Abstract
Hybridization and abiotic stress are natural agents hypothesized to influence activation and proliferation of transposable elements in wild populations. In this report, we examine the effects of these agents on expression dynamics of both quiescent and transcriptionally active sublineages of long terminal repeat (LTR) retrotransposons in wild sunflower species with a notable history of transposable element proliferation. For annual sunflower species Helianthus annuus and H. petiolaris, neither early generation hybridization nor abiotic stress, alone or in combination, induced transcriptional activation of quiescent sublineages of LTR retrotransposons. These treatments also failed to further induce expression of sublineages that are transcriptionally active; instead, expression of active sublineages in F1 and backcross hybrids was nondistinguishable from, or intermediate relative to, parental lines, and abiotic stress generally decreased normalized expression relative to controls. In contrast to findings for early generation hybridization between H. annuus and H. petiolaris, ancient sunflower hybrid species derived from these same two species and which have undergone massive proliferation events of LTR retrotransposons display 2× to 6× higher expression levels of transcriptionally active sublineages relative to parental sunflower species H. annuus and H. petiolaris. Implications and possible explanations for these findings are discussed.
Keywords: LTR retrotransposon, TE activation, hybridization, abiotic stress, gypsy, copia
Introduction
Much of genome size variation among plant lineages can be ascribed to copy number differences of mobile genetic elements known as long terminal repeat (LTR) retrotransposons. T3/gypsy and Ty1/copia elements represent the two main superfamilies of autonomous LTR retrotransposons in plants (Kumar and Bennetzen 1999; Wicker et al. 2007) and are typically 4 to approximately 15 kb in length and possess an interior coding region normally harboring two genes and multiple domains involved in different steps of element transposition (Llorens et al. 2011). Flanking the coding region are terminal repeats (the LTRs) that are identical in sequence at the time of element insertion and that harbor regulatory sequences involved in element activation (Grandbastien et al. 1997; Takeda et al. 1999; Beguiristain et al. 2001).
The LTR retrotransposon replication cycle involves mobilization via a copy-and-paste mechanism whereby individual elements serve as transcriptional templates for the synthesis of mRNAs that are reverse transcribed before integration at new chromosomal positions as double-stranded DNA molecules (Kumar and Bennetzen 1999; Sabot and Schulman 2006). LTR retrotransposons thus can achieve exceptionally high copy numbers and have come to represent the majority of nuclear DNA of many organisms.
Because of mutational effects of transposable element insertions near or within native genes, host genomes have evolved mechanisms to disrupt steps of the LTR retrotransposon replication cycle (Zilberman and Henikoff 2004; Slotkin and Martienssen 2007; Lisch 2009). Notwithstanding these mechanisms of repression, several biotic and abiotic phenomena have been linked to LTR retrotransposon activation and/or proliferation. Hybridization has been demonstrated to activate LTR retrotransposons in several species groups (Waugh O'Neill et al. 1998; Labrador et al. 1999; Liu and Wendel 2000), presumably due to the disruption of host repression mechanisms as a consequence of merging differentiated genomes. Exposure to biotic and abiotic stressors also has been implicated in element derepression and copy number amplification (Wessler 1996; Grandbastien 1998). Interestingly, regulatory sequences in LTRs bear strong resemblance to endogenous stress-responsive plant genes, thus providing a potential mechanism through which stress-induced transcriptional activation might occur (Takeda et al. 1999; Salazar et al. 2007; Woodrow et al. 2011). The relative importance of hybridization and stress as triggers of LTR retrotransposon activation in natural populations is not well known, however, nor have these potential triggers been simultaneously evaluated experimentally.
Wild sunflower species in the genus Helianthus provide an excellent system in which to examine ecological and evolutionary dynamics of LTR retrotransposon derepression and proliferation and the potential roles of hybridization and abiotic stress in these processes. Among annuals of Helianthus, three species (Helianthus anomalus, H. deserticola, and H. paradoxus) independently experienced large-scale proliferation events of T3/gypsy elements (Ungerer et al. 2006, 2009; Staton et al. 2009) and to a lesser extent Ty1/copia elements (Kawakami et al. 2010). Both hybridization and abiotic stress have played important roles in the evolutionary history of these sunflower hybrid taxa. The three hybrid taxa derive from hybridization events hypothesized to have occurred 0.5–1 Myr ago between two annual sunflower species, H. annuus and H. petiolaris, and are locally adapted to harsh abiotic conditions: H. anomalus and H. deserticola inhabit desert-like conditions of the southwestern United States, whereas H. paradoxus is restricted to marsh environments mostly in New Mexico and Texas with high concentrations of salt and other ions (Welch and Rieseberg 2002; Karrenberg et al. 2006).
In this report, we examine transcriptional dynamics of multiple distinct sublineages of T3/gypsy and a single sublineage of Ty1/copia in greenhouse synthesized, early generation H. annuus × H. petiolaris hybrid genotypes under both control and abiotic stress treatments and in the ancient hybrid species under control conditions. We demonstrate that, unlike many previous reports, hybridization and abiotic stress treatments do not facilitate or further induce LTR retrotransposon expression. Sublineage-specific elements of Ty3/gypsy that proliferated in the sunflower hybrid taxa were transcriptionally active in all samples assayed but expressed at far higher levels in the ancient hybrid taxa.
