Abstract
Aromatic amine drugs have been associated with agranulocytosis (neutrophil depletion) for which the mechanism is unknown. We have previously shown that the metabolism of two aromatic amine drugs by human myeloperoxidase (MPO) results phenyl radical metabolite formation and also in protein free radical formation on MPO (Siraki et al., 2007; 2008). Since the concentration of drug required to produce a maximum signal for MPO protein free radical (MPO•) detection was different for each drug, this prompted us to consider that other aromatic amines may also show varying degrees of ability to induce MPO• formation. Immunoassay experiments (using the immuno-spin trapping technique) were performed which evaluated the potency of 21 different aromatic amines (containing the aniline substructure) to generate the MPO•. Each reaction contained equal amounts of H2O2, 5,5-dimethyl-1-pyrroline-N-oxide (DMPO), MPO, and variable concentrations of aniline derivatives. Several physicochemical parameters for aniline derivatives were used to derive QSAR equations, which showed that the Hammett constant (σ) best correlated with the MPO• formation for all aniline derivatives. More statistically robust equations were derived if the anilines were separated into mono- and di-substituted groups. However, some aniline derivatives did not induce MPO• formation. Using electron spin resonance (ESR) spectroscopy, we evaluated the ability of all aniline derivatives tested to produce phenyl radical metabolites, as previously shown for the aromatic amine drugs by spin trapping. Interestingly, we found that only those aniline derivatives that produced a phenyl radical also formed MPO•. We propose that the phenyl radical is the reactive free radical metabolite responsible for generating the MPO•.
Keywords: aniline, free radical, myeloperoxidase, immune-spin trapping, QSAR
Introduction
Aromatic amine xenobiotics are associated with numerous acute toxicities, including methemoglobinemia and hemolytic anemia, while chronic exposure to certain aromatic amines (such as 4-aminobiphenyl) is associated with cancer. Occupational exposure has been a focal point for the determination of biomarkers to evaluate human exposure. Drug molecules containing the aromatic amine or aniline substructure are also associated with toxic side-effect, including agranulocytosis – the depletion of neutrophils (<500/ μl) – which increases the risk of sepsis. The mechanism of drug-induced agranulocytosis is not clear; however, it is believed that reactions resulting from the formation of reactive drug metabolites are involved (Christie et al., 1989; Uetrecht, 1989b). The reaction center for aromatic amines is the aniline moiety. In the instance of procainamide-induced lupus, N-acetylprocainamide, which also has pharmacological activity, was shown to significantly delay the onset of anti-nuclear antibody formation (Lahita et al., 1979). The N-acetyl group is electron withdrawing, and therefore deactivating with respect to oxidation. suggesting that oxidation of the aromatic amine of procainamide to a reactive metabolite may play an etiological role. Furthermore, procainamide-induced lupus developed more rapidly in slow acetylator phenotype individuals (Reidenberg, 1983). No similar findings exist for drug-induced agranulocytosis, which is due in part to the low frequency of this side-effect as well as the lack of suitable models of the condition.
The possibility of the involvement of MPO1-generated free radical metabolites in drug-induced agranulocytosis has been proposed for some time (Fischer et al., 1991). Peroxidase enzymes are capable of oxidizing numerous substrates to free radical metabolites (Dunford, 1999). Since MPO is located in neutrophils, it has been suggested as a catalyst for reactive metabolite generation that could lead to neutrophil death (Uetrecht, 1992). It was recently found that in a case of dapsone-induced agranulocytosis, the patient presented an absence of neutrophils, eosinophils, and CD34+ progenitor cells, but basophil levels were unchanged (Besser et al., 2009); this finding was attributed to reactive metabolite formation by peroxidases (myeloperoxidase and eosinophil peroxidases), since basophils are typically low in peroxidase activity.
We have previously found that myeloperoxidase catalyzes the oxidation of two aromatic amine drugs, which leads to the formation of protein free radicals on myeloperoxidase in the HL-60 cell line as well as in purified enzyme preparations (Siraki et al., 2007; Siraki et al., 2008). In this study, we build upon our previous findings to investigate the factors responsible for these observations. We have focused on the physicochemical properties of aniline derivatives that lead them to generate protein free radicals, and developed quantitative structure activity relationships (QSARs, Hansch analysis) that relate their potency in generating protein free radicals to their physicochemical properties. In addition, we have examined the relationship between the detection of aryl radicals formed from aromatic amines and their ability to form protein free radicals.
Materials and Methods
Reagents
Human MPO was purchased from Athens Research & Technology (Athens, GA) and was dissolved in 0.1 M phosphate buffer pH 7.4 and dialyzed twice for 12 hours each time. MPO concentration was determined by its extinction coefficient (178 mM−1 cm−1 at 429 nm) (Hsuanyu and Dunford, 1999). Horseradish peroxidase type VI (HRP, Sigma Chemical co., St. Louis, MO) was desalted in a PD-10 desalting column (GE Healthcare, Waukesha, WI) prior to use and its concentration was determined by 102 mM−1 cm−1 at 402 nm. 5,5-Dimethyl-1-pyrroline N-oxide (DMPO) was purchased from Alexis Biochemicals (San Diego, CA), purified twice by vacuum distillation at room temperature, and stored under argon atmosphere at −80 °C until use. Hydrogen peroxide (30% v/v, H2O2; Fisher Scientific Co., Fair Lawn, NJ) was assayed by its extinction coefficient of 43.6 M−1cm−1 at 240 nm (Beers and Sizer, 1952). All aniline compounds, 2-methyl-2-nitrosopropane (MNP), and diethylenetriaminepentaacetic acid were purchased from Sigma Chemical Co. (St. Louis, MO) at the highest grade available. MNP was dissolved in buffer (2 mg/mL), protected from light, sealed, and shaken overnight at 32 °C. Chelex-100 resin (Bio-Rad Laboratories, Hercules, CA) was mixed with purified water overnight, and filtered before use. Phosphate buffer used in experiments was mixed with Chelex-100 resin overnight to remove metal contaminants, and the resin was removed by vacuum filtration. Diethylenetriaminepentaacetic acid (100 mM) was included in the buffer to prevent metal-catalyzed reactions.
