Abstract
Whole-genome duplications (WGDs) have recurred in the evolution of angiosperms, resulting in many duplicated chromosomal segments. Local gene duplications are also widespread in angiosperms. WGD-derived duplicates, that is, ohnologs, and local duplicates often show contrasting patterns of gene retention and evolution. However, many genes in angiosperms underwent multiple gene duplication events, possibly by different modes, indicating that different modes of gene duplication are not mutually exclusive. In two representative angiosperm genomes, Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), we found that 9.6% and 11.3% of unique ohnologs, corresponding to 15.5% and 17.1% of ohnolog pairs, were also involved in local duplications, respectively. Locally duplicated ohnologs are widely distributed in different duplicated chromosomal segments and functionally biased. Coding sequence divergence between duplicated genes is denoted by nonsynonymous (Ka) and synonymous (Ks) substitution rates. Locally duplicated ohnolog pairs tend to have higher Ka, Ka/Ks, and gene expression divergence than nonlocally duplicated ohnolog pairs. Locally duplicated ohnologs also tend to have higher interspecies sequence divergence. These observations indicate that locally duplicated ohnologs evolve faster than nonlocally duplicated ohnologs. This study highlights the necessity to take local duplications into account when analyzing the evolutionary dynamics of ohnologs.
Keywords: local gene duplication, whole-genome duplication, ohnolog, divergence, colinearity
Introduction
With tens of plant genomes sequenced, it is now clear that whole-genome duplications (WGDs) are widespread in angiosperm (flowering plant) genomes (Bowers et al. 2003; Paterson et al. 2010). WGDs resulted in many duplicated chromosomal segments. However, computational identification of duplicated chromosomal segments is a challenging task. Besides phylogenomics methods (Bowers et al. 2003), detection of synteny (homologous genes remaining in corresponding chromosomal regions) is often adopted (Wei et al. 2007; Tang et al. 2008a, 2008b, 2010). One general method for synteny detection is the clustering of neighboring matching gene pairs (Vandepoele et al. 2002; Hampson et al. 2003; Luc et al. 2003; Ling et al. 2009; Vergara and Chen 2009). However, gene losses, tandem duplicates, and chromosomal rearrangements often complicate synteny detection. Collinearity, a more specific form of synteny that requires conserved gene order, can facilitate more accurate detection of synteny. Thus, another general method for synteny detection uses dynamic programming to chain pair-wise collinear genes, often based on a certain scoring scheme that rewards adjacent collinear genes and penalizes the distance difference between them (Haas et al. 2004; Soderlund et al. 2006; Wang et al. 2006, 2012; Tang et al. 2008b; Rodelsperger and Dieterich 2010).
In angiosperm genomes, the duplicated genes located at syntenic/collinear positions between duplicated chromosomal segments were thought to arise from WGDs (Bowers et al. 2003; Paterson et al. 2004) and often referred to as ohnologs (Wolfe 2000). Single-gene duplications including local (tandem or proximal), transposed, and dispersed duplications are also widespread in angiosperms (Freeling 2009; Wang Y, et al. 2011; Wang Y, Wang X, et al. 2012). Indeed, the different modes of gene duplication represent different evolutionary origins of duplicated genes, depending on which, duplicated genes often show different evolutionary patterns and consequences (Wang Y, Wang X, et al. 2012). The retention of ohnologs and local duplicates is functionally biased and often reciprocal in many gene families: WGDs tend to retain transcription factors, whereas local duplicates tend to retain the genes functioning in stress response (Freeling 2009; Rodgers-Melnick et al. 2012). Single-gene duplications frequently result in more divergent gene expression between duplicated genes than WGDs (Casneuf et al. 2006; Ganko et al. 2007; Wang Y, et al. 2011). Moreover, local duplicates were suggested to be important for lineage-specific adaptive evolution to rapidly changing environments (Hanada et al. 2008).
