Abstract
Activity of the polyamine biosynthetic enzyme ornithine decarboxylase (ODC), and intracellular levels of ODC protein are controlled very tightly. Numerous studies have described ODC regulation at the levels of transcription, translation and protein degradation in normal cells, and dysregulation of these processes in response to oncogenic stimuli. Although post-transcriptional regulation of ODC has been well-documented, the RNA binding proteins (RBPs) that interact with ODC mRNA and control synthesis of the ODC protein have not been defined. Using Ras-transformed rat intestinal epithelial cells (Ras12V cells) as a model, we have begun identifying the RBPs that associate with the ODC transcript. Binding of RBPs could potentially regulate ODC synthesis by either changing mRNA stability or rate of mRNA translation. Techniques for measuring RBP binding and translation initiation are described here. Targeting control of ODC translation or mRNA decay could be a valuable method of limiting polyamine accumulation and subsequent tumor development in a variety of cancers.
Keywords: ornithine decarboxylase, polyamines, RNA stability, protein synthesis, translational regulation, polysome profiles, mRNP assay, AU-rich region, HuR
1. Introduction
Ornithine decarboxylase (ODC) is the first rate-limiting enzyme in the polyamine biosynthetic pathway, converting the amino acid ornithine to the diamine putrescine, which is subsequently used to synthesize the higher polyamines spermidine and spermine (1). Polyamine content, as well as ODC enzyme activity, is tightly regulated in the cell, and ODC is regulated at the levels of transcription, translation, and degradation (1–6). It has been shown that ODC enzyme activity is induced in numerous epithelial cancers, including skin, breast, and colon (7–10). Understanding how ODC synthesis is controlled is crucial in defining the role of high ODC levels in maintaining the transformed phenotype. Our recent studies have used a Ras-transformed rat epithelial cell line (Ras12V cells) to study post-transcriptional regulation of the ODC mRNA (11). These cells will be used as a model in the methods described here.
Cap-dependent translational regulation of ODC through its long, structured 5'-untranslated region (5'UTR) has been well-established, and ODC activity and translation are induced in eIF4E-overexpressing cells (4E-P2 cells) (12, 13). It has also been shown that the presence of the ODC 3'UTR results in decreased synthesis of the ODC protein (14–16). Interestingly, despite extensive study, the RNA-binding proteins (RBPs) that control either ODC translation or stability of the ODC transcript have yet to be described. However, Wang and colleagues have reported a link between changes in intracellular polyamines and post-transcriptional regulation of a variety of mRNAs. It has been found that the RBP human antigen R (HuR) binds to and stabilizes several mRNA's encoding proteins essential for growth control, including p53 and ATF-2, in response to polyamine depletion (17, 18). RBPs generally regulate labile mRNA transcripts by binding to adenosine and uridine-rich elements defined as AREs. These sequences are typically located within the 3'UTR of mRNA (19). One of the best-characterized RBP families is the Hu/elav family of proteins, including the ubiquitously expressed HuR protein. HuR binding generally leads to stabilization of its target mRNAs (20). Binding of a second class of proteins, including the zinc finger protein tristetraprolin (TTP) and TIA-1, promotes instability of target messages (20, 21). A third class of RBPs, for example AUF1, can play a role in both stabilization and destabilization (19). In addition to control of mRNA decay, several RBPs, including HuR and TIA-1, have been shown to modify translation efficiency of their target RNAs as well (22, 23).
Given the extensive post-transcriptional regulation of ODC, and the response of RBPs to changes in polyamines, we have undertaken experiments to determine whether RBPs interact with the ODC mRNA itself, and the consequences of this interaction. In order to assay for endogenous binding of RBPs to the ODC transcipt, we conduct mRNP assays, in which RBPs are immunoprecipitated under conditions that preserve their association with target mRNAs (17). To examine changes in translation initiation of the ODC mRNA brought about by RBP binding, polysome profiles are performed (24). We show examples of results obtained using both of these techniques in Ras12V cells.
2. Materials
2.1. Cell culture and cell extract preparation
1× phosphate buffer: 14 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH buffer to 7.4, and sterilize by autoclaving. Store at 4°C.
Cycloheximide stock: dissolve 100 mg cycloheximide (Calbiochem, San Diego, CA) in 1 ml 100% ethanol; Store at −20°C.