Enzyme-Linked Immunosorbent Assay (ELISA) and MPO protein free radical detection
Ninety-six-well ELISA plates (Greiner Labortechnik, Germany) were used to carry out reactions in order to detect the DMPO-MPO adducts. These reactions were carried out in buffer and contained 100 mM DMPO, 100 μM H2O2, 50 nM MPO, and varying concentrations of anilines. The reactants were mixed in 1.5 ml centrifuge tubes, then 100 μl of this reaction was transferred to each of three wells containing 200 μl water. This reaction was carried out for 2 hours in an Eppendorf Thermomixer R (Germany) at 37 C with mixing at 500 rpm. At the end of the incubation period, the plate was washed and blocked overnight with 4 % cold fish gelatin and anti-DMPO detection of MPO-DMPO was carried out as described previously (Siraki et al., 2007). The concentration of aniline derivative necessary to induce a two-fold increase above background for detection of MPO• (MPO-DMPO) was designated the EC2 value
ESR spectroscopy detection of aniline free radical metabolites
Free radical metabolites were detected by ESR spectroscopy using the spin trapping technique (Lagercrantz, 1971). The spin trap we used was MNP, which we previously found provided significant detail for revealing the structure of the free radical that was formed (Siraki et al., 2007; Siraki et al., 2008). ESR spectra were recorded using an Elexsys E500 spectrometer (Billerica, MA) using an ER 4122 SHQ cavity. The settings for most spectra were recorded at 9.78 GHz, 100 kHz modulation field, 20 mW power, 0.4 G modulation amplitude, 1024 points, and 6.32×105 receiver gain. In certain cases where the spectra were not well resolved, we used the following modifications to these settings: 1 mW power, 0.07 G modulation amplitude, conversion time and time constant of 2621.44 ms, 2048 data points, 5368 s sweep time. In order to generate free radical metabolites, we added 10 mM MNP, 5 mM aniline, 1.7 μM HRP, and 1 mM H2O2 in 200 μL buffer (described above), briefly vortexed the reaction, and added it to a quartz flat cell for recording spectra. We chose these concentrations based on preliminary experiments.
Physicochemical Parameters and QSAR derivation
The Hammett sigma (σ) values were obtained from (Perrin et al., 1981), lipophilicity (log P) was obtained from (Tetko et al., 2005); and ionization potential of the parent aniline (IP-P), electron affinity of the parent aniline (EA-P), and ionization potential and electron affinity of the corresponding phenyl radical metabolites (IP-R, EA-R, respectively), were calculated using MOPAC with PM3 parameters in Sybyl 7.1. QSAR equations were derived using SigmaPlot 11.0. Best subset regression was used to identify the physicochemical parameter that best correlated with the EC2 values, and it was evaluated whether or not the inclusion of additional parameters would improve the statistical robustness of the equation. In the case of outliers, we identified them both by standardized residuals, as well as visually. Outliers were removed only if they increased the correlation (r2) by at least 0.05.
Results
Aniline-induced MPO free radical formation
The aniline derivatives used in this study were assessed for their ability to induce anti-DMPO detection (formation of MPO•) that was two-fold greater than control levels (defined as the EC2 value). As shown in Table 1, we were able to derive EC2 values for most aniline compounds used in this study. The EC2 values ranged from 2.4 μM to 733 μM. The most potent inducers of protein free radical formation were 2,4-dimethylaniline, and 4-ethylaniline, and the least potent was 2,6-dichloroaniline. We were not able to derive EC2 values for p-anisidine, 3,4-dimethoxyaniline, 2,4-dimethoxyaniline, or 3-nitro-or 2-nitroaniline, because for these compounds there was no increase in anti-DMPO detection significantly greater than control found in the concentration range between 0.1 μM – 500 μM of aniline. The physicochemical parameters were obtained as described in the legend and Materials & Methods.
Table 1.
EC2 values for aniline-induced MPO free radical formation and physicochemical parameters for anilines.
| EC2 | log EC2 | σ 1 | log P2 | EA-R3 | IP-R3 | IP-P3 | EA-P3 | |
|---|---|---|---|---|---|---|---|---|
| aniline | 20.5 | 1.31 | 0 | 0.9 | 4.96 | 9.97 | 8.53 | −0.64 |
| p-toluidine | 3.1 | 0.49 | −0.14 | 1.39 | 4.92 | 9.95 | 8.35 | −0.61 |
| m-toluidine | 8.9 | 0.95 | −0.06 | 1.4 | 4.90 | 9.89 | 8.48 | −0.59 |
| o-toluidine | 3.6 | 0.56 | 0.1 | 1.32 | 4.91 | 9.93 | 8.44 | −0.60 |
| p-anisidine | --* | -- | −0.28 | 0.95 | ||||
| m-anisidine | 6.8 | 0.83 | 0.11 | 0.93 | 5.03 | 10.01 | 8.50 | −0.62 |
| o-anisidine | 3.6 | 0.56 | 0 | 1.18 | 5.04 | 10.06 | 8.26 | −0.57 |
| 3,4-dimethylaniline | 2.7 | 0.43 | −0.2 | 1.84 | 4.88 | 9.88 | 8.32 | −0.61 |
| 2,4-dimethylaniline | 2.4 | 0.38 | −0.04 | 1.68 | 4.89 | 9.92 | 8.28 | −0.61 |
| 2,6-dimethylaniline | 7.3 | 0.86 | 0.2 | 1.84 | 4.89 | 9.92 | 8.35 | −0.60 |
| 3,4-dimethoxyaniline | -- | -- | −0.17 | |||||
| 2,4-dimethoxyaniline | -- | -- | −0.28 | |||||
| 4-chloroaniline | 19.3 | 1.29 | 0.24 | 1.83 | 5.24 | 10.26 | 8.60 | −0.28 |
| 3-chloroaniline | 248 | 2.39 | 0.37 | 1.88 | 5.26 | 10.22 | 8.73 | −0.26 |
| 2-chloroaniline | 61.2 | 1.79 | 0.67 | 1.9 | 5.32 | 10.31 | 8.62 | −0.29 |
| 4-fluoroaniline | 16.3 | 1.21 | 0.06 | 1.15 | 5.27 | 10.34 | 8.55 | −0.29 |
| 2,4-dichloroaniline | 67.6 | 1.83 | 0.91 | 2.78 | 5.58 | 10.58 | 8.68 | −0.01 |
| 3,4-dichloroaniline | 455 | 2.66 | 0.61 | 2.69 | 5.49 | 10.46 | 8.74 | −0.00 |
| 2,6-dichloroaniline | 733 | 2.87 | 1.34 | 2.76 | 5.67 | 10.64 | 8.73 | 0.01 |
| 4-nitroaniline | 345 | 2.54 | 0.78 | 1.39 | 5.89 | 10.93 | 9.27 | 0.78 |
| 3-nitroaniline | -- | -- | 0.74 | 1.37 | ||||
| 2-nitroaniline | -- | -- | 1.72 | 1.85 | ||||
| 4-ethylaniline | 2.4 | 0.38 | −0.15 | 1.96 | 4.92 | 9.94 | 8.33 | −0.65 |
| 4-aminosalicylate | 29.8 | 1.47 | 0.57 | 0.89 | 6.00 | 11.1 | 9.47 | 0.52 |
| 4-chloro-2-methylaniline | 3.64 | 0.56 | 0.34 | 2.1 | 5.20 | 10.23 | 8.51 | −0.29 |
| 4-aminobenzonitrile | 279 | 2.45 | 0.7 | 0.9 | 5.49 | 10.53 | 8.87 | 0.19 |
|
| ||||||||
| aminoglutethimide | 1.5 | 0.176 | −0.154 | 1.1 | 8.30 | |||
| procainamide | 3.2 | 0.505 | 0.365 | 0.88 | 8.43 | |||
From Perrin et al., 1981.