Results
Transcriptional Dynamics in H. annuus, H. petiolaris, and Interspecific F1 and Backcross Hybrid Genotypes under Control and Stress Conditions
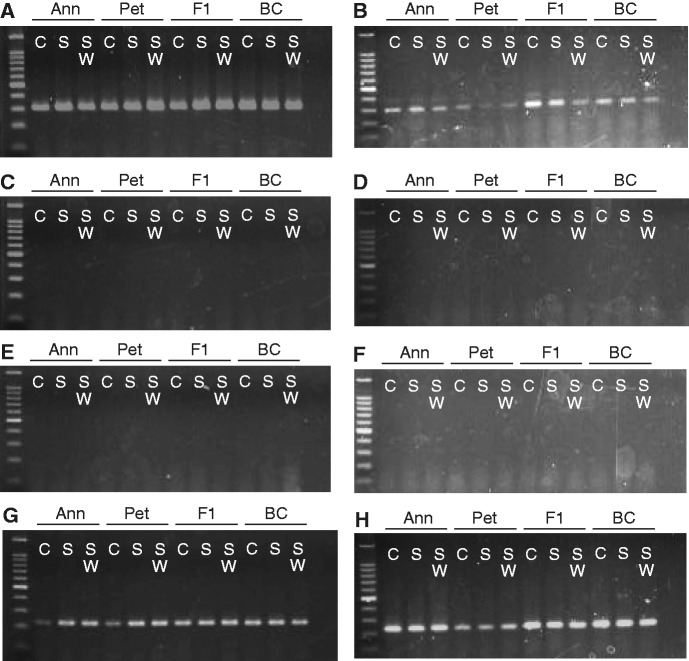
Transcriptional activity of three Ty3/gypsy sublineages and a single Ty1/copia sublineage was assayed via RT-PCR for each of 10–12 H. annuus, H. petiolaris, and interspecific F1, and BC hybrid individuals (supplementary tables S1 and S2 and fig. S1, Supplementary Material online) under control (C) and two experimental stress conditions 42 and 70 days postgermination. The Ty3/gypsy sublineages correspond to phylogenetically well-supported sublineages A, C, and E reported in Ungerer et al. (2009), and the Ty1/copia sublineage corresponds to sublineage B′ reported in Kawakami et al. (2010). Transcriptional activity was observed for one of the Ty3/gypsy sublineages (Sublineage A; fig. 1A and B) and the single Ty1/copia sublineage (fig. 1G and H) across all treatments and at both sampling time points. These same lineages were shown previously to be transcriptionally active in natural H. annuus × H. petiolaris hybrid zones and adjacent stands of H. annuus and H. petiolaris (Kawakami et al. 2011) and to have undergone proliferation events independently in three diploid hybrid species derived from H. annuus and H. petiolaris (Ungerer et al. 2006; Kawakami et al. 2010). The remaining T3/gypsy sublineages assayed (sublineages C and E) were transcriptionally inactive under all treatments and at both sampling time points (fig. 1C–F). These transcriptional patterns were invariant across all individuals sampled regardless of genotype. For brevity, results for single individuals of each genotype/treatment/time point combination are shown in figure 1. Representative positive and negative controls reactions for RT-PCR are shown in supplementary figure S2, Supplementary Material online.
Fig. 1.—
RT-PCR results for three sublineages of Ty3/gypsy and one sublineage of Ty1/copia LTR retrotransposons for H. annuus (Ann), H. petiolaris (Pet), and their F1and backcross (BC) hybrids. Lane C, control treatment; lane S, salt treatment; and lane SW, salt + wound treatment. (A) and (B), Ty3/gypsy sublineage A; (C) and (D), Ty3/gypsy sublineage C; (E) and (F), Ty3/gypsy sublineage E; and (G) and (H), Ty1/copia. (A), (C), (E), and (G): 42 days postgermination and (B), (D), (F), and (H): 70 days postgermination.
Transcriptionally active Ty3/gypsy and Ty1/copia lineages were assayed by quantitative PCR to determine the effects of experimental treatments on expression levels. Analysis of variance revealed significant effects of Treatment, Time point, Genotype × Time point, and Treatment × Time point for both Ty3/gypsy and Ty1/copia expression responses (table 1); a significant interaction effect of Genotype × Treatment was additionally observed for Ty3/gypsy expression (table 1), and a significant effect of Genotype was additionally observed for Ty1/copia expression (table 1). Figures 2 and 3 illustrate patterns of quantitative transcriptional response for Ty3/gypsy and Ty1/copia, respectively, under different experimental treatments and at different sampling time points. Ty3/gypsy and Ty1/copia expression levels were higher, on average, at the second sampling time point (figs. 2B and 3B). Exposure to stress reduced Ty3/gypsy and Ty1/copia expression although this response only was evident at the second sampling time point (figs. 2B and 3B). The significant effect of Genotype on Ty1/copia expression (table 1) was manifest as higher expression in H. annuus versus H. petiolaris for most Time point × Treatment combinations with expression in interspecific hybrids generally nondistinguishable from or intermediate relative to parental lines (fig. 3). In no instances were expression levels in hybrid genotypes significantly higher than those measured for parental lines.
Table 1.
