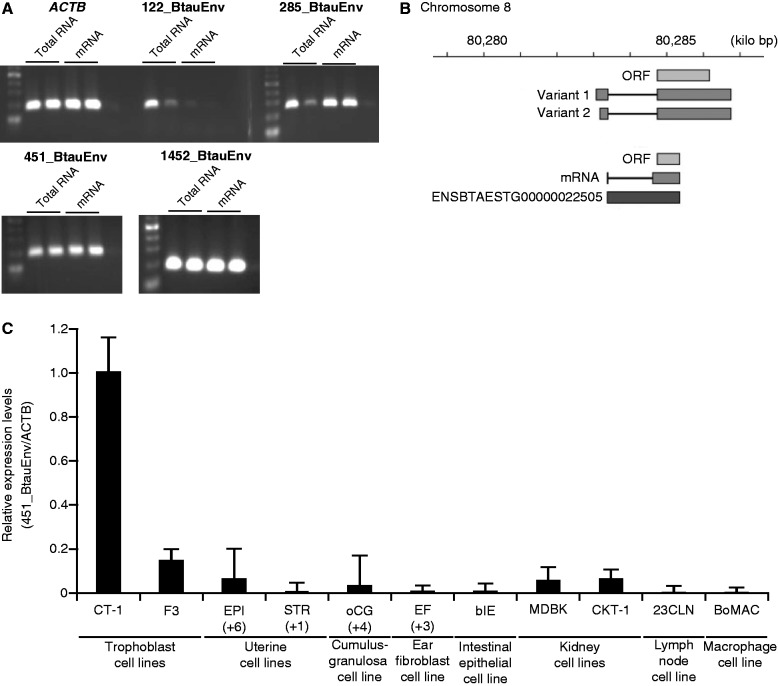
We experimentally validated the expression of several env-derived sequences that meet all the following criteria: 1) was identified as an env-derived sequence by RetroTector, 2) possessed an ORF of at least 100 amino acids, 3) was detected on at least 2 days by the SOLiD sequencing, 4) had the TLV coat motif with an E value less than 0.01, and 5) had at least one membrane domain identified through the TMHMM search. Using these five criteria, four env-derived genes expressed in conceptuses before placental development were identified (table 3). mRNAs corresponding to these candidates were characterized by means of PCR with primers presented in supplementary table S8, Supplementary Material online, and RNA/mRNA isolated from bovine trophoblast CT-1 cell lines. Although numerous PCR products existed when total RNA was examined, three candidates (285_BtauEnv, 451_BtauEnv, and 1452_BtauEnv) clearly showed specific bands when poly(A)+ RNA was subjected to PCR analysis (fig. 3A). The nucleotide sequences of the three PCR products were verified by homology search against the bovine genome (supplementary table S8, Supplementary Material online), although similar sequences were also found in other genomic loci. The gene with the longest ORF, 451_BtauEnv was subjected to further analyses.
Table 3.
env-Derived Sequences that Are Selected for Experimental Validation
| ID | Chromosome | Position | Length (aa) | HMMa (E Value) | TMHMMb |
|---|---|---|---|---|---|
| 122_BtauEnv | 1 | 131,679,101–131,680,526 | 218 | 8.5E-18 | 1 |
| 285_BtauEnv | 6 | 109,530,334–109,531,763 | 170 | 8.6E-21 | 1 |
| 451_BtauEnv (BERV-P env) | 8 | 80,284,742–80,286,268 | 477 | 2.5E-17 | 2 |
| 1452_BtauEnv | 29 | 40,102,194–40,103,478 | 103 | 0.000051 | 1 |
aAn E value was calculated by hmmsearch in HMMER3 with the TLV coat motif profile (PF00429) in the Pfam database.
bThe number of TM regions identified by TMHMM with default parameters.
Fig. 3.—
Validation of env-derived sequences detected by SOLiD sequencing. Transcripts of env-derived genes were validated by PCR (A). Total RNA (1,000 ng, n = 2 each) or poly(A)+ RNA (30 ng, n = 2 each), isolated from CT-1 cells, was reverse transcribed and subjected to PCR analysis with primers presented in supplementary table S8, Supplementary Material online. ACTB mRNA was used as an internal control. (B) Chromosomal locations of BERV-P mRNA variants. 5′- and 3′-RACE was used to clone the entire BERV-P cDNA. There are two variants of BERV-P, resulting from difference in lengths of 5′-noncoding regions. BERV-P transcripts were found in various bovine cell lines and primary cells (C). Total RNAs (1,000 ng, n = 2 each), isolated from trophoblast cell lines (CT-1 or F3), endometrial epithelial (EPI) or stromal (STR) primary cells, ovarian cumulus granulosa (oCG) primary cells, ear-derived fibroblast (EF) primary cells, intestinal epithelial cell line (bIE), kidney cell lines (MDBK or CKT-1), lymph node cell line (23CLN), or macrophage cell line (BoMAC), were subjected to real-time PCR for BERV-P transcripts with primers presented in supplementary table S8, Supplementary Material online. ACTB transcript was used as endogenous reference gene product. The expression of the transcript was normalized to the expression of ACTB measured in the same cDNA preparation. The values are shown as the mean ± standard error of the mean.