From Tetko, I. V.; Gasteiger, J.; Todeschini, R.; Mauri, A.; Livingstone, D.; Ertl, P.; Palyulin, V. A.; Radchenko, E. V.; Zefirov, N. S.; Makarenko, A. S.; Tanchuk, V. Y.; Prokopenko, V. V. Virtual computational chemistry laboratory - design and description, J. Comput. Aid. Mol. Des., 2005, 19, 453–63 (http://www.vcclab.org).
Values were determined by MOPAC calculations as described in Materials & Methods. EA-R, electron affinity of the radical metabolite; IP-R, ionization potential of the radical metabolite; IP-P, ionization potential of the parent aniline; EA-P, electron affinity of the parent aniline.
Value for –C(CH3)3.
Value for –CONHCH3.
Derivation of QSAR equations
Before derivation of QSARs it is important to determine the relationship between the physicochemical parameters to be used in QSAR derivation (intercorrelation). In Table 2 we show the correlation between different physicochemical parameters. This analysis is important because if two parameters are collinear, they cannot be used in the same QSAR equation. Table 2 revealed that σ was significantly correlated with EA-R, IP-R, IP-P, and EA-P. The quantum chemical parameters were highly collinear, and only log P was not significantly correlated with the other parameters. QSAR derivation will also reveal whether a parameter contributes significantly to the equation. Table 3 shows the QSAR equations derived from the anilines used in this study. Interestingly, the best overall fit was obtained using σ not the calculated quantum chemical parameters (Figure 1).
Table 2.
Pearson correlation (correlation matrix) for the physicochemical parameters used to derive QSAR equations in this study.
| log P | EA-R | IP-R | IP-P | EA-P | |
|---|---|---|---|---|---|
| σ | 0.47a 0.033 |
0.83
3.1×10−7 |
0.795
1.7×10−6 |
0.648
0.00150 |
0.754
7.8×10−5 |
| log P | 0.16 0.48 |
0.107 0.643 |
−0.109 0.638 |
0.0736 0.751 |
|
| EA-R |
0.996
3.7 ×10−21 |
0.918
4.8×10−9 |
0.962
4.0×10−12 |
||
| IP-R |
0.925
2.1×10−9 |
0.965
1.5×10−12 |
|||
| IP-P |
0.93
6.4×10−10 |
Each pair of values corresponds to the correlation (r) and significance (p) between the physicochemical parameters that are compared. Values in bold typeface show highly significant correlation.
Table 3.
QSAR Equations for aniline-induced MPO free radical formation.
| Eqn | All anilines | n | r 2 | F | s |
|---|---|---|---|---|---|
| 1 | log EC2 = 0.794(±0.126) + 1.737σ(±0.249) | 21 | 0.719 | 48.6 | 0.458 |
| 2 | log EC2 = 0.751(±0.114) + 1.721σ(±0.223) Outlier: 3-chloroaniline | 20 | 0.767 | 59.4 | 0.410 |
| 3 | log EC2 = 0.789(±0.106) + 1.730σ(±0.204) Outliers: 3-chloroaniline, 4-chloro-2-methylaniline | 19 | 0.808 | 71.7 | 0.375 |
|
| |||||
| Mono-substituted anilines | |||||
| 4 | log EC2 = 0.851(±0.126) + 2.117σ(±0.338) | 13 | 0.781 | 39.3 | 0.379 |
| 5 | log EC2 = 0.809(±0.103) + 2.008σ(±0.277) outlier: 3-Cl | 12 | 0.841 | 52.7 | 0.307 |
| 6 | log EC2 = −21.252(±3.457) + 2.627IP-P(±0.403) | 13 | 0.795 | 42.5 | 0.368 |
| 7 | log EC2 = −32.238(±3.730) + 3.921IP-P(±0.438) outlier: 4-nitroaniline | 12 | 0.889 | 80.3 | 0.248 |
|
| |||||
| Disubstituted anilines | |||||
| 8 | log EC2 = 0.578(±0.256) + 1.726σ(±0.386) | 8 | 0.769 | 20.0 | 0.515 |
| 9 | log EC2 = 0.474(±0.137) + 1.630σ(±0.205) Outlier: 3,4-dichloroaniline removed | 7 | 0.927 | 63.4 | 0.271 |
Figure 1.