However, the evolutionary origins of many duplicated genes are not unique. In many synteny detection tools, tandem duplicates are collapsed before synteny detection (Lyons et al. 2008; Tang et al. 2008b; Proost et al. 2012; Wang et al. 2012), which indicates that in a genome, duplicated genes may be simultaneously located within syntenic positions and tandem arrays, further suggesting that duplicated genes may have undergone both WGDs and local duplications. The MCScanX software has enabled the recovery of “collinear tandem arrays” after synteny detection (Wang et al. 2012). However, a genome-wide evolutionary analysis of the duplicated genes with multiple origins has yet to be reported.
In two representative angiosperm genomes, Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), we identified the ohnologs that were also locally duplicated, which were named locally duplicated ohnologs (the remaining ohnologs were named nonlocally duplicated ohnologs). We examined whether locally duplicated and nonlocally duplicated ohnologs have different levels of coding sequence and expression divergence. The evolutionary impact of local duplications of ohnologs was investigated.
Materials and Methods
Genome Annotations
Genome annotations for A. thaliana (Swarbreck et al. 2008), B. rapa (Wang X, et al. 2011), O. sativa (Ouyang et al. 2007), and Bra. distachyon (International Brachypodium Initiative 2010) were obtained from Phytozome 8.0 (http://www.phytozome.net, last accessed February 6, 2013). Transposable element-related genes in rice were removed. If a gene has multiple transcripts, only the first transcript in the genome annotation was used in related analyses.
Homology Search
To search for potential duplicated genes (i.e., the homology input of MCScanX) in Arabidopsis and rice, the protein-coding genes were compared against themselves within their genomes using BLASTP (Altschul et al. 1990). For a protein sequence, the best five nonself-hits that met an E-value threshold of 10−10 were kept. For each Arabidopsis or rice gene, to search for its closest ortholog in an outgroup species, the best BLASTP hit with E value ≤ 10−10 was retained.
Coding Sequence Divergence
Coding sequence divergence was measured by nonsynonymous (Ka) and synonymous (Ks) substitution rates. To generate a coding sequence alignment for a pair of homologous genes, an alignment of their protein sequences was first generated using Clustalw (Thompson et al. 1994) with default parameters. Then, the protein alignment was transformed into a coding sequence alignment using the “Bio::Align::Utilities” module of the BioPerl package (http://www.bioperl.org/, last accessed February 6, 2013). Ka and Ks were computed using Nei–Gojobori method (Nei and Gojobori 1986), available in the “Bio::Align::DNAStatistics” module of the BioPerl package. Note that extremely high levels of sequence divergence between homologous genes may cause the “Bio::Align::DNAStatistics” module to generate invalid Ka or Ks, which were excluded from related analysis. Ks < 3.0 were involved in related statistical analyses.
GO Term Enrichment Analysis
GO term enrichment analysis was performed using Fisher's exact test. The P value was corrected with the total number of terms (i.e., Bonferroni correction) to account for multiple comparisons. GO terms with corrected P value < 0.05 were returned.
Gene Expression Data
Processed Arabidopsis and rice microarray data measured by the Affymetrix Arabidopsis ATH1 Genome Array (GEO platform GPL198) and GeneChip Rice Genome Array (GEO platform GPL2020), respectively, were obtained from a previous study (Wang Y, et al. 2011). The microarray data contained 4,566 and 508 samples in Arabidopsis and rice, respectively. To avoid the expression values generated by cross-hybridization, probe sets that were mapped to multiple genes were excluded from analysis. (Note that the probe sets that were mapped to multiple transcripts of the same gene were kept.) For a gene, there may be multiple probe sets available on the array, and the first probe set according to alphabetic sorting was used to represent its expression profile. In Arabidopsis, 448 (60.4%) locally duplicated ohnolog pairs and 3,015 (74.7%) nonlocally duplicated ohnolog pairs have expression profiles. In rice, 419 (70.4%) locally duplicated ohnolog pairs and 2,373 (82.1%) nonlocally duplicated ohnolog pairs have expression profiles. Similarity between the expression profiles of two genes was initially measured by Pearson's correlation coefficient (denoted by r). Their corresponding expression divergence was measured by 1 − r (Liao and Zhang 2008; Liao et al. 2010).