Heparin stock: Dissolve 50 mg Heparin (Grade 1, Sigma, St. Louis, MO) in 1 ml RNAse-free water; Store at 4°C.
mRNP lysis buffer (RLB): 100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.0, 0.5% Nonidet P-40a; additions to be added at time of use: 1mM dithiorthrectol (DTT), 100 units/ml RNase OUT (Invitrogen), and 1× complete protease inhibitor cocktail (Roche, Nutley, NJ) (store in aliquots at −20°C).
Polysome lysis buffer (PLB): Make before each use at the following final concentrations in 0.2 μ-filtered, RNAse-free water: 15mM Tris-HCl pH 7.6, 15mM MgCl2, 0.3M NaCl, 1% Triton X-100, 0.1mg/ml Cycloheximide and 1mg/ml Heparin.
2.2. mRNP immunoprecipitation
Mouse IgG1 antibody (Invitrogen, Carlsbad, CA).
HuR antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA).
Protein A Sepharose (PAS) beads (Sigma).
Bovine serum albumin (BSA): 5% (w/v) in RNAse free water (Invitrogen).
NT2 buffer: 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40. Store at 4°C.
RNA OUT (Invitrogen).
Proteinase K (New England BioLabs, Ipswich, MA).
Phenol:Chloroform:Isoamylacohol (25:24:1) (VWR, West Chester, PA).
Dnase, 10× Dnase reaction buffer (Ambion, Austin, TX).
Chloroform (J.T. Baker Chemical Company, Phillipsburg, NJ).
Glycoblue (Qiagen, Valencia, CA).
2.3. Polysome gradient
20% (w/w) Sucrose (STEP 1 for Sucrose gradient): Combine the following: 21.6 g ultra-pure sucrose; 10 ml 3M NaCl; 3 ml 500 mM MgCl2; 1.5 ml 1 M Tris-HCl, pH 7.6; 100 μl 100 mg/ml Cycloheximide; 2 ml 50mg/ml Heparin. Bring to 100 ml final volume with RNAse-free water and filter through a 0.2 μ filter. Store at 4°C.
47% (w/w) Sucrose (STEP 9 for Sucrose gradient): Combine the following: 57 g ultra-pure Sucrose; 10 ml 3M NaCl; 3 ml 500 mM MgCl2; 1.5 ml 1M Tris-HCl, pH 7.6; 100 μl 100 mg/ml Cycloheximide; 2 ml 50 mg/ml Heparin. Bring to 100 ml final volume with RNAse-free water and filter through a 0.2 μ filter. Store at 4°C.
8 M Guanidine-HCl made with RNAse-free water (solution will be very cold; if problems dissolving occur, leave at 4°C for 1 h). Store at R.T.
3 M Sodium Acetate made with RNAse-free water; adjust pH to 5.2 with glacial acetic acid. Store at R.T.
TE, pH 8.0: 10 mM Tris-HCl, pH 8.0 and 0.1 mM EDTA in RNAse-free water. Store at 4°C
2.7 Northern blot analysis
Hybod N+ membrane (GE Healthcare, Piscatawy, NJ).
Hybridization buffer: 0.5 M NaH2PO4, pH 7.2, 7% SDS, 1 mM EDTA, and 1% BSA in RNAse-free water; store at −20°C in aliquots.
20× SSPE: Combine the following: 175.3 g NaCl; 27.6 g NaH2PO4; 7.4 g EDTA; bring to a final volume of 1000 ml with 0.2 μ-filtered, RNAse-free water, adjust pH to 7.4 with NaOH, sterilize by autoclaving and store at R.T.
10× MOPS: Combine the following: 46.3 g MOPS – Na salt; 6.8 g sodium acetate; 3.4 g EDTA; bring to a final volume of 1000 ml with 0.2 μ-filtered, RNAse-free water, adjust pH to 7.0 with glacial acetic acid, sterilize by autoclaving and store at R.T.
20× SSC: Combine the following: 175.3 g NaCl; 88.2 g sodium citrate; bring to a final volume of 1000 ml with 0.2 μ-filtered, RNAse-free water, adjust pH to 7.0 with NaOH, sterilize by autoclaving and store at R.T.
3. Methods
3.1. Preparation of mRNP lysates from cell culture
Grow and harvest tissue culture cells (confluent) by washing two times with ice-cold PBS.