ANOVA Results for Transcriptional Response of Gypsy and Copia LTR Retrotransposons under Control and Stress Treatments
| Source | df | SS | F | P |
|---|---|---|---|---|
| Gypsy | ||||
| Genotype | 4 | 11.151731 | 2.1848 | 0.0717 |
| Treatment | 2 | 98.534610 | 38.6081 | <0.0001 |
| Time point | 1 | 18.316558 | 14.3537 | 0.0002 |
| Genotype × Treatment | 8 | 30.482701 | 2.9860 | 0.0034 |
| Genotype × Time point | 4 | 14.950350 | 2.9289 | 0.0218 |
| Treatment × Time point | 2 | 35.788266 | 14.0227 | <0.0001 |
| Genotype × Treatment × Time point | 8 | 11.266268 | 1.1036 | 0.3619 |
| Error | 215 | 274.35869 | ||
| Copia | ||||
| Genotype | 4 | 73.049805 | 29.6429 | <0.0001 |
| Treatment | 2 | 14.513961 | 11.7793 | <0.0001 |
| Time point | 1 | 16.502056 | 26.7855 | <0.0001 |
| Genotype × Treatment | 8 | 6.999687 | 1.4202 | 0.1893 |
| Genotype × Time point | 4 | 10.145782 | 4.1171 | 0.0031 |
| Treatment × Time point | 2 | 15.253429 | 12.3794 | <0.0001 |
| Genotype × Treatment × Time point | 8 | 3.245660 | 0.6585 | 0.7276 |
| Error | 215 | 132.45746 | ||
Note.—ANOVA, analysis of variance.
Fig. 2.—
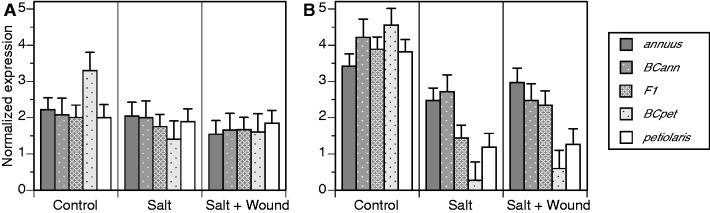
Quantitative PCR results for Ty3/gypsy sublineage A expression in H. annuus, H. petiolaris, and their F1 and backcross hybrids under control, salt, and SW treatments. (A) Assays performed 42 days postgermination and (B) assays performed 70 days postgermination. Sample sizes are as follows: H. annuus, n = 8–12; BCann, n = 5–6; F1, n = 8–11; BCpet, n = 5–6; and H. petiolaris, n = 7–10. Error bars indicate 1 SE.
Fig. 3.—
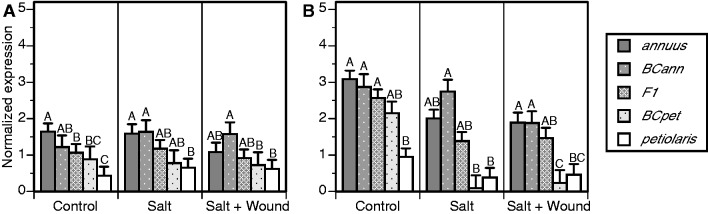
Quantitative PCR results for Ty1/copia expression in H. annuus, H. petiolaris, and their F1and backcross hybrids under control, salt, and SW treatments. (A) Assays performed 42 days postgermination and (B) assays performed 70 days postgermination. Sample sizes are as follows: H. annuus, n = 8–12; BCann, n = 5–6; F1, n = 8–11; BCpet, n = 5–6; and H. petiolaris, n = 7–10. Error bars indicate 1 SE. For each time point/treatment combination, bars not connected by the same letter are significantly different (Tukey’s HSD). Similar post hoc tests were not performed for Ty3/gypsy expression data (fig. 2) as significant effects of genotype (P ≤ 0.01) were not detected for any of the time point/treatment combinations.
Transcriptional Dynamics in Stabilized Hybrid Species H. anomalus, H. deserticola, and H. paradoxus
RT-PCR assays of the same Ty3/gypsy and Ty1/copia sublineages in four individuals of each hybrid derivative species yielded similar qualitative expression patterns to those observed for the parental taxa and early generation interspecific F1 and backcross hybrids, whereby Ty3/gypsy sublineage A and Ty1/copia sublineage B′ were transcriptionally active in all samples (fig. 4A and D, respectively) and the remaining Ty3/gypsy sublineages were transcriptionally inactive (fig. 4B and C). Although RT-PCR bands indicating Ty3/gypsy sublineage A expression were visible for all individuals in all species, those for H. anomalus samples appeared weaker in staining intensity, suggesting potentially lower expression levels in that species. Individuals of the hybrid derivative species were not grown under salt (S) or salt + wounding (SW) stress, and thus transcriptional activity was not assayed under those experimental treatment conditions. Representative positive and negative control reactions for the RT-PCR assays in the hybrid sunflower taxa are provided in supplementary figure S3, Supplementary Material online.