Results
Identification of Locally Duplicated Ohnologs
BLASTP (Altschul et al. 1990) was used to generate the whole set of duplicates in Arabidopsis and rice (see Materials and Methods). MCScanX (Wang et al. 2012) was then implemented (parameters set as default except maximum gaps = 40) to detect gene synteny/collinearity and duplicated chromosomal segments. A total of 4,775 and 3,484 ohnolog pairs, corresponding to 7,839 and 5,968 unique ohnologs, were identified in Arabidopsis and rice, respectively.
To detect locally duplicated ohnologs, local gene duplications, defined as two duplicated genes within 20 annotated genes of each other on the same chromosomes without any paralog (BLASTP hit) between them, were identified. If an ohnolog was involved in a local duplication, it was classified as a locally duplicated ohnolog. If at least one ohnolog in an ohnolog pair was locally duplicated, this ohnolog pair was deemed a locally duplicated ohnolog pair. A total of 754 (9.6%) and 675 (11.3%) ohnologs, corresponding to 741 (15.5%) and 595 (17.1%) ohnolog pairs, were identified as locally duplicated ohnologs in Arabidopsis and rice, respectively (listed in supplementary table S1, Supplementary Material online).
For locally duplicated ohnologs, it would be interesting to distinguish between ohnologs that were locally duplicated after WGDs and locally duplicated genes that were retained following WGDs. Synonymous (Ks) substitution rates are often used as a proxy of time since divergence (Li 1997). However, using Ks as a proxy of time has great uncertainty. For example, although ohnologs belonging to the same WGD event were created simultaneously, their Ks indeed have a wide range (Zhang et al. 2002; Blanc and Wolfe 2004b). In addition, Ks increase approximately linearly with time only for relatively low levels of sequence divergence (Li 1997). Thus, for two gene duplications, the order of their Ks levels is not a reliable indicator of the sequence of the duplications. However, it may be reasonable to compare the relative ages between large groups of genes based on Ks because the variances of Ks can be estimated. For locally duplicated ohnologs, local duplications have significantly smaller Ks than ohnolog pairs in both Arabidopsis and rice (P value ≤ 1.875 × 10−6, two-sample t-test), suggesting that local duplications are relatively younger than WGDs, which is consistent with previous studies (Blanc and Wolfe 2004b; Yu et al. 2005). Note that this observation may not suggest that the rate of local duplications increases after WGDs but rather confirm the fact that the turnover rate of duplicates following local duplications is much higher than that following WGDs, making a very small portion of local duplicates survive over long evolutionary times (Lynch and Conery 2000; Blanc and Wolfe 2004b).