Trypinize cells according to the standard protocol for the cell line used. Once the cells detach from the plates, add PBS to each plate in order to dilute the trypsin. The cells are transferred to a 50 ml Falcon tube and pelleted by centrifugation at 3000 rpm for 5 min at 4°C.
The supernatant is discarded and the pellet is loosened by gently flicking the bottom of the Falcon tube.
One pellet volume of RLB buffer that has been fully supplemented with RNAse, DTT, and protease inhibitor is added to the pellet. The cells are resuspended by pippetting, NOT by vortexing.
Place the resuspended pellet/RLB mixture on ice for 10 min.
Freeze in liquid nitrogen and store in −80°C until use.
3.2. Preparing the Protein A Sepharose (PAS) beads for mRNP assay
In order to prepare the beads for use, swell the beads in a 50 ml Falcon tube overnight in 5% BSA solution at 4°C. Extra solution is added to the 15 ml mark of the Falcon tube.
The next morning, pour off the excess solution so that the beads are in a 50% (v/v) slurry.
Add 0.1% sodium azide to the bead slurry and place at 4°C for long-term storage.
3.3. Coating Protein A Sepharose beads with antibody for mRNP assay
Place a 100 μl volume of PAS beads into a clean, RNAse-free 1.5 ml microfuge tube.
Add 30 μg of antibody, in our case HuR, to the beads. Make a separate tube with beads plus the appropriate IgG control antibody (see Note 2). Add between 100 and 200 μl more NT-2 buffer to each tube.
Allow the antibody to bind to the beads overnight by placing the tubes in a table top, end-over-end rotator on medium speed at 4°C.
The next morning, centrifuge the beads at 18,000 × g for 5 min at 4°C. Remove the supernatant.
Wash the beads with 1 ml of NT-2 buffer, vortex the tubes for 10 seconds, and centrifuge at 18,000 × g for 5 min at 4°C.
Remove the supernatant, and repeat step 8 for a total of 5 times.
After the fifth wash, remove the NT-2 buffer. The beads are now ready to be used for the pulldown (proceed immediately to section 3.4).
3.4. Pre-clearing the mRNP lysate (see Note 3)
Thaw the mRNP lysate (step 7, Section 3.1) and centrifuge for 18,000 × g for 30 min at 4°C. Transfer the supernatant to a clean, RNAse-free 1.5 ml tube.
Pre-clear the lysate by adding 15 μg IgG1 antibody (see Note 4) to the lysate and allowing it to incubate for 30 min on ice.
Add 50 μl non-coated PAS beads to the lysate (step 2), and incubate the mixture on a table top, end-over-end rotator on medium speed for 30 min at 4°C.
Centrifuge the PAS beads/lysate mix (step 3) at 18,000 × g for 1 min at 4°C.
Place the supernatant into a clean, RNAse-free 1.5 ml microfuge tube. This is the pre-cleared lysate.
Conduct a protein assay to determine the protein concentration. We use the method of Bradford (25). The pre-cleared lysate can be stored at −80°C or can subsequently used for the immunoprecipation step (see Note 5).
3.5. Immunoprecipitation of mRNPs
Add 700 μl NT-2 buffer to the pre-coated PAS/antibody beads (from Section 3.3). Subsequently, add 10 μl 100 mM DTT (see Note 6), 10 μl RNAout, and 33 μl 0.5 M EDTA. Vortex the tubes briefly for 5–10 sec.
Add pre-cleared lysate (from Section 3.4). Typically, we use enough cells to obtain 2–3 mg of lysate for each immunoprecipitation reaction.
Bring the total volume of each tube to 1 ml with NT-2 buffer.
Mix the contents of the tube by flicking the bottom of the 1.5 ml tube.
Incubate the tube for 2 h at room temperature on a table top, end-over-end rotator.
Centrifuge the tubes at 18,000 × g for 5 min at 4°C.
Remove the supernatant and wash the pellet with 1 ml ice-cold NT-2 buffer. Vortex the tube for 5 sec, and then centrifuge at 18,000 × g for 5 min at 4°C.
Repeat step 7 four additional times.
After the final wash, discard the supernatant. Add the following to the pellet: 2.5 μl of Proteinase K (10mg/ml), 1 μl of 10% SDS, and 100 μl of NT-2 buffer.
Incubate the tubes at 55°C for 30 min in a Thermomix (Eppendorf, Westbury, NY) set at 700 rpm.
Centrifuge the tubes at 18,000 × g for 2 min at 4°C.