Fig. 4.—
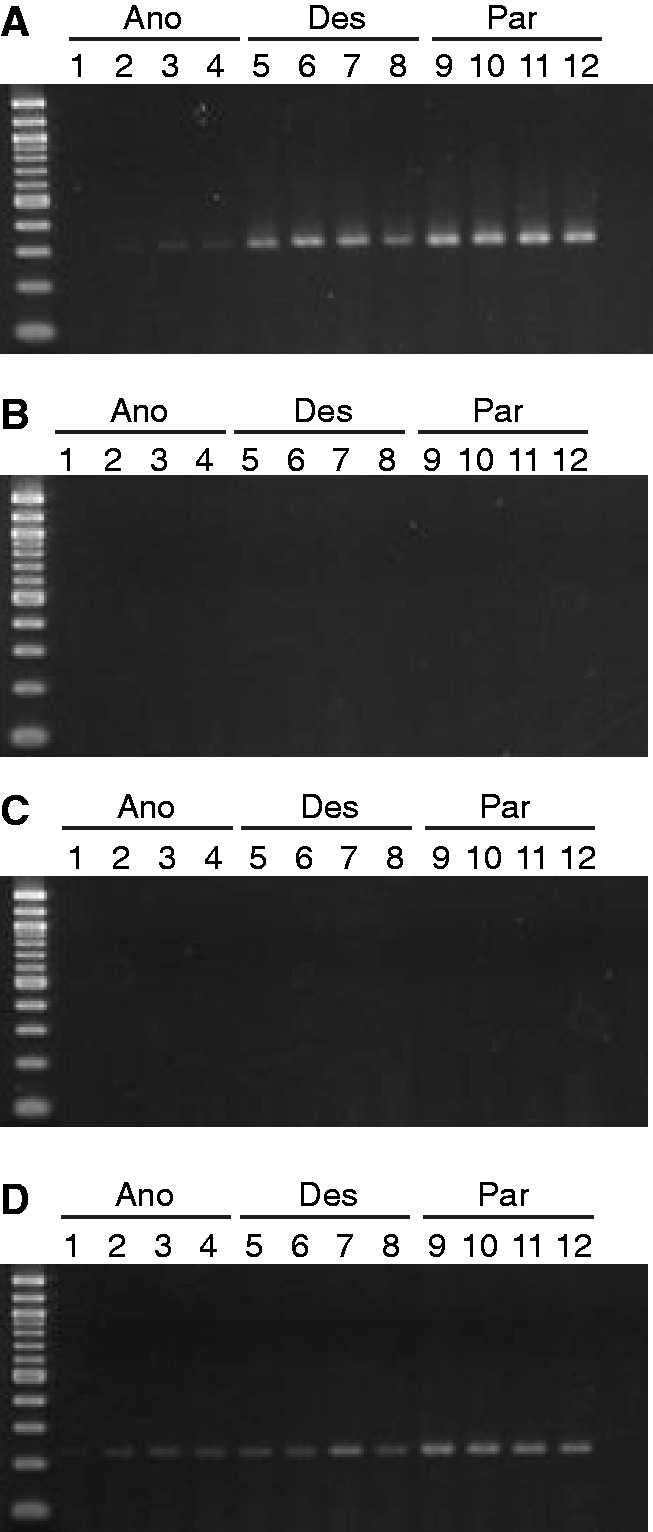
RT-PCR results for three sublineages of Ty3/gypsy and one sublineage of Ty1/copia LTR retrotransposons for sunflower hybrid species H. anomalus (Ano), lanes 1–4; H. deserticola (Des), lanes 5–8; and H. paradoxus (Par), lanes 9–12. (A) Ty3/gypsy sublineage A; (B) Ty3/gypsy sublineage C; (C) Ty3/gypsy sublineage E; and (D) Ty1/copia. Assays were performed 42 days postgermination.
Quantitative RT-PCR assays of transcriptionally active Ty3/gypsy and Ty1/copia sublineages were performed for 9–11 individuals of each hybrid species and compared with results for the parental species H. annuus (n = 12) and H. petiolaris (n = 10) obtained under control conditions at the 42-day harvest time point (fig. 5). Ty3/gypsy sublineage A transcriptional levels were approximately 2× to approximately 6× higher in the hybrid species compared with the parental species; post hoc tests revealed significant differences between parental species and two of the hybrid derivative species (H. deserticola and H. paradoxus), with the third hybrid species (H. anomalus) displaying a trend of increased expression relative to H. annuus and H. petiolaris. No notable expression differences were observed between parental and hybrid derivative species for Ty1/copia although significantly higher expression was observed for H. annuus and H. paradoxus compared with H. petiolaris (fig. 5).
Fig. 5.—
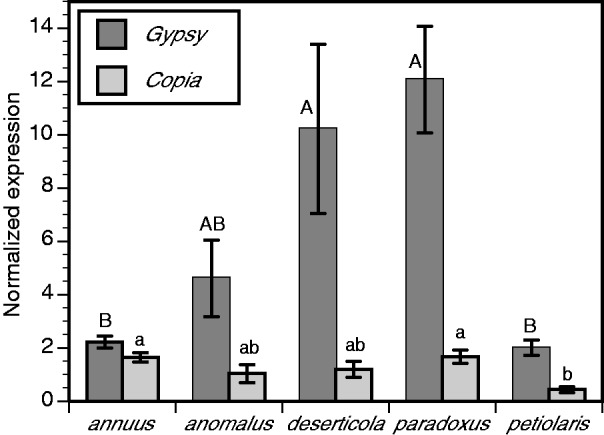
Quantitative PCR results for transcriptionally active Ty3/gypsy sublineage A and Ty1/copia in H. annuus, H. petiolaris, H. anomalus, H. deserticola, and H. paradoxus under control conditions 42 days postgermination. Sample sizes are as follows: H. annuus, n = 12; H. anomalus, n = 9; H. deserticola, n = 11; H. paradoxus, n = 10; and H. petiolaris, n = 10. Bars not connected by the same uppercase letter (Ty3/gypsy sublineage A) or lowercase letter (Ty1/copia) are significantly different (Tukey’s HSD). Error bars indicate 1 SE.