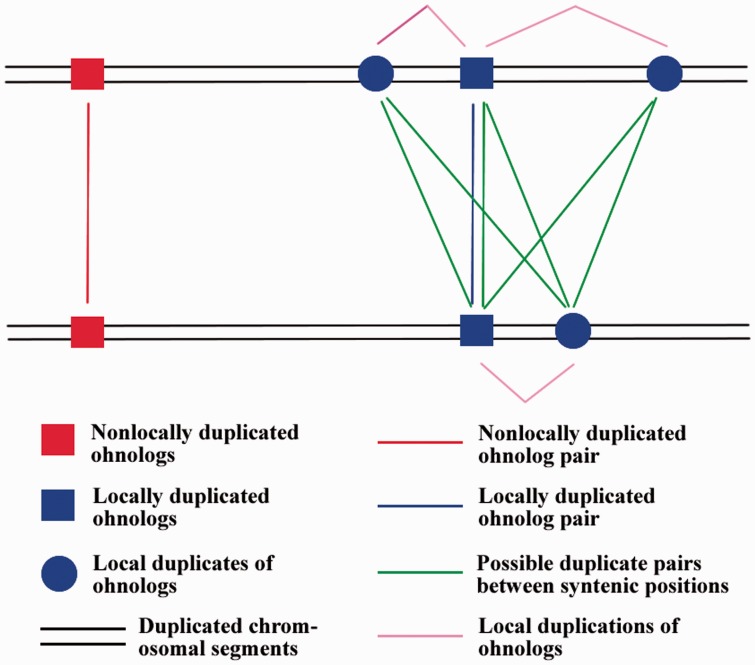
Between the syntenic positions defined by a locally duplicated ohnolog pair, there may be multiple duplicate pairs. For each locally duplicated ohnolog pair, the best duplicate pair, defined as the one with the smallest Ks among all possible duplicate pairs between its syntenic positions, was also recorded. The terms describing different relationships among the duplicated genes in duplicated chromosomal segments are explained in figure 1.
Fig. 1.—
Different relationships among the duplicated genes in duplicated chromosomal segments and their terms in this study.
Chromosomal Distributions of Locally Duplicated Ohnologs
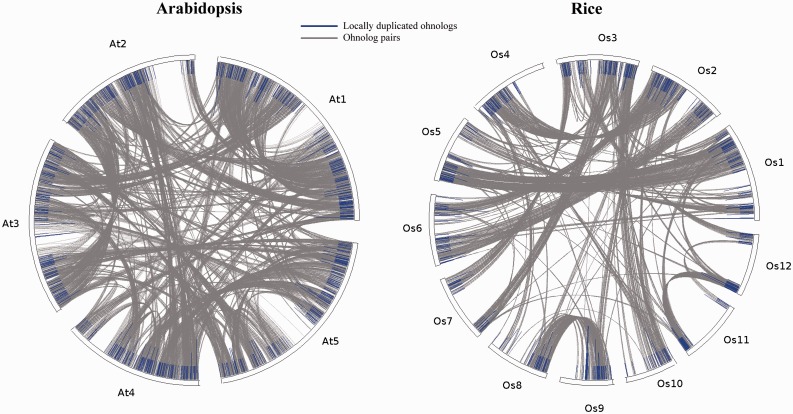
Genome circle plots were made to visualize the chromosomal distributions of locally duplicated ohnologs. In both Arabidopsis and rice, locally duplicated ohnologs are widely distributed in different duplicated chromosomal segments that cover all chromosomes (fig. 2), suggesting that local duplications of ohnologs are not specific to certain chromosomal regions.
Fig. 2.—
Genomic distributions of locally duplicated ohnologs in Arabidopsis and rice. At, Arabidopsis thaliana; Os, Oryza sativa.
Locally Duplicated Ohnologs Are Functionally Biased
The retention of duplicated genes following WGDs in terms of gene functions has been extensively studied. There is a consensus that WGDs tend to retain genes involved in transcription regulation and signal transduction (Blanc and Wolfe 2004a; Maere et al. 2005). Some studies also reported that metabolic genes are likely to be over-retained following WGDs (Rensing et al. 2007; Gout et al. 2009). To evaluate the functional characteristics of locally duplicated ohnologs, GO term enrichment analysis was performed using all ohnologs as the reference database. In Arabidopsis and rice, there were 22 and 26 enriched GO terms in locally duplicated ohnologs, respectively (supplementary table S2, Supplementary Material online). The common enriched GO terms between Arabidopsis and rice included heme binding (GO:0020037), electron carrier activity (GO:0009055), iron ion binding (GO:0005506), oxidation–reduction process (GO:0055114), monooxygenase activity (GO:0004497), and recognition of pollen (GO:0048544). This analysis suggests that locally duplicated ohnologs are functionally biased.