Collect the supernatant (approximately 100 μl) and place into a clean, RNAse-free 1.5 ml microfuge tube.
Add 200 μl NT-2 buffer to the beads and vortex the tubes for 5 seconds.
Centrifuge the tubes at 18,000 × g for 2 min at 4°C.
Collect the supernatant from step 14 (approximately 200 μl), and combine with the supernatant collected in step 12. A total of 300 μl should now be in a 1.5 ml tube. The beads can now be discarded.
Add 800 μl phenol:chloroform:isoamylalcohol (25:24:1) to the 300 μl supernatant (see Note 7).
Vortex the tubes for 1 min.
Centrifuge the tubes at 18,000 × g for 1 min at room temperature.
Collect the upper phase (approximately 300 μl) and add 30 μl of 3 M sodium acetate, 650 μl 100 % ethanol, and 5 μl glycoblue to each tube. Mix the contents of the tube well and store overnight at −20°C.
The next day, spin the tubes at 18,000 × g for 30 min at 4°C.
Discard the supernatant.
Add 1 ml ice-cold 70% ethanol to the pellet and mix by inversion.
Centrifuge the tubes at 18,000 × g for 2 min at 4°C.
Discard the supernatant.
Centrifuge the pellet at 18,000 × g for 1 min at 4°C.
Remove any residual ethanol from the pellet carefully with a pipette.
Allow the pellet to air dry for 5 min at room temperature.
Resuspend the pellet in 20 μl of water that has been heated to 65°C. Place the tubes at 65°C for 5 min.
Add 2 μl 10× DNAse reaction buffer and 1 μl DNAse 1 to each tube.
Mix the contents of the tube and incubate the tubes at 37°C for 30 min.
Add 80 μl of RNAse-free water and 100 μl phenol:chloroform:isoamylalcohol to the tubes and vortex for 2 min.
Centrifuge the tubes at 18,000 × g for 10 min at 4°C.
Place the upper phase into a clean, RNAse-free 1.5 ml microfuge tube.
To the upper phase (from step 33), add 100 μl chloroform and vortex for 20 sec.
Centrifuge the tubes at 18,000 × g for 10 min at 4°C.
Remove the upper phase and place into a clean, RNAse-free 1.5 ml microfuge tube.
To the upper phase (from step 36), add 10 μl of 3 M sodium acetate, 250 μl 100% ethanol, and 2.5 μl glycoblue.
Mix the contents of the tube well and place at −20°C for at least 1 h.
Centrifuge the tubes at 18,000 × g for 30 min at 4°C.
Discard the supernatant.
Wash the pellet with 70% ice-cold ethanol.
Centrifuge the tubes at 18,000 × g for 5 min at 4°C.
Discard the supernatant.
Centrifuge the tubes at 18,000 × g for 1 min at 4°C.
Remove any residual ethanol from the pellet.
Allow the pellet to air dry for 5 min at room temperature.
Resuspend the pellet in 15 μl of water that has been heated to 65°C.
Incubate the tubes at 65°C for 5 min.
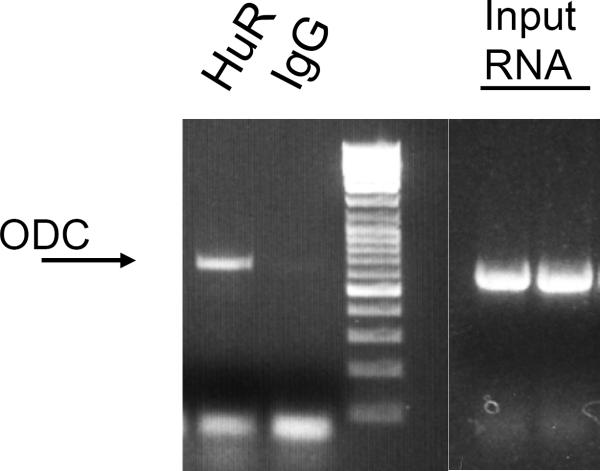
This is your pulldown RNA. cDNA can be made for RT PCR analysis or qRTPCR analysis using one of several commercially available kits. We use the Transcripter 1st Strand cDNA Synthesis Kit (Roche). PCR primers to detect ODC RNA bound to HuR are listed in Note 8. Typical results are shown in Figure 1.
Figure 1. mRNP assay of Ras12V cell lysate to measure endogenous RBP binding to ODC RNA.