Discussion
Hybridization and abiotic stress have been implicated as important triggers of transposable element derepression and proliferation in many species groups (Wessler 1996; Grandbastien 1998; Waugh O'Neill et al. 1998; Labrador et al. 1999; Beguiristain et al. 2001; Kimura et al. 2001; Kashkush et al. 2002; Tapia et al. 2005; Wang et al. 2010; Guerreiro 2012). In this report, we evaluated the impact of these factors on transcriptional dynamics of Ty3/gyspy and Ty1/copia LTR retrotransposons in a group of wild sunflowers with a notable history of LTR retrotransposon proliferation following, or associated with, speciation via hybridization and adaptation to abiotically stressful environments (Rieseberg et al. 2003; Lai et al. 2005; Ungerer et al. 2006). Knowledge of the parental species that gave rise to the hybrid taxa and the ability to generate and study experimentally their early generation hybrids alongside the established hybrid derivative taxa provide a powerful setting for studying activational dynamics of transposable elements in a relevant evolutionary context. Utilization of plant materials from geographically diverse populations of the parental species H. annuus and H. petiolaris and a diversity of greenhouse synthesized hybrid genotypes suggest our experimental findings are likely to be general for this group of plants.
Transcriptional Activity of Ty3/gypsy and Ty1/copia Sublineages
Transcriptionally active sublineages of Ty3/gypsy and Ty1/copia identified in this study represent the same variants shown previously to be active in natural H. annuus × H. petiolaris hybrid zones (Kawakami et al. 2011) and are closely related to variants that massively proliferated in the sunflower hybrid species (Ungerer et al. 2006; Kawakami et al. 2010). Consistent with previous findings (Kawakami et al. 2011), these sublineages remain transcriptionally active in samples of H. annuus and H. petiolaris from multiple populations (supplementary table S1, Supplementary Material online) and in all synthesized hybrid genotypes (supplementary table S2, Supplementary Material online) under both control and experimental stress conditions. Transcriptional activity of highly similar variants has been detected in an inbred line of cultivated H. annuus by other investigators (Vukich et al. 2009).
Two additional sublineages of Ty3/gypsy that were investigated were transcriptionally inactive under control conditions in all samples and uninducible under S and SW stress conditions in samples of H. annuus, H. petiolaris, and their interspecific F1 and backcross hybrids. Abiotic stress treatments involving exposure to salt were used for the current experiments because one of the hybrid species (H. paradoxus) is locally adapted to saline marshes (Lexer et al. 2003; Karrenberg et al. 2006) and is the sole hybrid species to have undergone massive proliferation events of both Ty3/gypsy and Ty1/copia LTR retrotransposons (Ungerer et al. 2006; Kawakami et al. 2010).
Although uninducible under stress conditions examined herein, it is unlikely these sublineages lack the capacity for transcriptional/transpositional activation given intact and thus presumably functional GAG and POL genes and near-identical flanking 5′- and 3′-LTRs, indicating recent insertional activity of these elements. A phylogenetic signal of proliferation from within one of these sublineages (i.e., sublineage E) was reported previously for the three sunflower hybrid species (Ungerer et al. 2009), raising the possibility that more than a single sublineage of Ty3/gypsy has amplified in the sunflower hybrid taxa since their origins.
Although mounting evidence suggests LTR retrotransposon transcriptional and transpositional activation in response to hybridization and numerous biotic and abiotic stressors in other species, the universality of such phenomena is unclear as negative results are less likely to be published. Moreover, not all stressors tested experimentally induce transcriptional responses of normally quiescent transposable elements (Tapia et al. 2005; Ramallo et al. 2008). RT-PCR assays in this report were conducted for leaf tissue only; similar assays on reproductive (bud) tissue could reveal different results, especially given changes in gene expression and epigenetic phenomena that occur during the development of reproductive tissue (Boavida et al. 2011; Gutierrez-Marcos and Dickinson 2012).
We also have not assayed samples for element insertional activity in this study. We cannot rule out the possibility that parental and hybrid genotypes experience comparable transcriptional levels but hybrids experience higher insertion rates. In a previous report, however, we examined multiple, geographically diverse H. annuus × H. petiolaris hybrid populations for evidence of genome size expansion and LTR retrotransposon copy number increases (Kawakami et al. 2011). Genome size and element copy number estimates in these natural hybrid populations typically were intermediate relative to those of nearby stands of pure parental species, suggesting that insertion rates in hybrid genotypes are not elevated relative to parental lines.
Hybridization and Abiotic Stress Do Not Further Induce Transcriptionally Active Sublineages
Quantitative RT-PCR assays revealed that transcriptionally active Ty3/gypsy and Ty1/copia elements are not further induced as a consequence of hybridization or exposure to abiotic stress. To the contrary, H. annuus × H. petiolaris F1 and backcross hybrids exhibited normalized expression levels similar to or intermediate relative to parental species, and S and SW stress treatments had either no effect or resulted in reduced normalized expression compared with controls for all genotypes assayed. Results reported here and previously (Kawakami et al. 2011) fail to support earlier assertions regarding the potential roles of these phenomena in element proliferation events in the sunflower hybrid taxa (Ungerer et al. 2006), although we cannot rule out other potential stressors (i.e., water deficit, light stress, and biotic factors) as having contributed to natural proliferation events in hybrid species genomes. More severe stress (i.e., higher NaCl concentrations) also seem unlikely to trigger or further induce element activity as NaCl concentrations much beyond 90 mM have highly adverse effects on H. annuus and H. petiolaris survivorship, biomass, and other fitness-related characters (Welch and Rieseberg 2002).
Elevated Expression of Ty3/gypsy in Hybrid versus Parental Sunflower Species
The 2× to 6× higher expression of Ty3/gypsy sublineage A in the hybrid species versus parental taxa raises several questions pertaining to the historical amplification of these sequences in the hybrid species genomes and the molecular mechanism through which elevated transcriptional levels arose and are maintained. Whether higher transcriptional rates of Ty3/gypsy in the sunflower hybrid taxa translate to elevated element insertional rates and thus continuing proliferation in the hybrid sunflower species currently is unknown. Methods to detect individual insertion events (Waugh et al. 1997; Van den Broeck et al. 1998; Melayah et al. 2001) should allow this question to be addressed experimentally.