When all local duplicates were used as the reference database, there were no enriched GO terms in Arabidopsis locally duplicated ohnologs but 21 enriched GO terms in rice locally duplicated ohnologs (supplementary table S3, Supplementary Material online). Thus, in Arabidopsis, the functional biases of locally duplicated ohnologs reflect the general trend of local duplicates, while in rice, locally duplicated ohnologs show a high level of functional specificity.
Locally Duplicated Ohnologs Evolve Faster Than Nonlocally Duplicated Ohnologs
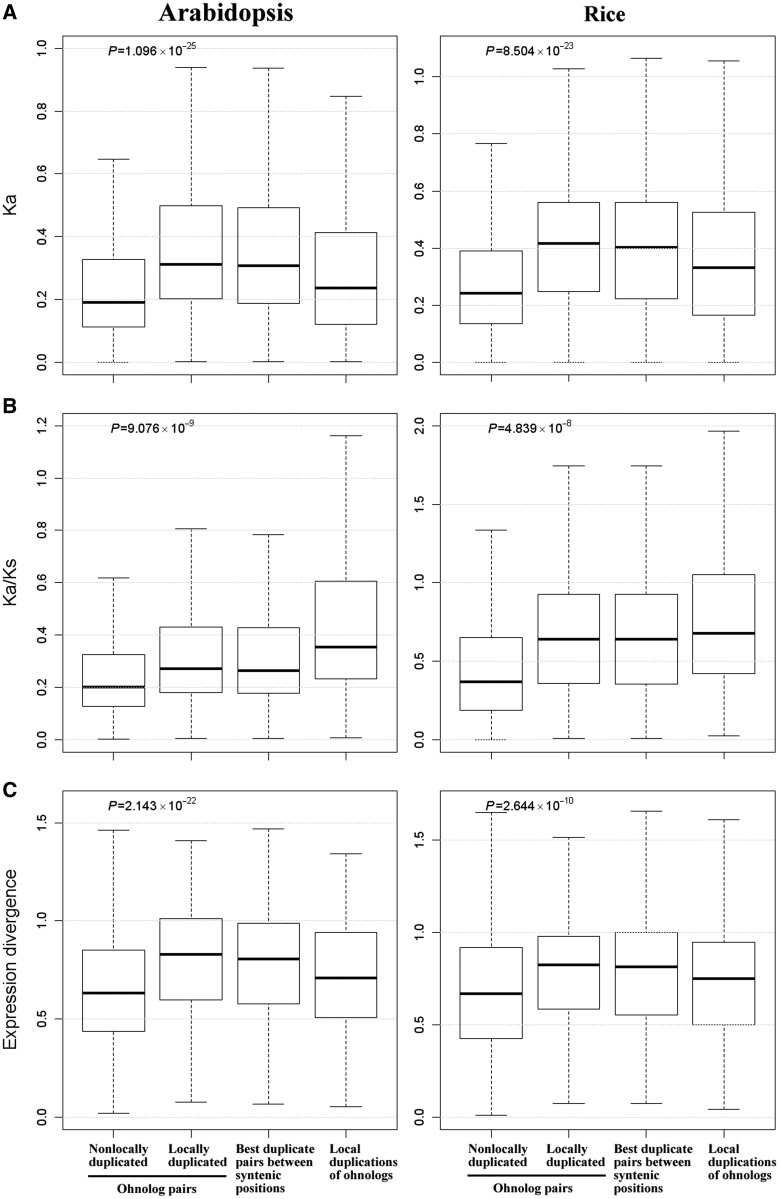
To study the evolutionary impact of local duplications of ohnologs, we first compared the divergence of ohnologs between locally duplicated and nonlocally duplicated ohnolog pairs. To ensure reliable comparison, the divergence of duplicates in the best duplicate pairs between the syntenic positions defined by locally duplicated ohnolog pairs and the local duplications of ohnologs was also assessed. In both Arabidopsis and rice, locally duplicated ohnolog pairs have significantly higher nonsynonymous (Ka) substitution rates than nonlocally duplicated ohnolog pairs (fig. 3A), indicating that locally duplicated ohnolog pairs tend to have more divergent protein sequences. Locally duplicated ohnolog pairs also show higher Ka/Ks (fig. 3B), suggesting that purifying selection acting on locally duplicated ohnologs is relatively relaxed. Comparison of gene expression divergence shows that locally duplicated ohnolog pairs have more divergent expression profiles than nonlocally duplicated ohnolog pairs (fig. 3C). The divergence of duplicates is comparable between locally duplicated ohnolog pairs and the best duplicate pairs between the syntenic positions defined by locally duplicated ohnolog pairs (fig. 3A–C), indicating that the higher divergence between locally duplicated ohnologs was not caused by incorrect selection of ohnolog pairs between the syntenic positions with local duplications. The divergence of duplicates in local duplications of ohnologs is higher than that in nonlocally duplicated ohnolog pairs (fig. 3A–C), implying that local duplicates, though are in general younger, tend to evolve faster than ohnologs.