To determine endogenous binding of HuR to the ODC transcript, HuR was immunoprecipitated from Ras12V cells as described in the Methods. Cell lysates isolated in RLB were incubated with a 50% (v/v) suspension of Protein A-sepharose beads precoated with either IgG1 or anti-HuR (Santa Cruz Biotech). After washing in NP40-based buffer, beads were treated with RNase-free DNase I and proteinase K, then RNA was extracted and reverse transcribed to obtain cDNA. The presence of ODC was analyzed by conventional PCR using primer pairs corresponding to a 635 bp region within the ODC transcript (see Note 8). Lanes labeled “Input RNA” represent duplicate PCR reactions performed with the same ODC primers on cDNA prepared from cell lysate without immunoprecipitation, indicating the presence of ODC RNA in the lysate.
3.6. Pouring sucrose step gradients for polysome analysis
Use Step 1 (20% sucrose) and Step 9 (47% sucrose) solutions prepared as described in Section 2; all other steps are combinations of these two.
- Pouring gradients: First make steps as follows:
40 ml Step 1 + 40 ml Step 9 = 80 ml Step 5
20 ml Step 1 + 20 ml Step 5 = 40 ml Step 3
10 ml Step 1 + 10 ml Step 3 = 20 ml Step 2
10 ml Step 3 + 10 ml Step 5 = 20 ml Step 4
20 ml Step 5 + 20 ml Step 9 = 40 ml Step 7
10 ml Step 5 + 10 ml Step 7 = 20 ml Step 6
10 ml Step 7 + 10 ml Step 9 = 20 ml Step 8 - Layering gradients:
- • Start with Step 9 and end with Step 1
- • Add 1.25 ml of Step 9 into a disposable ultracentrifuge tube (we use Beckman polyallomer tubes cat # 344059)
- • After adding Step 9, freeze in liquid nitrogen, then continue with each additional step (add 1.25ml Step 8, freeze, etc. until Step 1 is added to the top)
- • Store Step gradients at −80°C until use.
- • Thaw gradients overnight at 4°C the day before your experiment (once gradients are thawed be very careful when moving the tubes).
3.7. Preparation of cell lysates for polysome analysis
Treat cells as desired for experiment. We use about 1 × 107 cells per gradient.
For extraction, aspirate medium, add 5 ml pre-warmed PBS containing 0.1 mg/ml cycloheximide to each plate and incubate for 5 min at 37°C.
Aspirate and wash 2 times with ice cold PBS/cycloheximide (100 μg/ml). Be sure to remove all traces of PBS when aspirating,
Scrape cells into 600 μl total volume PLB (300 μl twice) into a sterile microfuge tube on ice.
Rock samples gently for 10 min at 4°C.
Centrifuge samples at 3000 × g for 15 min at 4°C.
Load 600 μl supernatant directly onto the top of a 20–47% sucrose gradient prepared as described in section 3.7. Layer gently so the gradient is not disturbed. Balance tubes if necessary with PLB.
Cool centrifuge and rotor at least 1 h prior to use. Centrifuge gradients at 34,000 rpm for 160 min at 4°C using a Beckman SW41Ti rotor (Beckman Coulter, Fullerton, CA) in a L90K Beckman ultracentifuge or equivalent.
3.8. Collection of fractions and RNA isolation (see Note 9)
Gradients are fractionated immediately after removing from the centrifuge.
The entire gradient can be split into many fractions. We split into 11 fractions every 30 sec.
Each fraction is collected directly into 2.5 ml of 8M guanidine-HCl plus 250 μl RNAse-free water.
Shake each fraction vigorously, add 2× volume of 100% ethanol, and precipitate overnight at −20°C.
Next day (after at least 16 h), centrifuge fractions at 10,000 × g and 4°C for 20 min to pellet RNA.
Aspirate supernatant, wash pellet with 75% ethanol (made with RNAse-free water) and centrifuge at 10,000 × g and 4°C for 10 min.
Dissolve pellet in 400 μl TE, pH 8.0.
Add 50 μl 3M Sodium Acetate, pH 5.2 and 1ml 100% ethanol. Precipitate overnight at −20°C.
Next day, centrifuge at 10,000 × g for 20 min at 4°C.
Remove supernatant and wash pellet one time with 75% ethanol. Centrifuge at 10,000 × g for 10 min at 4°C.