We first identified the sequence of their PBS of 451_BtauEnv as TGGCTCGTCCGGGAT that was complementary to the 3′-sequence of proline tRNAs. This PBS sequence is nearly identical to that of bovine leukemia virus (Sagata et al. 1984), but their env sequences are entirely different (data shown later). Therefore, 451_BtauEnv was named bovine ERV-P (BERV-P) env. Neither gag nor pol ORF sequences were found in BERV-P. We applied RACE to obtain the full-length sequence of the RNA transcript found within CT-1 cell line, identifying two mRNA variants at 1) 80,283,071–80,283,394 and 80,284,743–80,286,755 and 2) 80,283,174–80,283,394 and 80,284,743–80,286,755 on chromosome 8 (fig. 3B). Although an EST annotated in Ensembl database as ENSBTAESTT00000022505 overlapped with BERV-P, its ORF is different from that of the BERV-P env sequence. We also verified the expression of BERV-P env in trophoblast cell lines (fig. 3C). These results indicate that BERV-P env genes can be transcribed during trophectoderm development before placentation.
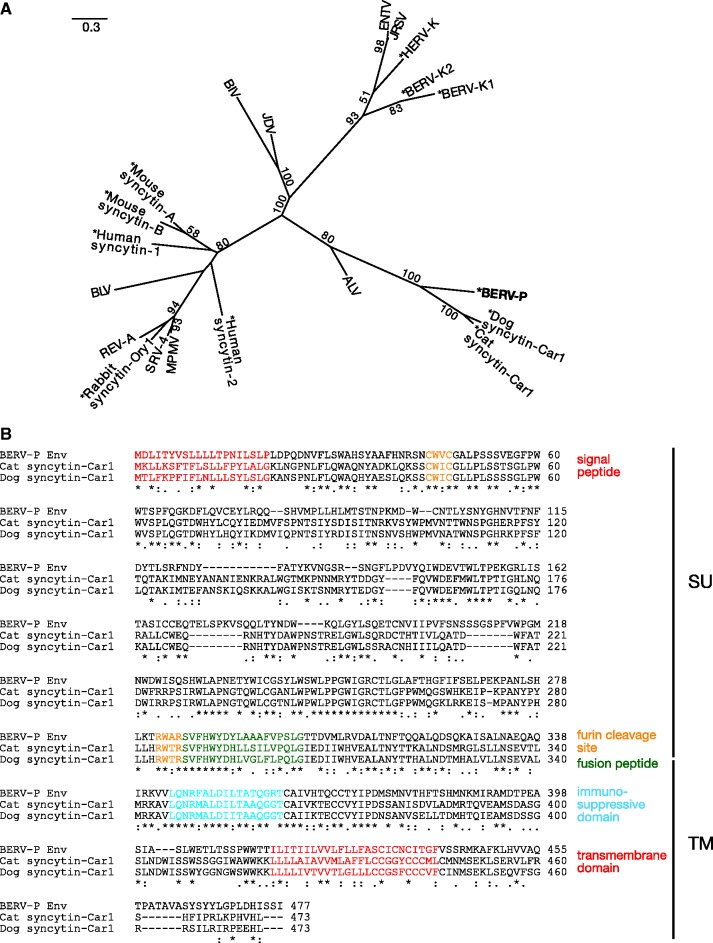
We conducted the molecular phylogenetic analysis of BERV-P Env with the 10 ERV-derived Env amino acid sequences, including BERV-K1 and K2, which are known to be expressed in the bovine placenta (Baba et al. 2011), and eight Env sequences of exogenous retroviruses (fig. 4A, see Materials and Methods). The multiple alignment of the sequence of the TM subunits used in this study was shown in supplementary figure S5, Supplementary Material online. The phylogenetic tree indicated that the BERV-P env is a member of the clade containing syncytin-Car1 in dogs and cats, which is supported by its high bootstrap value. Indeed, all regions, especially for functionally important regions such as signal peptide, furin cleavage site, fusion peptide, immunosuppressive domain, and TM domain, are quite similar among them (fig. 4B).