Fig. 3.—
Comparisons of divergence of duplicates between locally duplicated and nonlocally duplicated ohnolog pairs in Arabidopsis and rice. Statistical comparisons between locally duplicated and nonlocally duplicated ohnolog pairs were performed using two-sample t-test, whose P values are indicated. The divergence of duplicates in the best duplicate pairs between the syntenic positions defined by locally duplicated ohnolog pairs and the local duplications of ohnologs is also shown. (A) Comparison of Ka. (B) Comparison of Ka/Ks. (C) Comparison of expression divergence.
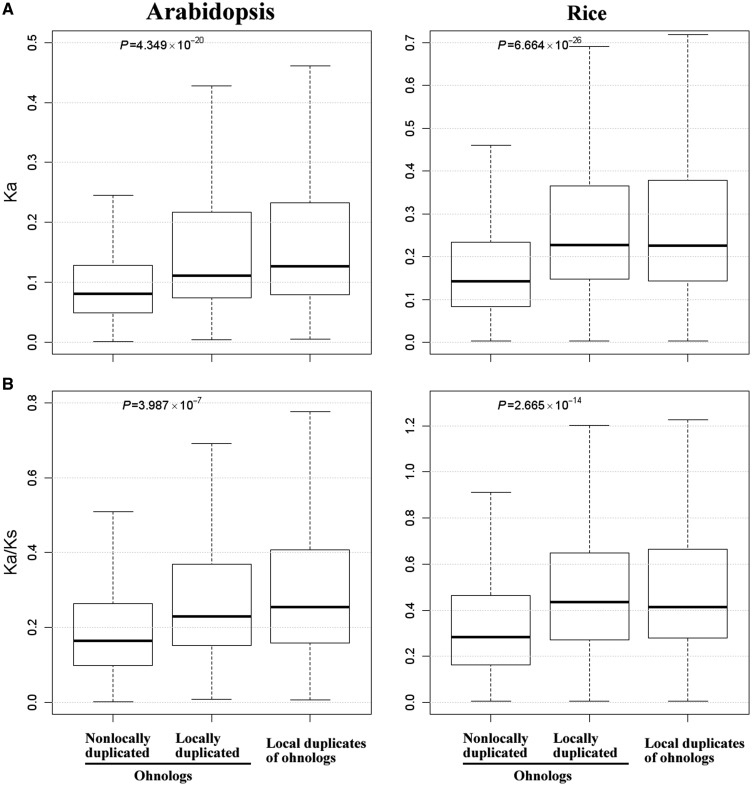
Next, we compared interspecies divergence between locally duplicated and nonlocally duplicated ohnologs. To this end, the Ka and Ks values between each Arabidopsis or rice ohnolog and its closest ortholog in Brassica rapa or Brachypodium distachyon, respectively, were computed. In both Arabidopsis and rice, locally duplicated ohnologs have significantly higher Ka and Ka/Ks than nonlocally duplicated ohnologs (fig. 4), suggesting that locally duplicated ohnologs also show higher levels of interspecies sequence divergence. In partial summary, locally duplicated ohnologs tend to have higher levels of sequence and expression divergence than nonlocally duplicated ohnologs, indicating that locally duplicated ohnologs evolve faster than nonlocally duplicated ohnologs.