Remove supernatant and dry pellet at room temperature for 5 min.
Add 15 μl nuclease-free water. If pellets do not dissolve, wait 3–5 min and carefully resuspend using a pipette.
Measure concentration using a spectrophotometer at 260 nm. Store at −80°C until use.
3.9. Northern blot analysis of polysome gradient fractions
For each fraction, use 15 μg RNA in 6 μl nuclease-free water. Fractions may need to be concentrated by lyophilizing before use.
- To each sample add:
- 12.5 μl Formamide
- 2.5 μl 10× MOPS
- 4.0 μl Formaldehyde (37% solution)
Incubate at 65°C for 5 min, then chill on ice.
Add 3 μl of 50% glycerol (in nuclease-free water) containing 0.1% bromphenol blue and 1 mM EDTA to each sample.
Prepare a 1.2% Formaldehyde gel as follows (prepare in fume hood): Combine 1.8 g agarose, 15 ml 10× MOPS, and 109.5 ml water. Dissolve agarose by heating in a microwave (do not allow the solution to boil). Cool to 50°C. Add 25.5 ml of Formaldehyde (37% solution). Mix and pour immediately (volumes may need to be adjusted depending on the gel apparatus used). Allow gel to polymerize at room temperature.
For each fraction, load the entire sample onto the gel. Run gel in 1× MOPS overnight at 20V.
Before transfer, soak gel in 100 ml nuclease-free water containing 4 μl of 10 mg/ml ethidium bromide (Caution: mutagen. Wear appropriate protection) for 20 min, then destain 1 h in water to visualize 18S and 28S bands.
Transfer to Hybond N+ membrane fro 4.5 h in 20× SSPE using a Turboblotter (Whatman Schleicher and Schuell, St Louis, MO) or similar apparatus, following the instructions of the manufacturer.
Wash membrane in 5× SSPE for 5 min, then remove from wash and place membrane on a piece of filter paper or paper towel.
Crosslink RNA to membrane using a Stratalinker (Agilent Technologies Stratagene, La Jolla, CA) or similar apparatus at a setting of 120,000 microjoules/cm2 (“auto-crosslink” on the Stratalinker).
Pre-hybridize membrane for 1–2 h at 65°C in 5 ml hybridization buffer.
- Probe preparation requires a DNA template. For ODC we use pGEM-ODC (26). The entire ODC coding region is removed by restriction digest as follows:
pGEM-ODC 10 μg BamHI (Invitrogen) 10 U EcoRI (Invitrogen) 10 U 100 × BSA (Invitrogen) 1 μl Invitrogen React 2 Buffer 10 μl Nuclease-free water to a final volume of 100 μl Allow reaction to proceed at 37°C for 1 h.
Run reaction mix on 0.8% agarose gel containing 1 μg/ml ethidium bromide. Visualize the bands by placing the gel on a transilluminator box and cut the 1.8 Kb insert from the gel with a scalpel.
Purify DNA from the agarose using Quick-spin columns (Sigma) according to the manufacturer's instructions.
Several commercial kits are available for probe preparation. We use 25 ng of the purified insert as a template to make the [32-P]-labeled probe with the Ladderman kit (Takara Bio Inc., Shiga, JAPAN), following the manufacturer's instructions exactly (see Note 10).
After probe synthesis, add 20 μl of probe to a sterile 1.2 ml microfuge tube, and place in a boiling water bath for 5 min.
Add 1 μl boiled probe per ml of hybridization buffer (we generally use 5 ml hybridization buffer).
Hybridize overnight at 65°C.
Next day, wash membrane once with 1× SSC + 0.1% SDS at R.T. for 10 min.
Wash membrane 3 times with 0.5× SSC + 0.1% SDS at 65°C for 10 min.
Expose membrane (wrapped in plastic) to Autoradiography film (e.g. HyBlot, Denville Scientific, Metuchen, NJ) in a cassette placed at −80°C overnight.