Fig. 4.—
The feature of BERV-P. Maximum-likelihood tree was constructed based on the amino acid sequences of TM regions of Env from BERV-P (bold) and 19 endogenous/exogenous retroviruses (A, see Materials and Methods). The scale of tree was shown in upper right considering the evolutionary distances used to infer the phylogenetic tree. An asterisk indicates an endogenous retroviral element. Percent bootstrap values obtained from 1,000 replicates are indicated (≥50) at branch junctions. The multiple alignment of BERV-P and syncytin-Car1 in dogs and cats (B). Characteristic structural features were shown in a color at each site. An asterisk indicates amino acid identity, and a colon indicates amino acid similarity observed at each site.
It has been reported that the syncytin-Car1 is not found in the bovine genome (Cornelis et al. 2012). However, to confirm that the origin of BERV-P env is different from that of syncytin-Car1, we examined 1) whether the BERV-P env exists in the genomes of dogs and cats and 2) whether syncytin-Car1 exists in the genome of bovines. For this purpose, we identified the orthologous genes located upstream and downstream of BERV-P env in dogs and cats, as well as those of syncytin-Car1 in bovines (supplementary fig. S6, Supplementary Material online). From the locations of these genes, we successfully identified syntenic regions of BERV-P env in the dog and cat genomes, as well as that of syncytin-Car1 in the bovine genome. We then conducted homology searches (SSEARCH, Goujon et al. 2010) with BERV-P env or syncytin-Car1 sequences against each corresponding synteny sequences. However, as a result, no corresponding regions were found in either synteny regions of BERV-P env in the genomes of dogs and cats or syncytin-Car1 in the bovine genome. The details of the homology searches were summarized in supplementary table S9, Supplementary Material online. Therefore, the origins are different between BERV-P env and syncytin-Car1. On the other hand, high similarities observed between BERV-P env and syncytin-Car1 indicated that the molecular function of BERV-P env might be related to trophectoderm development in the bovine species.
Discussion
In an attempt to identify transcripts possibly derived from retroelements, BERV-P env was found to be expressed in placenta in this study. Although we anticipated that a BERV-P env would exhibit trophoblast-specific expression, this mRNA was expressed in various organs at low levels relative to those in the trophoblast (fig. 3C). Similarly, it was recently found that syncytin-1 elements were transcribed in various human tissues and cells (Li et al. 2011). These results suggest that diversified expression of the syncytin-1 family results from both LTR-directed transcription and leaky transcription of syncytin-1 genes in normal human tissues. The same explanation could be applied to the bovine element, BERV-P env, identified in this study. Alternatively, the BERV-P env could require a specific receptor for its action. Although studies on receptor identification of BERV-P env are in progress, a specific receptor to this env-derived element has not been identified. Together with receptor identification and its expression, potential influences of genomic structure and orientation on the expression levels of individual env-derived elements in various bovine tissues require further systematic investigation.
Although syncytins have been characterized as having fusogenic and immunosuppressive activities in conceptuses in several mammals, a bovine counterpart has not been identified. One of the reasons is due to different origins of syncytins in various mammals. In terms of evolution, however, it is quite unlikely that the placenta of different mammals emerged independently in a convergent manner considering its essentiality in placental mammals. Indeed, coding sequences of syncytin-Car1 genes found in Carnivora families are under purifying selection, suggesting their functional importance in placentation (Cornelis et al. 2012). On the other hand, the morphological features of the placenta are also quite diversified among mammals, although their functions are similar (Wildman et al. 2006; Rawn and Cross 2008; Bazer et al. 2009). These evolutionarily complicated relationships could be explained simply by the fact that on several occasions, retroviral infections have resulted in the independent capture and replacement of genes for a convergent physiological role in placental development (Harris 1998; Palmarini 2004; Dupressoir et al. 2005; Heidmann et al. 2009). For example, the morphology of placenta is cotyledons or discoids for cows or humans, respectively. That may be because various genes involved in placenta development differ in different species.