Fig. 4.—
Comparisons of interspecies divergence between locally duplicated and nonlocally duplicated ohnologs. Arabidopsis and rice ohnologs were compared with their closest orthologs in Brassica rapa and Brachypodium distachyon, respectively. Statistical comparisons between locally duplicated and nonlocally duplicated ohnologs were performed using two-sample t-test, whose P values are indicated. (A) Comparison of Ka. (B) Comparison of Ka/Ks.
Discussion
Previous studies for comparing different modes of gene duplication often assigned a unique origin to each duplicate gene (Guan et al. 2007; Hakes et al. 2007; Wang Y, et al. 2011). Although WGDs and local duplications were found to have different functional biases and evolutionary patterns (Freeling 2009; Rodgers-Melnick et al. 2012), this study showed that approximately 10% of ohnologs have also experienced local duplications and the ohnologs that underwent local duplications have different evolutionary patterns than those that did not. In Arabidopsis and rice, local duplicates account for approximately 18.5% of the genes in the genome. The fact that approximately 10% of ohnologs also experienced local duplications might not indicate that ohnologs are less likely to be locally duplicated. If all duplicates in the local duplications of ohnologs are counted in ohnologs similar to a previous study (Bowers et al. 2003), the percentage of locally duplicated ohnologs increases to approximately 18%, suggesting that ohnologs and other types of genes may have comparable probabilities of being locally duplicated. Thus, to analyze the evolutionary dynamics of ohnologs, it is necessary to take local duplications into account.
Higher levels of Ka/Ks in locally duplicated ohnologs than nonlocally duplicated ohnologs can be explained by the gene balance hypothesis, which postulates that any successful genome has evolved an optimal balance of gene products that bind with one another to form protein complexes or are involved in multiple steps of biological pathways (Birchler and Veitia 2007; Freeling 2009). WGDs duplicate all nuclear genes at once, and dosage balance among genes is not altered. To maintain proper gene networks following WGDs, the dosage balance constraints impose strong purifying selection on dosage-sensitive ohnologs. Thus, among different types of duplicated genes, ohnologs tend to have smaller Ka/Ks values. In contrast, local duplications create only one more duplicated gene each time, and dosage imbalance is likely to occur. Thus, the local duplicate has a higher chance of being removed through accumulation of degenerative mutations, which implies a higher level of Ka/Ks. However, during accumulation of degenerative mutations, the local duplicate may occasionally evolve modified/new functions, rendering it finally preserved. If an ohnolog is locally duplicated, its dosage balance is broken and the evolutionary fate of the ohnolog becomes similar to local duplicates, resulting in an elevated level of Ka/Ks. Elevated Ka/Ks levels for local duplicates may provide them with greater potential of evolving lineage-specific functions (Hanada et al. 2008). Thus, there is a possibility that a part of ohnologs contribute to lineage-specific evolution by undergoing additional local duplications.
Supplementary Material
Supplementary tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org).