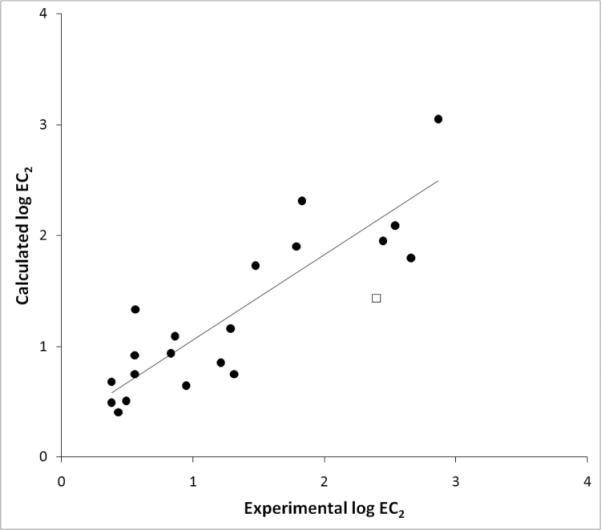
Experimental vs. Calculated log EC2 values for anilines. Each data point was derived by applying equation 2, Table 3, that contained both mono- and di-substituted anilines. Circles represent data points that were used in the regression, and the square shows the data point for 3-chloroaniline, which was excluded from this equation.
In addition, we performed a best subset regression that revealed whether more than one parameter would contribute to improving the statistics of the QSAR expression. However, only a one-parameter QSAR equation was significant. This implies that amongst the physico-chemical properties selected, there is one major component (parameter) that is most significant for protein free radical formation induced by anilines. The QSAR equation derived was also checked for statistical outliers by examining the standardized residuals as well as visual inspection. We did not find a standardized residual of 2.5 (the typical value for an outlier), however, 3-chloroaniline had a standardized residual of 2.0. When 3-chloroaniline was excluded, a significant improvement in regression statistics were observed (Eqn 2, Table 3), and when two compounds were excluded (3-chloroaniline and 4-chloro-2-methylaniline) the regression was improved again.
When we separately analyzed mono- and di-substituted anilines, the QSAR equations improved statistically compared to including all compounds to derive the QSAR equation. For mono substituted anilines, a QSAR equation was derived with σ (Eqn 4, Table 3), but a statistically improved equation was derived using IP-P (Eqn 6, Table 3). The removal of 3-chloroaniline from equation 4 and derivation of a QSAR (Eqn 5, Table 3) produced a better statistical fit. If 4-nitroaniline was excluded from equation 6, an improved QSAR was derived (Eqn 7, Table 3; Figure 2). For disubstituted anilines, σ was the best parameter for this subset of compounds (Eqn 8, Table 3). The removal of 3,4-dichloroaniline produced a robust correlation (Eqn 9, Table 3; Figure 3).
Figure 2.
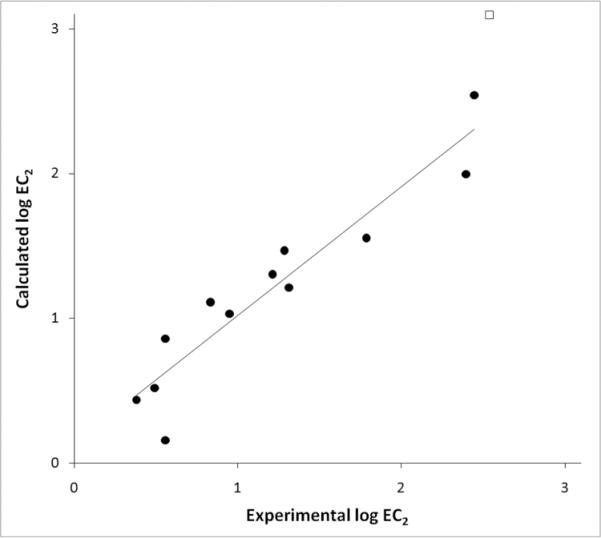
Experimental vs. Calculated log EC2 values for monosubstituted anilines. Each data point was derived by applying equation 7, Table 3. Circles represent anilines that were used in derivation of the QSAR equation. The square represents the outlier (4-nitroaniline) excluded from this equation.
Figure 3.
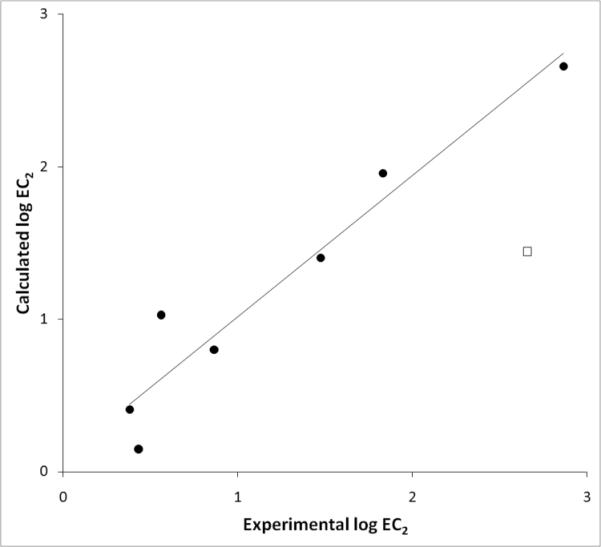
Experimental vs. calculated log EC2 values for di-substituted anilines. Circles represent the data points derived using equation 9 from Table 3, and the square is the outlier (3,4-dichloroaniline) excluded from the equation.
We tested the derived QSAR equations in their ability to predict the EC2 values for the aromatic amine drugs aminoglutethimide and procainamide, which we have previously studied. Since these drugs are monosubsituted aniline derivatives, we used equations 4 and 5 (Table 3) to predict the EC2 values of these drugs. As shown in Table 4, we found that aminoglutethimide was well predicted by both equations 4 & 5, but procainamide was not well predicted. The calculated EC2 value for procainamide was one log unit greater than the experimental value, which reflects a 10-fold difference in EC2 value. However, when we applied equations 6 & 7 a much better prediction was found for procainamide. Equation 7 also best predicted the EC2 for aminoglutethimide. This suggests that the ionization potential is a more suitable predictor for monosubstituted aniline drug induced protein free radical formation.
Table 4.