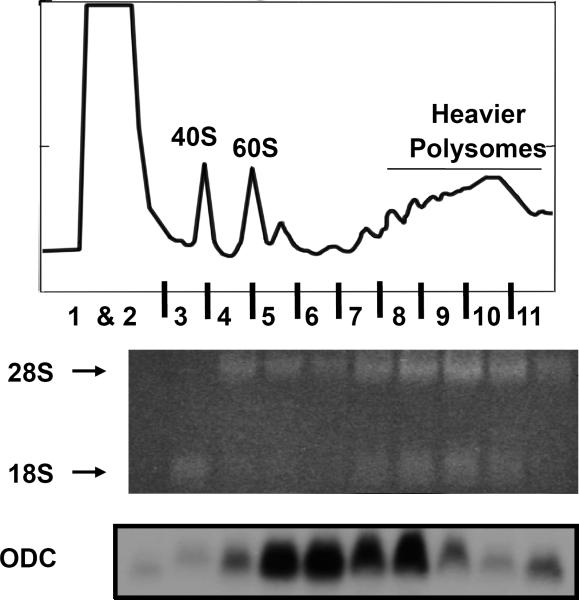
Next day, process film to reveal bands corresponding to ODC mRNA in each polysome fraction. Exposure time may need to be adjusted to get an optimal signal. Typical results of both agarose gel staining with ethidium bromide and polysome profile of ODC mRNA are shown in FIG 2.
Figure 2. Analysis of polysomal RNA integrity and polysomal association of ODC RNA in Ras12V cells.
Sucrose gradient centrifugation and polysome analysis of Ras12V cells were carried out as described in Methods. Eleven gradient fractions were collected and RNA was isolated from each fraction and analyzed on a 1.2% formaldehyde-MOPS agarose gel stained with Ethidium bromide. Equal volumes of total RNA were loaded for each fraction and the agarose gel image shows total RNA corresponding to each fraction. Arrows indicate the position of 18S rRNA and 28S rRNA. Fraction 1 and 2 do not show any detectable levels of RNA as they represent the non-polysomal fraction. Only 18S rRNA is visible in fraction 3, suggesting that it consists predominantly of the 40S ribosomal subunit. Fraction 4 represents the 60S ribosomal subunit that consists mostly of 28S rRNA. Fractions 5 and 6 represent the 80S and disome fractions showing mostly 28S rRNA and faint levels of 18S rRNA. Fractions 7 through 11 represent the polysomal peak, displaying similar levels of 18S rRNA in comparison to 28 rRNA. To visualize ODC RNA in each fraction, the gel was de-stained and Northern blot analysis was performed. The membrane was probed with a [32-P] radiolabeled cDNA complementary to the full length ODC open reading frame. The polysome profile shows that ODC RNA is associated mostly in the disome and small polysome fractions, indicating ODC is poorly translated in these cells.
4. Notes
For all of these procedures, it is important to use RNAse-free pipette tips, RNAse-free water, RNAse-free microfuge tubes, and to wear gloves.
For each antibody used, an IgG control must be added. For example, the HuR antibody is anti-mouse and thus for each cell line used we have a mouse IgG negative control.
This step can be completed the day before the mRNP is conducted or can be completed while the coated beads are being washed.
Make sure the IgG pre-clear is for the correct species. For example, theHuR antibody is anti-mouse therefore we use a mouse IgG to pre-clear the supernatant.
Clearing of the lysate can be accomplished before the beads are coated and the mRNP is conducted. If you pre-clear the lyaste before you are able to move forward with the experiment, store the pre-cleared lysates in −80°C.
Do not add DTT directly to the pellet. This reduces antibody efficiency.
Make sure the phenol chloroform solution is mixed well prior to use.
The following primers are used for ODC PCR following the mRNP assay (Note that these primers are to the mouse ODC sequence and may need to be altered for other species): Sense: 5'-CGAGAACCATGAGCAGCTTTAC-3' Antisense: 5'-CTACACATTGATCCTAGCAGCC-3'
This step requires a fractionator (such as an ISCO sucrose gradient absorbance profile analyzer, Teledyne Isco, Lincoln, NE) and uv detector at 254 nm. Parameters we use while collecting: Flow rate 2 ml/min, chart speed 150 cm/h, sensitivity 0.5. This must be determined experimentally for the fractionator used.
The Ladderman kit is designed for use with [α-32P], [α-35S] or [3H] dCTP labels. Generally, a probe with specific activity of > 109 dpm/μg DNA will be obtained with [a-32P] dCTP (~3000 Ci/mmol). Although kits designed for non-radioactive probe labeling are also available (e.g. Biotin-labeling kits) it is our experience that [32-P]-labeled probes give better-quality results.
Acknowledgements
The authors would like to thank Suzanne Sass-Kuhn for excellent technical assistance. This work was supported by R01 CA82768 (to LMS).