Acknowledgments
The author thanks Dr Andrew Paterson (PGML, University of Georgia) for direction during his Ph.D. studies, Drs Qi Sun and Hsiao-Pei Yang (CBSU, Cornell University) for helpful discussion, and Dr Katie Hyma (CBSU, Cornell University) for critical reading of the manuscript. This work was supported by a National Science Foundation grant (IOS #1127017) to Qi Sun.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004a;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004b;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong JK, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006;7:R13. doi: 10.1186/gb-2006-7-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ. Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol. 2007;24:2298–2309. doi: 10.1093/molbev/msm158. [DOI] [PubMed] [Google Scholar]
- Gout JF, Duret L, Kahn D. Differential retention of metabolic genes following whole-genome duplication. Mol Biol Evol. 2009;26:1067–1072. doi: 10.1093/molbev/msp026. [DOI] [PubMed] [Google Scholar]
- Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. 2007;175:933–943. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- Hakes L, Pinney JW, Lovell SC, Oliver SG, Robertson DL. All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 2007;8:R209. doi: 10.1186/gb-2007-8-10-r209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson S, McLysaght A, Gaut B, Baldi P. LineUp: statistical detection of chromosomal homology with application to plant comparative genomics. Genome Res. 2003;13:999–1010. doi: 10.1101/gr.814403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008;148:993–1003. doi: 10.1104/pp.108.122457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Li WH. Molecular evolution. Sunderland (MA): Sinauer Associates; 1997. [Google Scholar]
- Liao BY, Weng MP, Zhang J. Contrasting genetic paths to morphological and physiological evolution. Proc Natl Acad Sci U S A. 2010;107:7353–7358. doi: 10.1073/pnas.0910339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling X, He X, Xin D. Detecting gene clusters under evolutionary constraint in a large number of genomes. Bioinformatics. 2009;25:571–577. doi: 10.1093/bioinformatics/btp027. [DOI] [PubMed] [Google Scholar]
- Luc N, Risler JL, Bergeron A, Raffinot M. Gene teams: a new formalization of gene clusters for comparative genomics. Comput Biol Chem. 2003;27:59–67. doi: 10.1016/s1476-9271(02)00097-x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lyons E, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Ouyang S, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Freeling M, Tang H, Wang X. Insights from the comparison of plant genome sequences. Annu Rev Plant Biol. 2010;61:349–372. doi: 10.1146/annurev-arplant-042809-112235. [DOI] [PubMed] [Google Scholar]
- Proost S, et al. i-ADHoRe 3.0—fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 2012;40:e11. doi: 10.1093/nar/gkr955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, et al. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol Biol. 2007;7:130. doi: 10.1186/1471-2148-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodelsperger C, Dieterich C. CYNTENATOR: progressive gene order alignment of 17 vertebrate genomes. PLoS One. 2010;5:e8861. doi: 10.1371/journal.pone.0008861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E, et al. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 2012;22:95–105. doi: 10.1101/gr.125146.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: a system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16:1159–1168. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci U S A. 2010;107:472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, et al. Synteny and collinearity in plant genomes. Science. 2008a;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Tang H, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008b;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Saeys Y, Simillion C, Raes J, Van De Peer Y. The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res. 2002;12:1792–1801. doi: 10.1101/gr.400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara IA, Chen N. Using OrthoCluster for the detection of synteny blocks among multiple genomes. Curr Protoc Bioinformatics. 2009;27:6.10.11–16.10.18. doi: 10.1002/0471250953.bi0610s27. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinformatics. 2006;7:447. doi: 10.1186/1471-2105-7-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS One. 2011;6:e28150. doi: 10.1371/journal.pone.0028150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, Paterson AH. Genome and gene duplications and gene expression divergence: a view from plants. Ann N Y Acad Sci. 2012;1256:1–14. doi: 10.1111/j.1749-6632.2011.06384.x. [DOI] [PubMed] [Google Scholar]
- Wei F, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3:e123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. Robustness—it's not where you think it is. Nat Genet. 2000;25:3–4. doi: 10.1038/75560. [DOI] [PubMed] [Google Scholar]
- Yu J, et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Vision TJ, Gaut BS. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol Biol Evol. 2002;19:1464–1473. doi: 10.1093/oxfordjournals.molbev.a004209. [DOI] [PubMed] [Google Scholar]