Indeed, the expressed env-derived gene from BERV-P found on chromosome 8 in the bovine genome may have been acquired specifically in the bovine species, supported by the following results: a) the BERV-P was not found in the dog and cat genomes of each synteny block on chromosome 11 and D4, respectively (supplementary fig. S6A, Supplementary Material online); b) only a 2.8% nucleotide differences was observed between 5′- and 3′-LTR sequences (15 of 530 bp) in the BERV-P (supplementary fig. S7A, Supplementary Material online); and c) all Env sequences containing the same furin recognition motif, which were found in the bovine genome, show high similarity (supplementary fig. S7B, Supplementary Material online). Our SOLiD data also suggests that, although many of the expressed env-derived genes identified in this study might be artifacts or transcriptional noise, some of them may have acquired functions for processes leading to placentation. In addition, we conducted a genomic PCR amplification of BERV-P env sequence using the closely related species such as B. javanicus (Bali cattle), Bub. bubalis (water buffalo), T. spekii (sitatunga), and O. aries (sheep). It was found that among the species examined, the BERV-P existed only in the Bali cattle genome (supplementary fig. S8, Supplementary Material online). These results suggest the possibility that BERV-P recently infected in the Bos lineage has been functional for placentation. It is also possible that BERV-P might be inserted into the genome of an ancestor of the Bos lineage but deleted in other lineages. In either case, multiple insertions and deletions of ERV may have resulted in the independent captures of genes involved in placental development among mammals.
The proportion of expressed pol-derived genes and env-derived ones was observed to have exceeded that of the entire ERV for each day (fig. 2), indicating potential involvement of pol-derived genes for placentation. Indeed, PEG10 and PEG11/RTL1 originally from gag and pol regions of Sushi-ichi retrotransposons (Ono et al. 2001) are also found to be essential for placental development, although details of the molecular mechanisms associated with placental formation are unknown (Ono et al. 2006; Sekita et al. 2008). Interestingly, both genes were inserted before divergence of the eutherians and marsupials and are conserved among various placental mammals including cattle (Suzuki et al. 2007; Edwards et al. 2008). These results suggest that various ERV-derived genes may be involved in placentation, although their molecular function and evolutionary pathways differ greatly.
In this study, we found that among 1,542 possible env-derived sequences predicted in silico, 284 env-derived sequences were identified through our next-generation sequencing analysis. On the other hand, because of lower global DNA methylation levels in the placenta compared with other tissues, various ERV-derived genes including nonfunctional ones can be expressed in placenta (Kudaka et al. 2008). Therefore, we experimentally validated the expression of several ERV-derived elements, sequences of which were captured by the SOLiD3. To the best of our knowledge, this is the first report on a whole-transcriptome analysis in bovine conceptuses during the periattachment period, although RNA-seq studies of human and rat placenta were recently reported with no details of endogenous retroviral elements (Kim et al. 2012; Shankar et al. 2012). This provides justification for further analyses on these retroelements in the bovine genome and is evident that comprehensively analyzing large amounts of expression data from conceptus tissues are important in understanding the evolutionary dynamics of genes involved in placental development.
Supplementary Material
Supplementary materials and methods, figures S1–S8, and tables S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Drs A. Ideta and Y. Aoyagi (Zen-noh ET center, Hokkaido, Japan) for generously providing bovine conceptus tissues and Dr Jun Zhou for the comments on this work. We also appreciate Mr. T. Ishikura, Ms. K. Azuma, and Dr H. Hanaoka (Life Technologies, Tokyo, Japan) for the execution of SOLiD3 analysis and for critical discussion throughout the course of the study. This work was supported by JSPS Research Fellowship for Young Scientists to S.N. and H.B. and the Program for Promotion of Basic Research Activities for Innovative Bioscience (BRAIN, http://www.naro.affrc.go.jp/brain/).
Literature Cited
- Andersson AC, et al. Developmental expression of HERV-R (ERV3) and HERV-K in human tissue. Virology. 2002;297:220–225. doi: 10.1006/viro.2002.1428. [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D, et al. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell. 2010;143:251–262. doi: 10.1016/j.cell.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Baba K, et al. Identification of novel endogenous betaretroviruses which are transcribed in the bovine placenta. J Virol. 2011;85:1237–1245. doi: 10.1128/JVI.01234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Bazer FW, Spencer TE, Johnson GA, Burghardt RC, Wu G. Comparative aspects of implantation. Reproduction. 2009;138:195–209. doi: 10.1530/REP-09-0158. [DOI] [PubMed] [Google Scholar]
- Bénit L, Dessen P, Heidmann T. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J Virol. 2001;75:11709–11719. doi: 10.1128/JVI.75.23.11709-11719.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovine Genome Sequencing and Analysis Consortium. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Chen PT, Chang GD, Huang CJ, Chen H. Functional characterization of the placental fusogenic membrane protein syncytin. Biol Reprod. 2004;71:1956–1962. doi: 10.1095/biolreprod.104.033340. [DOI] [PubMed] [Google Scholar]