Differential effectiveness of epigenetic TE silencing (Slotkin and Martienssen 2007; Lisch 2009) represents a plausible mechanism underlying variation in transcriptional levels among these sunflower species. Comparative genome-level analyses of methylation (Cokus et al. 2008) or small RNA content (Cantu et al. 2010) may provide insights into differential transcriptional patterns reported herein. A second possibility could involve expression differences that are a natural consequence of different copy number abundances of these elements in the parental versus hybrid species. In previous work, we estimated via quantitative PCR copy number abundances of these proliferative Ty3/gypsy and Ty1/copia elements in the parental versus hybrid sunflower species (Ungerer et al. 2006; Kawakami et al. 2010). When compared with mean copy number estimates for the parental species H. annuus and H. petiolaris, the hybrid species harbor from 7.7- to 10.6-fold more copies of Ty3/gypsy and from 1.7- to 3.7-fold more copies of Ty1/copia. Higher expression in the hybrid species thus simply could result from more copies that are active transcriptionally. This argument, however, assumes transcriptional activity of all (or most) physical copies of the proliferative variants and only could explain expression differences of Ty3/gypsy; systematic expression differences between parental and hybrid species were not observed for Ty1/copia. (However, we acknowledge that Ty1/copia copy number differences between parental and hybrid species also are of lower scale.)
Finally, a third possibility could involve differential expression based on cis and/or trans regulatory variation among species. It is difficult to invoke cis-regulatory differences between parental and hybrid taxa as all elements in the ancient hybrid species are necessarily derived from elements present in the parental species H. annuus and/or H. petiolaris. The occurrence of mutations in promoter regions causing enhanced expression seems unlikely as they would have needed to arise in each of the independently derived hybrid species. Differences in trans-regulation may be more plausible, however, and potentially could evolve as a result of population genetic processes if transcription factors regulating these LTR retrotransposon sublineages experienced different selection pressures in the parental versus hybrid sunflower species. Transcription factors governing expression of the Tto1 LTR retrotransposon in tobacco have been characterized and shown also to regulate defense-related tobacco genes (Sugimoto et al. 2000). Identification of the transcription factor(s) governing expression of the active Ty3/gypsy sublineages in sunflower, determining whether they regulate other sunflower genes, and examining historical patterns of selection on these regulatory genes could provide evidence for the trans-regulation hypothesis.
Materials and Methods
Plant Materials
Seeds of the five sunflower species under study were obtained from the USDA National Plant Germplasm System (supplementary table S1, Supplementary Material online) and represent original wild-collected materials. F1 and backcross individuals were generated in the Kansas State University greenhouses by crossing H. annuus and H. petiolaris individuals from multiple populations and by crossing resulting F1s to relevant parental species individuals (supplementary table S2, Supplementary Material online). The use of H. annuus and H. petiolaris individuals from multiple natural populations resulted in a diverse array of natural genetic variation among hybrid genotypes. Crosses to generate hybrid individuals were performed in 2008 and 2009.
Growing Conditions and Experimental Stress Treatments
Individuals of H. annuus, H. petiolaris, and their interspecific F1 and BC1 hybrids were subjected to S, SW, and a control (C) treatment. Salt solution was prepared following Karrenberg et al. (2006) to simulate the natural below-ground environment of the salt-tolerant hybrid species H. paradoxus. Individuals subjected to S and SW stress were exposed initially to a solution consisting of 25 mM NaCl, 8.0 mM CaCl2, and 5.5 mM MgSO4, with subsequent increases in NaCl concentration according to the following protocol: days 1–14, 25 mM NaCl; days 15–28, 40 mM NaCl; days 29–42, 65 mM NaCl; and days 43–70, 90 mM NaCl. For the SW treatment, leaf samples were wounded mechanically with razor blades 6 h before harvest.
All seeds were germinated in the dark on moist filter paper in Petri dishes using salt solution for S and SW stress treatment groups and tap water for the control group. Germinated seedlings were transferred to four-inch pots and grown in a Kansas State University greenhouse. All plants were watered daily or as needed and provided with a weak nutrient solution (N:P:K = 15:30:15) once per week. Leaf material from H. annuus, H. petiolaris, F1, and BC1 hybrids was harvested for assays of LTR retrotransposon transcriptional activity 42 and 70 days postgermination, marking the conclusions of the 65 mM and 90 mM periods of NaCl exposure. Hybrid species individuals (H. anomalus, H. deserticola, and H. paradoxus) were grown under control conditions only, with leaves of these individuals harvested 42 days postgermination. All plant positions were randomized across six 1.6 × 3.1 m greenhouse benches. Harvested leaf samples were immediately flash frozen with liquid nitrogen. Total RNA was extracted following protocols in Kawakami et al. (2011).
RT-PCR Assays
Three sublineages of Ty3/gypsy and a single sublineage of Ty1/copia were evaluated for transcriptional activity 1) in H. annuus, H. petiolaris, and their interspecific F1 and BC1 hybrids under two stress treatments and a control treatment and 2) in the sunflower hybrid species under the control treatment. One of the Ty3/gypsy sublineages and the Ty1/copia sublineage were shown in previous reports to have undergone proliferation events in the sunflower hybrid species (Ungerer et al. 2006; Kawakami et al. 2010) and correspond to Ty3/gypsy sublineage A (Ungerer et al. 2009) and Ty1/copia sublineage B′ (Kawakami et al. 2010), respectively. These sublineages also have been documented previously as transcriptionally active in natural populations (Kawakami et al. 2011).