Predicted EC2 values for aminoglutethimide and procainamide.
| Experimental | Equation 4 | Equation 5 | Equation 6 | Equation 7 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| log EC2 | EC2 | log EC2 | EC2 | log EC2 | EC2 | log EC2 | EC2 | log EC2 | EC2 | |
|
|
||||||||||
| Aminoglutethimide | 0.176 | 1.5 | 0.533 | 3.4 | 0.508 | 3.2 | 0.552 | 3.56 | 0.306 | 2.02 |
| Procainamide | 0.505 | 3.2 | 1.6 | 41.0 | 1.53 | 34.0 | 0.894 | 7.83 | 0.816 | 6.54 |
Detection of HRP-catalyzed phenyl radical metabolite formation from anilines
HRP/H2O2 catalyzed aniline free radical metabolites were spin-trapped using MNP, and the coupling constants obtained from their spectra were compared with the ESR coupling constants for tertiary butyl aryl nitroxide radicals (obtained from reference tables in the Landolt-Bornstein book series) (Forrester, 1979b; Forrester, 1979c; Forrester, 1979d; Forrester, 1979a; Forrester, 1989). As shown in Table 5, we were able to detect free radical metabolites of most anilines; however, the complete structure (i.e., what atom the free radical was located on and trapped by MNP) could not be determined for many of the anilines (indicated by “unresolved H-splittings”). In addition, some anilines did not produce any detectable adducts. The compounds that produced well-defined ESR spectra were aniline, p-toluidine, 4-chloroaniline, 4-fluoroaniline, 4-ethylaniline, 2-chloroaniline, and 4-aminobenzonitrile. These compounds all had substitutions on the benzene ring para to the amine, except for 2-chloroaniline. The ESR spectra from 4-, 3-, and 2-chloroaniline including their simulated spectra are shown in Figure 4.
Table 5.
Spin-trapping of aniline-derived free radicals formed by HRP/H2O2 using MNP.
| Coupling constants (aryl nitroxides) from references (G)* | Coupling constants using HRP/H2O2 (G) | |
|---|---|---|
| aniline | aN=11.86, aHo=2.16, aHm=0.92, aHp=2.21 (Nelsen et al., 1972) | aN=15, aHo,p = 1.6, aHm = 0.92 |
| p-toluidine | aN = 13.6, aHo= aH(CH3) = 1.86, aHm = 0.93, (Lemaire et al., 1965) | aN=15, aHm=0.99, aHo,p-CH3=1.7 |
| m-toluidine | aN=13.45, aHo,p=1.8, aHm=aH(CH3)=0.6 (Barbarella and Rassat, 1969) | aN=15.5, unresolved H-splittings |
| o-toluidine | aN=13.5, aHo=0.8, aHm=0.6, 0.7, aHp=0.39, aH(CH3)=0.24, aH(tbu)=0.23# (Calder and Forrester, 1967; Calder et al., 1970; Forrester and Hepburn, 1970; Torssell, 1970; Pedersen and Torssell, 1971) | aN=15.6, unresolved H-splittings |
| p-anisidine | aN=13.9, aHo=1.86, aHm=0.93 (Lemaire et al., 1965; Pedersen and Torssell, 1971; Razuvaev et al., 1973) | Not detected |
| m-anisidine | aN=12.8, aHo=1.8, aHp=1.8, aHm=0.8 (Barbarella and Rassat, 1969) | aN=15.6, unresolved H-splittings |
| o-anisidine | aN=14.5, aHo=0.99, aH(3)=0.61, aH(4)=0.41, aH(5)=0.87, aH(OCH3)=0.063, aH(tbu)=0.26# (Pedersen and Torssell, 1971) | aN=15.6, unresolved H-splittings |
| 3,4-dimethylaniline | aN=15.6, unresolved H-splittings | |
| 2,4-dimethylaniline | aN=13.5, aH(2, CH3)=0.24, aH3=0.685, aH(4,CH3)=0.41, aH5=0.685, aH6=0.8# (Calder et al., 1970; Forrester and Hepburn, 1970) | aN=15.6, unresolved H-splittings |
| 2,6-dimethylaniline | aN=13.4, aH(2, CH3)=(6,CH3)=0.17, aH(3,5)=0.68, aH4=0.15, aH(tbu)=0.34# (Lemaire and Rassat, 1964; Calder and Forrester, 1967; Calder et al., 1970; Forrester and Hepburn, 1970; Pedersen and Torssell, 1971) | aN=15.6, unresolved H-splittings |
| 3,4-dimethoxyaniline | aN=14.5 (Frangopol et al., 1975) | Not detected |
| 2,4-dimethoxyaniline | Not detected | |
| 4-chloroaniline | aN=13.0, aHo=2.01, aHm=0.98 (Lemaire et al., 1965) | aN=14.6, aHo=1.8, aHm=0.94 |
| 3-chloroaniline | aN=12.45, aHo=1.9, aHp=1.9, aHm=0.8 (Barbarella and Rassat, 1969) | aN=15.6, unresolved H-splittings |
| 2-chloroaniline | aN=13.9, aHo=0.74, aHm=0.47,0.82, aHp=0.29, aH(tbu)=0.25# (Pedersen and Torssell, 1971; Calder et al., 1973) | aN=15.3, aHo=0.83, aHm=0.50,0.87, aHp=0.37, aHtbu=0.23 |
| 4-fluoroaniline | aN=13.7, aHo=1.72, aHm=0.86, aF=3.5 (Lemaire et al., 1965) | aN=15.1, aHo=1.68, aHm=0.89, aF=3.1 |
| 2,4-dichloroaniline | aN=13.8# (Torssell, 1970; Pedersen and Torssell, 1971) | aN=15.2, unresolved H-splittings |
| 3,4-dichloroaniline | Not detected | |
| 2,6-dichloroaniline | aN=13.1, aHm=0.69, aHp=0.1, aH(tbu)=0.35 (Pedersen and Torssell, 1971) | aN=14.8, unresolved H-splittings |
| 4-nitroaniline | Not detected | |
| 3-nitroaniline | Not detected | |
| 2-nitroaniline | Not detected | |
| 4-ethylaniline | aN=13, aHo=1.9, aHm=0.9# (Calder and Forrester, 1967; Calder et al., 1969; Torssell et al., 1973; Forrester et al., 1974) | aN=15.0, aHo=1.87, aHm=1.1 |
| 4-aminosalicylic acid | aN=15.2, unresolved H-splittings | |
| 4-chloro-2-methylaniline | aN=15.4, unresolved H-splittings | |
| 4-aminobenzonitrile | aN=10.06, aHo=2.46, aHm=0.94, aN(CN)=0.41 (Nelsen et al., 1972) | aN = 13.3, aHo = 2.1, aHm = 0.97, aN(CN)=0.27 |
The aN is typically lower in organic solvents which was used in these references.
Value for 2-chloro-4-bromoaniline.