- Cornelis G, et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci U S A. 2012;109:E432–E441. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, et al. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 2008;6:e135. doi: 10.1371/journal.pbio.0060135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, et al. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc Natl Acad Sci U S A. 2008;105:17532–17537. doi: 10.1073/pnas.0807413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, et al. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JR. Placental endogenous retrovirus (ERV): structural, functional, and evolutionary significance. Bioessays. 1998;20:307–316. doi: 10.1002/(SICI)1521-1878(199804)20:4<307::AID-BIES7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new "syncytin" in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideta A, Urakawa M, Aoyagi Y, Saeki K. Early development in utero of bovine nuclear transfer embryosusing early G1 and G0 phase cells. Cloning Stem Cells. 2007;9:571–580. doi: 10.1089/clo.2007.0017. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kaminuma E, et al. DDBJ launches a new archive database with analytical tools for next-generation sequence data. Nucleic Acids Res. 2010;38:D33–D38. doi: 10.1093/nar/gkp847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. Transcriptome landscape of the human placenta. BMC Genomics. 2012;13:115. doi: 10.1186/1471-2164-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Krysan DJ, Rockwell NC, Fuller RS. Quantitative characterization of furin specificity. Energetics of substrate discrimination using an internally consistent set of hexapeptidyl methylcoumarinamides. J Biol Chem. 1999;274:23229–23234. doi: 10.1074/jbc.274.33.23229. [DOI] [PubMed] [Google Scholar]
- Kudaka W, Oda T, Jinno Y, Yoshimi N, Aoki Y. Cellular localization of placenta-specific human endogenous retrovirus (HERV) transcripts and their possible implication in pregnancy-induced hypertension. Placenta. 2008;29:282–289. doi: 10.1016/j.placenta.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Li F, Nellâker C, Yolken RH, Karlsson H. A systematic evaluation of expression of HERV-W elements; influence of genomic context, viral structure and orientation. BMC Genomics. 2011;12:22. doi: 10.1186/1471-2164-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nagaoka K, et al. Regulation of blastocyst migration, apposition, and initial adhesion by a chemokine, interferon gamma-inducible protein 10 kDa (IP-10), during early gestation. J Biol Chem. 2003;278:29048–29056. doi: 10.1074/jbc.M300470200. [DOI] [PubMed] [Google Scholar]
- Ng VL, Wood TG, Arlinghaus RB. Processing of the env gene products of Moloney murine leukaemia virus. J Gen Virol. 1982;59:329–343. doi: 10.1099/0022-1317-59-2-329. [DOI] [PubMed] [Google Scholar]
- Ono R, et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–237. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- Ono R, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38:101–106. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- Palmarini M, Mura M, Spencer TE. Endogenous betaretroviruses of sheep: teaching new lessons in retroviral interference and adaptation. J Gen Virol. 2004;85:1–13. doi: 10.1099/vir.0.19547-0. [DOI] [PubMed] [Google Scholar]
- Rawn SM, Cross JC. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol. 2008;24:159–181. doi: 10.1146/annurev.cellbio.24.110707.175418. [DOI] [PubMed] [Google Scholar]
- Sagata N, Yasunaga T, Ogawa Y, Tsuzuku-Kawamura J, Ikawa Y. Bovine leukemia virus: unique structural features of its long terminal repeats and its evolutionary relationship to human T-cell leukemia virus. Proc Natl Acad Sci U S A. 1984;81:4741–4745. doi: 10.1073/pnas.81.15.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, et al. Induction of endogenous interferon tau gene transcription by CDX2 and high acetylation in bovine nontrophoblast cells. Biol Reprod. 2009;80:1223–1231. doi: 10.1095/biolreprod.108.073916. [DOI] [PubMed] [Google Scholar]
- Schuster SC. Next-generation sequencing transforms today’s biology. Nat Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Sekita Y, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 2008;40:243–248. doi: 10.1038/ng.2007.51. [DOI] [PubMed] [Google Scholar]
- Shankar K, et al. RNA-seq analysis of the functional compartments within the rat placentation site. Endocrinology. 2012;153:1999–2011. doi: 10.1210/en.2011-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber GO, Airola T, Jern P, Blomberg J. Automated recognition of retroviral sequences in genomic data—RetroTector. Nucleic Acids Res. 2007;35:4964–4976. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Suzuki S, et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RA. The discovery of endogenous retroviruses. Retrovirology. 2006;3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildman DE, et al. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc Natl Acad Sci U S A. 2006;103:3203–3208. doi: 10.1073/pnas.0511344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, et al. Determination of genes involved in the process of implantation: application of GeneChip to scan 6500 genes. Biochem Biophys Res Commun. 2000;272:531–538. doi: 10.1006/bbrc.2000.2818. [DOI] [PubMed] [Google Scholar]