Sublineage-specific primers were developed for two additional Ty3/gypsy sublineages corresponding to phylogenetically well-supported lineages C and E in Ungerer et al. (2009). The RT-RNASEH-INT domains of six (lineage C) and two (lineage E) full-length and intact elements were aligned and primers designed based on regions of the INT domain that are conserved within, but differ among, sublineages. Ty3/gypsy elements of sublineages C and E have not previously been shown to be transcriptionally active in sunflower. These sublineages likely remain capable of transcriptional/transpositional activation, however, based on intact interior coding regions and their "young" age as determined by sequence analysis of their flanking 5′- and 3′-LTRs: each of the six sublineage C elements harbor 5′- and 3′-LTRs that are 99.2–99.7% identical and the two sublineage E elements harbor 5′- and 3′-LTRs that are 97.3% and 99.4% identical. All primers were designed with Primer3 (http://frodo.wi.mit.edu/, last accessed February 1, 2013) and are listed in supplementary table S3, Supplementary Material online.
RT-PCR assays were conducted using ImProm-II™ Reverse Transcriptase (Promega, Madison, WI). Primers targeting Actin (supplementary table S3, Supplementary Material online) were used as positive control reactions. To test for genomic DNA contamination of RNA, negative control PCR reactions were performed by withholding the reverse transcriptase enzyme. RT-PCR amplifications were conducted with an initial denaturing step of 94 °C for 2 min followed by 35 cycles of 94 °C for 15 s, 60 °C for 15 s, and 72 °C for 15 s, with a final incubation step of 72 °C for 6 min. The amplified products were size separated via electrophoresis in 2% agarose gels and stained with ethidium bromide for visualization. RT-PCR assays were performed for 10–12 individuals of H. annuus, H. petiolaris, and diverse F1 and backcross hybrid genotypes (supplementary table S2, Supplementary Material online), for each of the two stress treatments and the control treatment. Identical assays were performed for 4 individuals of each hybrid species, H. anomalus, H. deserticola, and H. paradoxus under control conditions only.
Quantitative PCR Assays
Expression levels of transcriptionally active sublineages (Ty3/gypsy sublineage A and Ty1/copia sublineage B′) were further evaluated by quantitative RT-PCR using an iCycler iQ quantitative PCR system (Bio-Rad, Hercules, CA). Cycling conditions consisted of initial denaturing at 94 °C for 90 s, followed by 40 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. Reactions were performed using the iQ SYBR Green Supermix kit (Bio-Rad) with three technical replicates per sample. All reactions were performed in 30 µl volumes using 5 µl of template cDNA and primers targeting Ty3/gypsy (sublineage A) and Ty1/copia (sublineage B′). Actin and UBC (supplementary table S3, Supplementary Material online) were used for expression normalization. Quantitative PCR melt curves revealed single and unique peaks for each primer pair, confirming high specificity to the target sequence fragments. In addition, high PCR efficiency (96–103%) was confirmed for each primer pair by standard curve assays using a dilution series of cloned cDNA fragments of Ty3/gypsy, Ty1/copia, Actin, and UBC (5.0, 1.0, 0.2, 0.04, 0.008, and 0.0016 ng/ml). Relative expression of Ty3/gypsy and Ty1/copia sublineages was standardized to the geometric mean of relative expression values of the two reference genes following Vandesompele et al. (2002). Quantitative RT-PCR for hybrid species individuals under control conditions was expanded to n = 9–11 per species. Cycle threshold (Ct) values for all samples were within the linear range of standard curves.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Preeti Dhakal, Bradley Hartman-Bakken, and Thomas Nelson for technical support. This work was supported by National Science Foundation grant DEB-0742993 to M.C.U.