Discussion
The mechanism of drug-induced agranulocytosis is unknown, but is believed to have many components, with reactive metabolites being a major one. We previously have determined that aminoglutethimide and procainamide, both of which contain aniline substructures, produce protein free radicals upon oxidation by myeloperoxidase. In addition, it was found that both drugs generated a phenyl radical metabolite. The relationship between the phenyl radical metabolite and protein radical formation is unknown. Moreover, the capacity for aniline compounds to induce MPO• formation has also not been known previously. We thus undertook this study to identify the physico-chemical parameters that may be involved in protein free radical formation, and to determine whether phenyl radical metabolites could be implicated.
We have now derived QSARs that demonstrate a relationship between two electronic parameters (σ, IP-P) of aniline compounds and their EC2 values for protein free radical formation. Aniline lipophilicity was not related, suggesting that hydrophobic binding is not important for protein free radical formation. It has been shown previously that σ was inversely related to the rate constants for HRP Compound I reduction (Job and Dunford, 1976) We derived an equation from this data but instead of the ratio of substituted aniline to aniline, we simply used the log of the rate constant (kX) and correlated this with σ:
Simply, this equation shows that anilines which reduce HRP Compound I the fastest (i.e., the best HRP substrates) have more negative values for σ. In our study, the derived QSAR equations showed a similar relationship, such that lower EC2 values (more potent MPO• inducers) were obtained from anilines that had more negative σ values. This implies that better substrates were better at generating MPO free radical formation.
In our experiments, however, we found that there were two groups of substrates that did not induce MPO free radical formation. One group, p-anisidine and 3,4-, and 2,4-dimethoxyaniline, are highly efficient donor substrates for peroxidases; p-anisidine has a σ = −0.28, 3,4-dimethoxyaniline σ = −0.17, and 2,4-dimethoxyaniline σ = −0.28. Interestingly, ptoluidine (σ = −0.14) and aminoglutethimide (σ = −0.15) both induced MPO free radical formation with great efficacy. This suggests that MPO• formation is not favoured by the most highly efficient peroxidase substrates. On the other hand, relatively inefficient peroxidase substrates appeared to be capable of generating MPO•, e.g., dichloroanilines and nitroanilines. However, they did so with much less efficacy as indicated by their relatively high EC2 values. Electronic factors may not solely be responsible for the low efficacy of MPO• generation, since we could not determine EC2 values for 2- and 3-nitroaniline (σ = 1.72, 0.74, respectively) but we could for 4-nitroaniline (σ = 0.78). The proximity of the nitro group to the amine could prevent generation of a reactive metabolite that could oxidize MPO.
Our initial studies on drug induced MPO• formation involved the peroxidase metabolism of aminoglutethimide and procainamide, which are both aniline-based drugs. Having derived QSAR equations to describe the physicochemical relationship between aniline compounds and MPO formation, we wished to determine whether these equations would reasonably predict the EC2 values of the drugs. The drugs were not used to derive the QSAR equations and therefore represented a small test set. Interestingly, equations using σ did not accurately predict the EC2 for procainamide, but did so for aminoglutethimide. One reason for this disparity could be that the σ values for the drugs are not known (we used σ values from structurally similar para substituents). Another reason may be that the group in the para position for procainamide is electron withdrawing (-CONH2CH2R, σ=0.36), and would be predicted to be a weak protein radical inducer, and a relatively poor peroxidase substrate. However, when the QSAR equation using IP was applied (Eqn. 6 & 7), a significantly better prediction was obtained. This suggests that σ may not be the best predictor of the ability to induce MPO• formation.
Identification of the presumed free radical metabolite that is responsible for generating the MPO• is technically challenging because it is difficult to differentiate the effects of the nitrogen-centered cation radical from those of the carbon-centered phenyl radical. Since we previously determined that aminoglutethimide and procainamide formed phenyl radicals, we wished to investigate whether the anilines used in this study also formed phenyl radicals. In addition, we wished to determine the relationship between the formation of a phenyl radical metabolites and MPO•. With two exceptions out of 26 aniline derivatives, it appeared that there was a qualitative relationship between phenyl radical metabolite formation and MPO•. More importantly, the aniline compounds for which we were unable to detect phenyl radicals did not form MPO•. This suggests that the phenyl radical is the metabolite responsible for MPO• formation.
The mechanisms of drug induced agranulocytosis are still unresolved, and are considered to be multifactoral, i.e., a particular alignment of many parameters must be present for the adverse drug reaction to occur. In the absence of suitable models, investigations of such parameters have been hindered. It has been proposed for some time that leukocyte (myeloperoxidase)-generated reactive metabolites may be involved in the etiology of drug-induced agranulocytosis (Uetrecht, 1989a); a role for myeloperoxidase-generated free radical metabolites has also been proposed (Fischer et al., 1991). We have focused on the physicochemical side of these reactions by evaluating the tendency of aromatic amines that generate phenyl radical metabolites to induce MPO•. This study established that the substitution on the aniline ring can determine the outcome of protein free radical formation on MPO. In the future, this research needs to establish the significance of MPO• in the etiology of agranulocytosis and identify potential in vivo pathways of inducing and altering this reaction.
Figure 4a.