References
- (1).Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2006;281:14529–32. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- (2).Zhao B, Butler AP. Core promoter involvement in the induction of rat ornithine decarboxylase by phorbol esters. Mol Carcinog. 2001;32:92–9. doi: 10.1002/mc.1068. [DOI] [PubMed] [Google Scholar]
- (3).Li R, Abrahamsen MS, Johnson RR, Morris DR. Complex interactions at a GC-rich domain regulate cell type-dependent activity of the ornithine decarboxylase promoter. J. Biol. Chem. 1994;269:7941–7949. [PubMed] [Google Scholar]
- (4).Wallon UM, Persson L, Heby O. Regulation of ornithine decarboyxlase during cell growth. Changes in the stability and translatability of the mRNA, and in the turnover of the protein. Mol. Cell. Biochem. 1995;146:39–44. doi: 10.1007/BF00926879. [DOI] [PubMed] [Google Scholar]
- (5).Shantz LM. Transcriptional and translational control of ornithine decarboxylase during Ras transformation. Biochem. J. 2004;377:257–264. doi: 10.1042/BJ20030778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Shantz LM, Pegg AE. Translational regulation of ornithine decarboxylase and other enzymes of the polyamine pathway. Int. J. Biochem. Cell Biol. 1999;31:107–122. doi: 10.1016/s1357-2725(98)00135-6. [DOI] [PubMed] [Google Scholar]
- (7).Huang Y, Pledgie A, Casero RA, Jr., Davidson NE. Molecular mechanisms of polyamine analogs in cancer cells. Anticancer Drugs. 2005;16:229–41. doi: 10.1097/00001813-200503000-00002. [DOI] [PubMed] [Google Scholar]
- (8).Gerner EW, Meyskens FL., Jr. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–92. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- (9).O'Brien TG, Megosh LC, Gilliard G, Peralta Soler A. Ornithine decarboxylase overexpression is a sufficent condition for tumor promotion in mouse skin. Cancer Res. 1997;57:2630–2637. [PubMed] [Google Scholar]
- (10).O'Brien TG. The induction of ornithine decarboxylase is an early, possibly obligatory event in mouse skin carcinogenesis. Cancer Res. 1976;36:2644–2653. [PubMed] [Google Scholar]
- (11).Origanti S, Shantz LM. Ras transformation of RIE-1 cells activates cap-independent translation of ornithine decarboxylase: regulation by the Raf/MEK/ERK and phosphatidylinositol 3-kinase pathways. Cancer Res. 2007;67:4834–42. doi: 10.1158/0008-5472.CAN-06-4627. [DOI] [PubMed] [Google Scholar]
- (12).Shantz LM, Pegg AE. Overproduction of ornithine decarboxylase caused by relief of translational repression is associated with neoplastic transformation. Cancer Res. 1994;54:2313–2316. [PubMed] [Google Scholar]
- (13).Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Grens A, Scheffler IE. The 5'- and 3'-untranslated regions of ornithine decarboxylase mRNA affect the translational efficiency. J. Biol. Chem. 1990;265:11810–11816. [PubMed] [Google Scholar]
- (15).Lorenzini EC, Scheffler IE. Co-operation of the 5' and 3' untranslated regions of ornithine decarboxylase mRNA and inhibitory role of its 3' untranslated region in regulating the translational efficiency of hybrid RNA species via cellular factors. Biochem. J. 1997;326:361–367. doi: 10.1042/bj3260361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lövkvist-Wallstrom E, Stjernborg-Ulvsbäck L, Scheffler IE, Persson L. Regulation of mammalian ornithine decarboxylase. Studies on the induction of the enzyme by hypotonic stress. Eur. J. Biochem. 1995;231:40–44. [PubMed] [Google Scholar]
- (17).Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–94. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- (18).Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Zhou H, Gorospe M, Wang JY. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol Biol Cell. 2007;18:4579–90. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- (20).Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem. 2007;100:1477–92. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- (22).Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- (23).Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- (24).Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci U S A. 1999;96:13118–23. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bradford M. A rapid sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- (26).Lu L, Stanley BA, Pegg AE. Identification of residues in ornithine decarboxylase essential for enzymatic activity and for rapid protein turnover. Biochem. J. 1991;277:671–675. doi: 10.1042/bj2770671. [DOI] [PMC free article] [PubMed] [Google Scholar]