Literature Cited
- Beguiristain T, Grandbastien MA, Puigdomenech P, Casacuberta JM. Three Tnt1 subfamilies show different stress-associated patterns of expression in tobacco. Consequences for retrotransposon control and evolution in plants. Plant Physiol. 2001;127:212–221. doi: 10.1104/pp.127.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida LC, Borges F, Becker JD, Feijo JA. Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 2011;155:2066–2080. doi: 10.1104/pp.110.169813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D, et al. Small RNAs, DNA methylation and transposable elements in wheat. BMC Genomics. 2010;11:408. doi: 10.1186/1471-2164-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien MA. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998;3:181–187. [Google Scholar]
- Grandbastien MA, et al. The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica. 1997;100:241–252. [PubMed] [Google Scholar]
- Guerreiro MP. What makes transposable elements move in the Drosophila genome? Heredity. 2012;108:461–468. doi: 10.1038/hdy.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Dickinson HG. Epigenetic reprogramming in plant reproductive lineages. Plant Cell Physiol. 2012;53:817–823. doi: 10.1093/pcp/pcs052. [DOI] [PubMed] [Google Scholar]
- Karrenberg S, Edelist C, Lexer C, Rieseberg L. Response to salinity in the homoploid hybrid species Helianthus paradoxus and its progenitors H. annuus and H. petiolaris. New Phytol. 2006;170:615–629. doi: 10.1111/j.1469-8137.2006.01687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Dhakal P, Katterhenry AN, Heatherington CA, Ungerer MC. Transposable element proliferation and genome expansion are rare in contemporary sunflower hybrid populations despite widespread transcriptional activity of LTR retrotransposons. Genome Biol Evol. 2011;3:156–167. doi: 10.1093/gbe/evr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Strakosh SC, Zhen Y, Ungerer MC. Different scales of Ty1/copia-like retrotransposon proliferation in the genomes of three diploid hybrid sunflower species. Heredity. 2010;104:341–350. doi: 10.1038/hdy.2009.182. [DOI] [PubMed] [Google Scholar]
- Kimura Y, et al. OARE-1, a Ty1-copia retrotransposon in oat activated by abiotic and biotic stresses. Plant Cell Physiol. 2001;42:1345–1354. doi: 10.1093/pcp/pce171. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Labrador M, Farre M, Utzet F, Fontdevila A. Interspecific hybridization increases transposition rates of Osvaldo. Mol Biol Evol. 1999;16:931–937. doi: 10.1093/oxfordjournals.molbev.a026182. [DOI] [PubMed] [Google Scholar]
- Lai Z, et al. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics. 2005;171:291–303. doi: 10.1534/genetics.105.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer C, Welch ME, Durphy JL, Rieseberg LH. Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. Mol Ecol. 2003;12:1225–1235. doi: 10.1046/j.1365-294x.2003.01803.x. [DOI] [PubMed] [Google Scholar]
- Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 2009;60:43–66. doi: 10.1146/annurev.arplant.59.032607.092744. [DOI] [PubMed] [Google Scholar]
- Liu B, Wendel JF. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome. 2000;43:874–880. [PubMed] [Google Scholar]
- Llorens C, et al. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 2011;39:D70–D74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melayah D, Bonnivard E, Chalhoub B, Audeon C, Grandbastien MA. The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J. 2001;28:159–168. doi: 10.1046/j.1365-313x.2001.01141.x. [DOI] [PubMed] [Google Scholar]
- Ramallo E, Kalendar R, Schulman AH, Martinez-Izquierdo JA. Reme1, a Copia retrotransposon in melon, is transcriptionally induced by UV light. Plant Mol Biol. 2008;66:137–150. doi: 10.1007/s11103-007-9258-4. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Sabot F, Schulman AH. Parasitism and the retrotransposon life cycle in plants: a hitchhiker's guide to the genome. Heredity. 2006;97:381–388. doi: 10.1038/sj.hdy.6800903. [DOI] [PubMed] [Google Scholar]
- Salazar M, González E, Casaretto JA, Casacuberta JM, Ruiz-Lara S. The promoter of the TLC1.1 retrotransposon from Solanum chilense is activated by multiple stress-related signaling molecules. Plant Cell Rep. 2007;26:1861–1868. doi: 10.1007/s00299-007-0375-y. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nature Reviews Genetics. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Staton SE, Ungerer MC, Moore RC. The genomic organization of Ty3/Gypsy-like retrotransposons in Helianthus (Asteraceae) homoploid hybrid species. Am J Botany. 2009;96:1646–1655. doi: 10.3732/ajb.0800337. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Takeda S, Hirochika H. MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell. 2000;12:2511–2527. doi: 10.1105/tpc.12.12.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Sugimoto K, Otsuki H, Hirochika H. A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 1999;18:383–393. doi: 10.1046/j.1365-313x.1999.00460.x. [DOI] [PubMed] [Google Scholar]
- Tapia G, et al. Involvement of ethylene in stress-induced expression of the TLC1.1 retrotransposon from Lycopersicon chilense Dun. Plant Physiol. 2005;138:2075–2086. doi: 10.1104/pp.105.059766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, Stimpson KM. Proliferation of Ty3/gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biol. 2009;7:40. doi: 10.1186/1741-7007-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, Zhen Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr Biol. 2006;16:R872–R873. doi: 10.1016/j.cub.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Van den Broeck D, et al. Transposon display identifies individual transposable elements in high copy number lines. Plant J. 1998;13:121–129. doi: 10.1046/j.1365-313X.1998.00004.x. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukich M, Giordani T, Natali L, Cavallini A. Copia and Gypsy retrotransposons activity in sunflower (Helianthus annuus L.) BMC Plant Biol. 2009;9:150. doi: 10.1186/1471-2229-9-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, et al. Transpositional reactivation of two LTR retrotransposons in rice-Zizania recombinant inbred lines (RILs) Hereditas. 2010;147:264–277. doi: 10.1111/j.1601-5223.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- Waugh O'Neill RJ, O'Neill MJ, Marshall Graves JA. Undermethylation associated with retroelement activation and chromosomal remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- Waugh R, et al. Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP) Mol Gen Genet. 1997;253:687–694. doi: 10.1007/s004380050372. [DOI] [PubMed] [Google Scholar]
- Welch ME, Rieseberg LH. Habitat divergence between a homoploid hybrid sunflower species, Helianthus paradoxus (Asteraceae), and its progenitors. Am J Bot. 2002;89:472–478. doi: 10.3732/ajb.89.3.472. [DOI] [PubMed] [Google Scholar]
- Wessler SR. Turned on by stress. Plant retrotransposons. Curr Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Woodrow P, Pontecorvo G, Ciarmiello LF, Fuggi A, Carillo P. Ttd1a promoter is involved in DNA-protein binding by salt and light stresses. Mol Biol Rep. 2011;38:3787–3794. doi: 10.1007/s11033-010-0494-3. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Silencing of transposons in plant genomes: kick them when they're down. Genome Biol. 2004;5:249. doi: 10.1186/gb-2004-5-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]