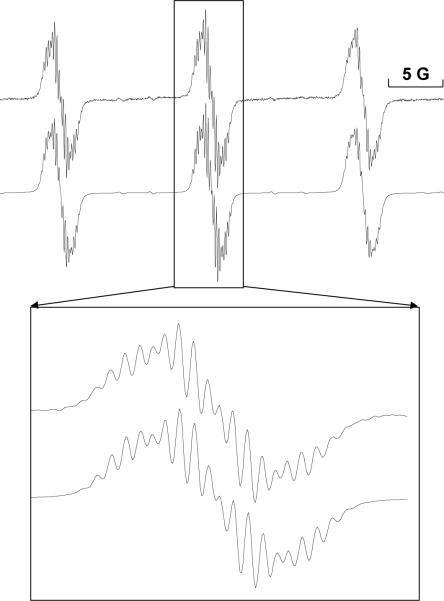
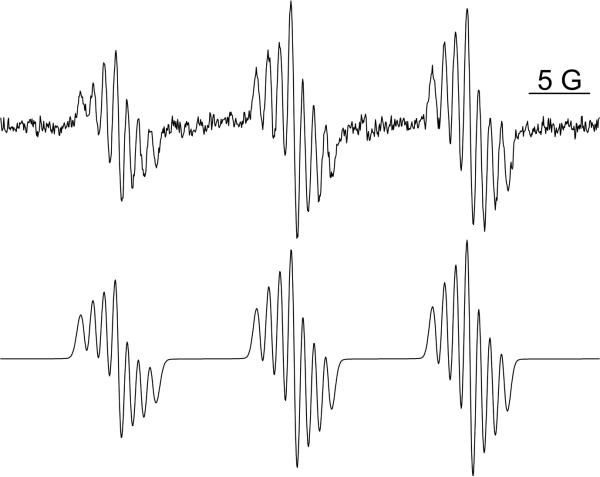
ESR spectrum of 2-chloroaniline when oxidized by HRP/H2O2. 2-chloroaniline, MNP, and H2O2 were mixed in Chelex-100 treated phosphate buffer and the reaction was initiated by adding HRP. The reaction was transferred to a flat cell and placed in the ESR cavity for recording. The spectrum for 2-chloroaniline was not well resolved, which required specific parameters to be used (see Materials and Methods). The simulation of this spectrum (correlation: r=0.99) is shown below the experimental spectrum (top), and the center peak is zoomed to show the overlay. The hyperfine splitting constants are shown in Table 5.
Figure 4b.
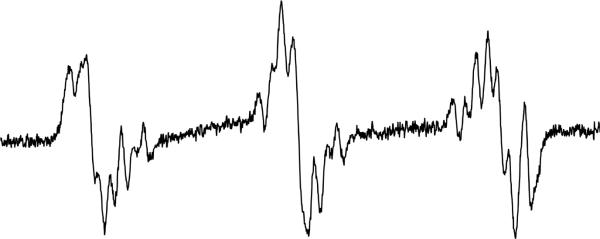
ESR spectrum of 3-chloroaniline oxidized by HRP/H2O2. The reaction was carried out as described in Figure 4a. In this case, the signal:noise was not as high as it was for 2-chloroaniline, which required 10 mW power to be used in order to obtain as resolved a spectrum. The other parameters were as described in the Materials and Methods. This spectrum could not be simulated, presumably because of mixed product formation.
Figure 4c.
ESR spectrum obtained upon 4-chloroaniline oxidation by HRP/H2O2. The reaction was carried out as described in Figure 4a; however, spectrum resolution was attained using the following instrument settings: 20 mW power, modulation amplitude of 0.4 G, and conversion and time constant at 655.36 ms. The simulation of the experimental spectrum (correlation: r=0.97) is shown below. The hyperfine splitting constants are shown in Table 5.
Acknowledgements
The authors thank Ms. Jean Corbett (NIEHS) for purification of DMPO and Dr. Ann Motten (NIEHS) for her careful review of the manuscript. This research was supported by the Intramural Research Program of the NIH, and NIH/NIEHS, as well as the University of Alberta Startup Fund.
Footnotes
Abbreviations: myeloperoxidase, MPO; myeloperoxidase protein free radical, MPO•; 5,5-dimethyl-1-pyrroline-N-oxide, DMPO; Hammett constant, σ; electron spin resonance, ESR; quantitative structure activity relationship, QSAR; horseradish peroxidases, HRP; 2-methyl-2-nitrosopropane, MNP; hydrogen peroxide, H2O2; Enzyme-Linked Immunosorbent Assay, ELISA; logarithm of the partition coefficient, log P; ionization potential of aniline, IP-P; electron affinity of aniline, EA-P; ionization potential of phenyl radical metabolite, IP-R; electron affinity of phenyl radical metabolite, EA-R; molecular orbital package, MOPAC; parametric method 3, PM3; concentration of aniline to induce a two-fold increase above background for MPO• detection, EC2; correlation coefficient, r2;
References
- Barbarella G, Rassat A. Nitroxides. XXXIV. Substituted tert-butyl phenyl nitroxide derivatives electron paramagnetic resonance. Ultrafine deviations due to nitrogen. Bulletin de la Societe Chimique de France. 1969;7:2378–2385. [Google Scholar]
- Beers RF, Jr., Sizer IW. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. Journal of Biological Chemistry. 1952;195:133–140. [PubMed] [Google Scholar]
- Besser M, Vera J, Clark J, Chitnavis D, Beatty C, Vassiliou G. Preservation of basophils in dapsone-induced agranulocytosis suggests a possible pathogenetic role for leucocyte peroxidases. International Journal of Laboratory Hematology. 2009;31:245–247. doi: 10.1111/j.1751-553X.2007.00983.x. [DOI] [PubMed] [Google Scholar]
- Calder A, Forrester AR. Stability of aryl-tert-butylnitroxides. Chemical Communications (London) 1967;14:682–684. [Google Scholar]
- Calder A, Forrester AR, Emsley JW, Luckhurst GR, Storey RA. Nitroxide radicals .7. Nuclear and electron resonance spectra of some aromatic tert-butyl nitroxides. Molecular Physics. 1970;18:481–489. [Google Scholar]
- Calder A, Forrester AR, Hepburn SP. Nitroxide radicals. XII. Decomposition of o-, m-, and p-halophenyl tert-butyl nitroxides. Journal of the Chemical Society, Perkin Transactions 1: Organic and Bio-Organic Chemistry. 1973;5:456–465. [Google Scholar]
- Calder A, Forrester AR, Thomson RH. Nitroxide radicals. IV. Oxidation of cyclic N-hydroxyimides. Journal of the Chemical Society C. 1969;3:512–516. [Google Scholar]
- Christie G, Breckenridge AM, Park BK. Drug-protein conjugates-XVIII. Detection of antibodies towards the antimalarial amodiaquine and its quinone imine metabolite in man and the rat. Biochemical Pharmacology. 1989;38:1451–1458. doi: 10.1016/0006-2952(89)90184-6. [DOI] [PubMed] [Google Scholar]
- Dunford HB. Heme peroxidases. John Wiley; New York: 1999. [Google Scholar]
- Fischer V, Haar JA, Greiner L, Lloyd RV, Mason RP. Possible role of free radical formation in clozapine (clozaril)-induced agranulocytosis. Molecular Pharmacology. 1991;40:846–853. [PubMed] [Google Scholar]
- Forrester A. Organic N-Centered Radicals and Nitroxide Radicals. 1979a. Other tert.-butyl-substituted phenyl nitroxides; pp. 682–693. [Google Scholar]
- Forrester A. Organic N-Centered Radicals and Nitroxide Radicals. 1979b. Part 1; pp. 651–661. [Google Scholar]
- Forrester A. Organic N-Centered Radicals and Nitroxide Radicals. 1979c. Part 2; pp. 662–672. [Google Scholar]
- Forrester A. Organic N-Centered Radicals and Nitroxide Radicals. 1979d. Part 3; pp. 673–682. [Google Scholar]
- Forrester A. Nitroxide Radicals. Part 1. 1989. 6.2.1.10 Aryl radicals; pp. 168–175. [Google Scholar]
- Forrester AR, Hepburn SP. Nitroxide radicals. VIII. Stability of ortho-alkyl-substituted phenyl tert-butyl nitroxides. Journal of the Chemical Society C. 1970;9:1277–1280. [Google Scholar]
- Forrester AR, Hepburn SP, McConnachie G. Nitroxide radicals. XVI. Unpaired electron distribution in para substituted aryl tert-butyl nitroxides and 2-naphthyl phenyl nitroxides. Journal of the Chemical Society, Perkin Transactions 1: Organic and Bio-Organic Chemistry. 1974;19:2213–2219. [Google Scholar]
- Frangopol PT, Frangopol M, Baikan R. New aryl tert-alkyl nitroxyls. Izvestiya Akademii Nauk SSSR, Seriya Khimicheskaya. 1975;11:2506–2511. [Google Scholar]
- Hsuanyu Y, Dunford HB. Oxidation of Clozapine and Ascorbate by Myeloperoxidase. Archives of Biochemistry and Biophysics. 1999;368:413–420. doi: 10.1006/abbi.1999.1328. [DOI] [PubMed] [Google Scholar]
- Job D, Dunford HB. Substituent effect on the oxidation of phenols and aromatic amines by horseradish peroxidase compound I. European Journal of Biochemistry. 1976;66:607–614. doi: 10.1111/j.1432-1033.1976.tb10588.x. [DOI] [PubMed] [Google Scholar]
- Lagercrantz C. Spin trapping of some short-lived radicals by the nitroxide method. The Journal of Physical Chemistry. 1971;75:3466–3475. [Google Scholar]
- Lahita R, Kluger J, Drayer DE, Koffler D, Reidenberg MM. Antibodies to nuclear antigens in patients treated with procainamide or acetylprocainamide. New England Journal of Medicine. 1979;301:1382–1385. doi: 10.1056/NEJM197912203012506. [DOI] [PubMed] [Google Scholar]
- Lemaire H, Marechal Y, Ramasseul R, Rassat A. Nitroxides. IX. Electron paramagnetic resonance of parasubstituted tert-butylphenylnitroxide. Bulletin de la Societe Chimique de France. 1965;2:372–378. [Google Scholar]
- Lemaire H, Rassat A. Hyperfine structure of nitrogen in nitroxide radicals. Journal de Chimie Physique et de Physico-Chimie Biologique. 1964;61:1580–1586. [Google Scholar]
- Nelsen SF, Landis RT, Kiehle LH, Leung TH. N-tert-Butylanilino Radicals. Journal of the American Chemical Society. 1972;94:1610–1614. [Google Scholar]
- Pedersen JA, Torssell K. Electron spin resonance and nuclear magnetic resonance spectra of sterically hindered aromatic nitroxide radicals - synthesis of stable nitroxide radicals. Acta Chemica Scandinavica. 1971;25:3151–3162. [Google Scholar]
- Perrin DD, Dempsey B, Serjeant EP. pKa prediction for organic acids and bases. Chapman and Hall; London: New York: 1981. [Google Scholar]
- Razuvaev GA, Abakumov GA, Sanaeva EP, Abakumova LG. 1. Synthesis and oxidative decomposition. Izvestiya Akademii Nauk SSSR, Seriya Khimicheskaya. 1973;1:68–71. [Google Scholar]
- Reidenberg MM. Aromatic amines and the pathogenesis of lupus erythematosus. American Journal of Medicine. 1983;75:1037–1042. doi: 10.1016/0002-9343(83)90885-9. [DOI] [PubMed] [Google Scholar]
- Siraki AG, Bonini MG, Jiang J, Ehrenshaft M, Mason RP. Aminoglutethimide-induced protein free radical formation on myeloperoxidase: a potential mechanism of agranulocytosis. Chemical Research in Toxicology. 2007;20:1038–1045. doi: 10.1021/tx6003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siraki AG, Deterding LJ, Bonini MG, Jiang J, Ehrenshaft M, Tomer KB, Mason RP. Procainamide, but not N-acetylprocainamide, induces protein free radical formation on myeloperoxidase: a potential mechanism of agranulocytosis. Chemical Research in Toxicology. 2008;21:1143–1153. doi: 10.1021/tx700415b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, Palyulin VA, Radchenko EV, Zefirov NS, Makarenko AS, Tanchuk VY, Prokopenko VV. Virtual computational chemistry laboratory--design and description. Jounal of Computer Aided Molecular Design. 2005;19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- Torssell K. Investigation of radical intermediates in organic reactions by use of nitroso compounds as scavengers the nitroxide method. Tetrahedron. 1970;26:2759–2773. [Google Scholar]
- Torssell K, Goldman J, Petersen TE. Spin delocalization of Group IV B elements in organic compounds. Justus Liebigs Annalen der Chemie. 1973;2:231–240. [Google Scholar]
- Uetrecht J. Drug metabolism by leukocytes and its role in drug-induced lupus and other idiosyncratic drug reactions. Critical Reviews in Toxicology. 1989a;20:213–235. doi: 10.3109/10408449009089863. [DOI] [PubMed] [Google Scholar]
- Uetrecht JP. Idiosyncratic drug reactions: Possible role of reactive metabolites generated by leukocytes. Pharmaceutical Research. 1989b;6:265–273. doi: 10.1023/a:1015934104984. [DOI] [PubMed] [Google Scholar]
- Uetrecht JP. The role of leukocyte-generated reactive metabolites in the pathogenesis of idiosyncratic drug reactions. Drug Metab Rev. 1992;24:299–366. doi: 10.3109/03602539208996297. [DOI] [PubMed] [Google Scholar]